SUMMARY
Simple surfactants achieve remarkable long-range order in aqueous environments. This organizing potential is seen most dramatically in biological membranes where phospholipid assemblies both define cell boundaries and provide a ubiquitous structural scaffold for controlling cellular chemistry. Here we consider simple peptides that also spontaneously assemble into exceptionally ordered scaffolds, and review early data suggesting that these structures maintain the functional diversity of proteins. We argue that such scaffolds can achieve the required molecular order and catalytic agility for the emergence of chemical evolution.
INTRODUCTION
Cell membranes organize life's chemistry through an intricate mosaic of lipids and proteins [1-3]. The order achieved by a phospholipid membrane may be important for all cellular functions, but nowhere is this need more apparent than in the multi-enzyme reaction complexes that achieve photosynthesis [4]. Oxidative phosphorylation similarly depends on the membrane to position appropriately the numerous electron transport chain components for burning food [5]. While the phospholipid membrane is quite naturally viewed as the barrier that buffers the cell against environmental fluctuations, this confined fluid, most importantly, provides the principle organizational scaffold in which functional proteins “float”[3] and self-assemble into large functional multi-component catalysts. Lipid rafts have now extended the role of the phospholipid membrane from a simple passive scaffold to a higher level of organization [6].
Nature's phospholipids then are known to self-assemble spontaneously into supramolecular structures in water, adopt diverse morphologies in response to their environment, survive harsh physical and chemical challenges, and maintain strong electrical and chemical potential gradients. Despite these essential physical capabilities of current cells, the structurally complex phospholipids are unlikely to have been present in the terrestrial prebiotic chemical inventory [7,8]. Other less complex amphiphiles more likely provided the thermodynamically accessible and functional superstructures necessary for the origins of chemical evolution. For example, simple fatty acid mixtures have been found in carbonaceous chondrites [9] and these structures can form stable [10-12] and functional protocell membranes [13-16].
Amino acids represent another likely component of prebiotic inventories. The Miller-Urey reactions [17,18] and meteorite deposition [19] both suggest potential sources for amino acids on a prebiotic earth. Simple polypeptides form via dehydration-rehydration cycles [20], at hydrothermal vents [21], in salts [22] on mineral surfaces [23,24], and under simple thermal heating cycles [25]. Peptides composed of alternating hydrophobic/hydrophilic residues can assemble into “membrane-like” β-sheet bilayers by dimerizing to sequester the hydrophobic faces and expose the hydrophilic residues to solution [26,27]. Four to 20 residue peptides can also assemble into a wide-range of morphologies, including fibers [28,29], hollow tubes [28,30-33], ribbons [34], and vesicles [30,33,35]; all structures reminiscent of the diversity seen in lipid surfactants. However, the peptide backbone and side chains carry much greater functionality than amphiphilic alkanes, and likely have very different structures from lipid membranes.
Can Peptide Assemblies Emulate the Functions of Lipid Membranes?
In aqueous solutions, simple amphiphile assembly is driven by dehydration of hydrophobic domains to create tail-to-tail bilayers stabilized by an exposed hydrophilic surface [2]. Zhang and co-workers have shown that amphiphilic peptide sequences with up to two charged residues at the C-terminus [36] and a stretch of 6-8 hydrophobic residues, including alanine, valine, leucine [30,37], and glycine [33], can assemble into worm-like micelles and nanovesicles with diameters ranging from 30 to 50nm [30]. These assemblies display a thickness similar to biological phospholipids, and structural models propose an extended peptide backbone conformations with the peptide axis oriented perpendicular to the tube wall [38,39] forming a bilayer [30] similar to lipid tubules [40].
The initial fluid mosaic model [3] of biological membranes viewed the lipid interior as a confined fluid that orients and stabilizes associated proteins by burial of the hydrophobic residues within the bilayer and directing the polar groups into the aqueous medium. The inherent fluidity of the structure [2] allows the dissolved components to diffuse laterally [2,41], interact within or outside the membrane, and associate into the assemblies required for such complex functions as photosynthesis and oxidative phosphorylation. Simple amphiphilic peptide surfactants (e.g. AAAAAAK and VVVVVVD) have now been shown to stabilize a variety of membrane bound proteins including glycerol-3-phosphate dehydrogenase [42], bovine rhodopsin [43], and the 36 protein complex photosystem I (PS-I)[36] as effectively as commercially available detergents. In fact, peptide stabilization of the PS-I complex showed a nearly 8-fold increase in O2 consumption and remained active for more than 2 months [36]. Interestingly, the degree of PS-I stabilization depends upon the amino acid sequence, specifically the position, number, and type of charged residues. Therefore, these early experiments suggest that simple peptide surfactants can be as effective as lipid surfactants in stabilizing typical membrane protein complexes, and the peptide sequence dependence may indicate that higher-order specific molecular interactions, beyond the simple fluid mosaic, are playing a role in these assemblies.
Can Peptide Membranes Access Typical Protein Architectures?
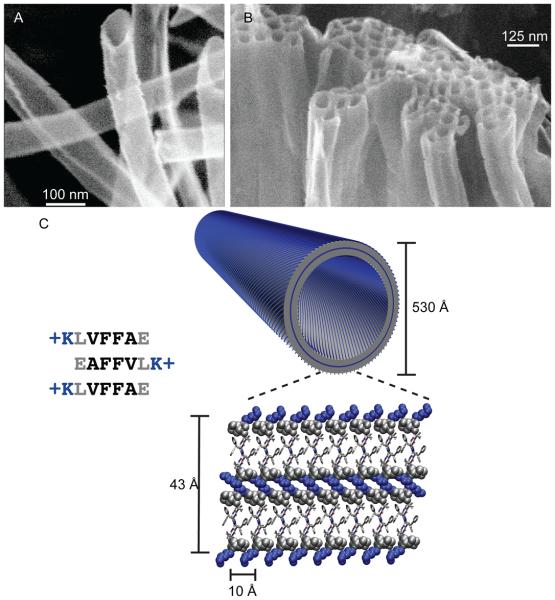
Others have argued that all proteins assemble into β-sheet rich amyloid under appropriate conditions [44,45]. This prediction is particularly relevant for short peptides, <15 residues, which can easily sample the extended β-sheet secondary structure in water [46]. Amyloidogenic peptides, such as the amphiphilic nucleating core of the Alzheimer's Disease Aβ peptide, Aβ(16-22) (KLVFFAE) [31], has also been shown to assemble into hollow nanotubes at pH 2 (Fig. 1A, B) with homogenous diameters of 52nm and wall thicknesses of ~4nm [31]. Since the extended peptide length of Aβ(16-22) is ~2 nm, accounting for roughly half the tube wall thickness, initial models predicted a membranous bilayer arrangement with the lysine ammonium ion side chain localized to the surfaces of the nanotube [31,47]. However, isotope-edited IR and NMR experiments on the nanotubes indicated that the peptides were precisely arranged as anti-parallel β-sheets with the registry of the strands within the sheets shifted by a single amino acid residue as illustrated in Fig. 1C [28].
Figure 1.
(A) Cryo-etch SEM image of KLVFFAE nanotubes assembled in acidic conditions[28]. (B) Bundling of positively charged nanotubes upon addition of sodium sulfate [60]. In the bilayer cartoon of KLVFFAE nanotubes, the peptides are organized as anti-parallel β-sheets placing hydrophilic lysine side chains (blue) at both the solvent exposed interface and buried in the bilayer interface. Leucine and protonated glutamic acids are drawn in grey space filling models. The remaining residues are drawn as sticks, with front peptide (grey) and back peptide (white).
These β-structural elements then appear to exist in the peptide membranes, suggesting that peptides utilize the forces of protein folding as opposed to the non-specific hydrophobic interactions of alkane phospholipid membrane bilayers. For example, while the Aβ(16-22) membranes discussed above expose half their lysine residues along the solvent exposed surface, the structural model predicts the other half of these positively charged side chains must be packed within the peptide bilayer interface. Non-covalent cross-strand pairing [28,48,49] interactions along the β-sheet surface, including phenylalanine π-π interactions [50-52], K-E salt bridges [28], and β-branched residue (Val, Ile) packing [48], must contribute not only to strand arrangements, but also to stabilize these leaflet packing morphologies.
Side-chain functionality also regulates assembly in the presence of metal ions in a manner very similar to proteins. Aβ(10-21), 10YEVHHQKLVFFA, which includes the nucleating core residues discussed above, assemblies very rapidly in the presence of Zn2+ [53]. In the absence of metals, Aβ(13-21)K16A 13HHQALVFFA, forms 5 nm fibers, but in the presence of Zn2+, the peptide assembles into ribbons and nanotubes with diameters two orders of magnitude larger than the metal-free fibers. His-ZnHis ligation between adjacent β-sheets is responsible for the observed increase in sheet stacking (Fig. 3B) [34,47]. Co-assembly of HAQKLVFFA with Cu2+ (Fig 3D) or Zn2+ accelerates assembly and each metal is specifically coordinated to side chain functionality [34,54]. Therefore, many of the architectures that are so prevalent in present day proteins, specific secondary structure elements, transition metal binding sites, and environmental switches, can be incorporated into these peptide bilayer membranes.
Figure 3.
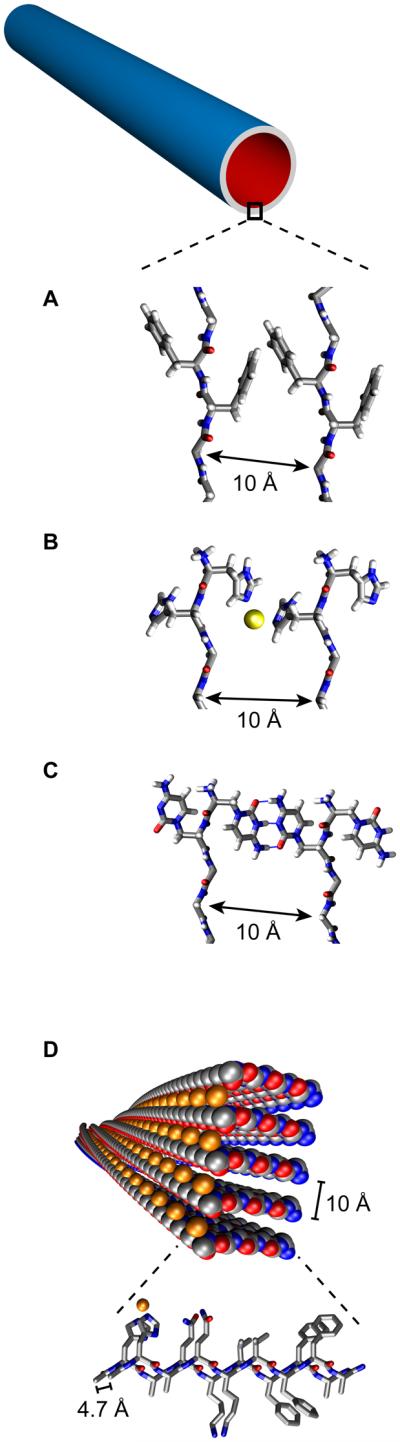
Peptide side-chain impact upon (A-C) nanotubes β-sheet lamination and (D) ability to display metal ions in fibers. (A) Two laminated β-sheets of KLVFFAE displaying F-F interactions[28], (B) HHQALVFFA displaying His-Zn-His side chain interaction between two laminated β-sheets[34] (C) ccQALVFFA displaying cytosine i-motif interaction between two laminated β-sheets[55] (D) HAQKLVFFA His-Cu-His interaction between hydrogen bonded β-strands[54].
Can Peptide Bilayers Acquire Function?
With the HHQALVFFA peptide nanotube[34] as a model (Fig. 3B), molecular dynamics simulations predicted cytosine i-motif base pairs as stabilizing elements between adjacent β-sheets. Indeed, when the base was incorporated as a β-(cytosine-1-yl)-alanine (c) into ccQALVFFA and allowed to assemble under conditions favorable for cytosine i-motif formation (Fig. 3C), fixed diameter hollow nanotubes formed rapidly [55]. Characterization by solid-state NMR positioned the peptides as parallel β-sheets, consistent with i-motif base pairing both stabilizing parallel strand arrangements in each sheet and parallel β-sheet lamination [55]. The 3.3 nm wall thickness, determined by small angle x-ray scattering [55], and the length of the ccQALVFFA peptide (3.1 nm), indicates a monolayer nanotube with distinct inner and outer surfaces; one surface lined with residues at the N-terminus and the other lined with residues at the C-terminus as shown in Figure 3C [55]. Such structures are not accessible in present day biological membranes, and could provide a simple mechanism of segregating functional entities on the inside or outside surface of the tube.
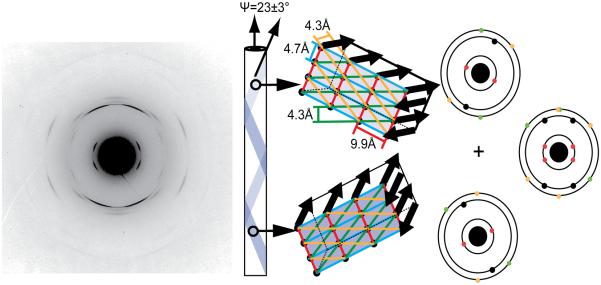
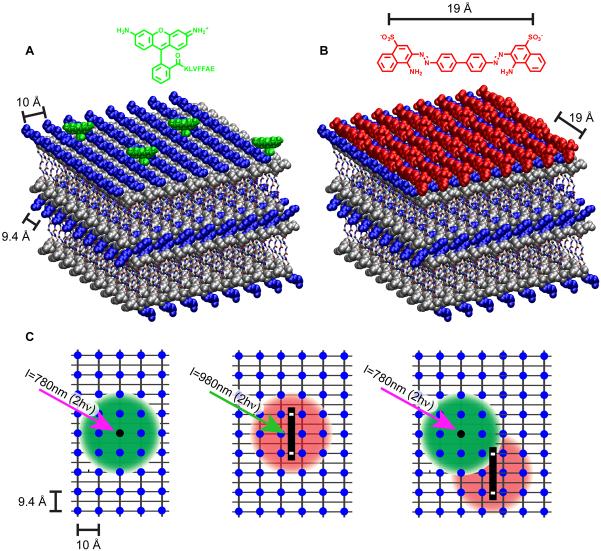
The analysis of oriented KLVFFAE peptide nanotubes and fibers by diffraction reveal their remarkably well-defined paracrystalline order. Reflections from the hollow Aβ(16-22) nanotube (Fig. 2B) showed a doubling of the traditional cross-β 4.7 Å and 9.9 Å d-spacings, arising from orthogonal H-bonded peptides and stacked β-sheets arrays. The doubling originates from distinct cross-β patterns from both top and bottom surfaces of flattened tubes, offset by 23±3° from the tube axis [28]. These repeating cross-β arrays of peptide membranes provide many closely packed binding sites for small molecules. Such arrays have been used to explain the unusual optical properties of the dyes Congo red (CR) [56,57] and Thioflavin T [58,59] bound to amyloid. The precisely patterned surfaces of the Aβ(16-22) nanotubes are composed of positively charged lysine (blue) knobs and leucine-rich hydrophobic groves (gray) as shown in Fig 4B [57]. An interesting outcome of this patterning is seen at CR saturation, which sufficiently passivates the surface and bundles the hollow nanotubes in a manner similar to sulfate [60] into higher order millimeter long supramacromolecular fibers similar to those shown in Fig. 1B.
Figure 2.
Electron diffraction of crystalline KLVFFAE nanotubes with corresponding d-spacings 9.9Å, 4.7Å, 4.3Å, and 4.0Å. Nanotubes are composed of β-sheets that helically coil around the nanotube axis.[28]
Figure 4.
Cartoon of nanotube surfaces. Positively charged lysine sidechains are shown as blue spheres and leucines and protonated glutamate sidechains are drawn as gray spheres. 9.4Å is the spacing along the H-bonding dimension between the lysines, which corresponds to three H-bonded peptides (2 × 4.7Å). (A) Covalent anchoring of Rhodamine 110 to KLVFFAE nanotubes, (B) Non-covalent binding of negatively charged Congo Red to KLVFFAL nanotubes. The distance between Congo Red sulfate groups is 19Å, which corresponds to 6 H-bonded peptides. (C) Conceptual figure of light antenna comprised of covalently attached Rhodamine110-KLVFFAE (black dot) and negatively charged, noncovalently bound Alexa555 (black rectangle) FRET pair on the tube surface. Each chromophore can be imaged via 2 photon fluorescence with Rhodamine110 (λEm = 520nm) and Alexa555 (λEm = 565nm). When both Rhodamine110 and Alexa555 are on the tube surface, two-photon excitation (λEx = 780nm) of Rhodamine110 results in energy transfer to nanotube bound Alexa555 and emission at 565nm.
Combining the insights gained from CR binding, the fluorescent dye Rhodamine 110, which does not bind to the cross-β template of Aβ(16-22), was covalently substituted for the N-terminal lysine of this peptide [61] and co-assembled with the unmodified KLVFFAE. The resulting fluorescent nanotubes (Fig 4A) readily bound the sulfonated Alexa 555 dye, a structure similar to CR. Co-localization was evaluated via fluorescence resonance energy transfer (FRET) across the well-defined grid of the nanotube surfaces (Fig. 4C) [61]. This experiment represents an early step in functional light energy capture and transfer along the peptide bilayer surface, in a manner similar to that seen in other amyloids [62,63].
Complexity and Chemical Evolution
The yeast prion Sup35, which serves naturally as a translational terminator, carries a conserved N-terminal amyloid domain that allows for epigenetic control of its function through fiber assembly [66]. This general concept of organizing active catalysts with simple amyloid domains has been synthetically tested with a variety of other proteins [64], including green fluorescent protein [65,66], barnase [66], carbonic anhydrase [66], glutathione S-transferase [66,67], and cytochrome C [68], confirming that amyloid can provide a robust scaffold for reversibly organizing large functional proteins. Is it possible then that this conserved Sup35 prion domain is an ancestral amyloid co-opted as a present day regulatory element? Might such assemblies be remnants of an earlier age rather than simply disease-causing protein misfolding errors?
We argued above that many peptides have the generic propensity to self-assemble and to be selected by an ability to form ordered peptide membranes. Building on insight provided by the CR binding site [58], such membranes could be selected for binding to and production of other membrane components, allowing the membranes to grow and “reproduce”. With such functional selection, autocatalytic cycles [69], like those seen for helical peptides [70-73] and coiled-coil fibers [74], could then efficiently produce these peptide membranes. Indeed, preliminary experiments have shown amyloid fibers can self-template the ligation of fragment in a similar manner [75,76]. Such a self-sustaining chemical system would benefit from very short peptides achieving micron scale organization of membranes containing typical protein catalytic features that include metal binding arrays [34,53-54], substrate binding sites [57,59], and even large catalytic domains [64-68]. Membrane and/or fibril localization of multiple copies of catalysts arrayed in close proximity could even grow to a “metabolism” of expanding structural complexity.
To the extent that chemical evolution is simply the nonrandom survival of randomly encoded information [77], these peptide membranes may represent a low fidelity strategy capable of emerging complexity from a well-recognized prebiotic inventory. The extent to which these molecular surface grids, arrays of catalytic protein domains, and functioning nanoscale molecular barriers can be selected is just now being explored. But just as RNA's catalytic function [78,79] suggested a common molecular ancestor that unified information storage and chemical catalysis for an origins of life hypothesis [78,80-82] the peptide membranes may reveal the earliest roots of a prebiotic chemical evolution.
ACKNOWLEDGEMENTS
We are indebted to Jeannette Taylor, Hong Yi, and the Robert P. Apkarian Microscopy Core, Emory University for training in TEM, electron diffraction, and Cryo-etch HR-SEM, and gratefully acknowledge funding from the U.S. DOE (ER15377), NSF CHE-0404677 and NSF-CBC-0739189.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
These ideas were first discussed at the 3rd Annual Advances in Biomolecular Engineering: Protein Design Symposium at the New York Academy of Sciences on June 12, 2009
REFERENCES
- 1.Jacobson K, Mouritsen OG, Anderson RGW. Lipid rafts: At a crossroad between cell biology and physics. Nat Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 2.Hianik T. Structure and physical properties of biomembranes and model membranes. Acta Phys Slovaca. 2006;56:687–806. [Google Scholar]
- 3.Singer SJ, Nicolson GL. Fluid mosaic model of structure of cell-membranes. Science. 1972;175:720–&. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 4.Amunts A, Nelson N. Plant photosystem I design in the light of evolution. Structure. 2009;17:637–650. doi: 10.1016/j.str.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Vonck J, Schafer E. Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim Biophys Acta-Mol Cell Res. 2009;1793:117–124. doi: 10.1016/j.bbamcr.2008.05.019. [DOI] [PubMed] [Google Scholar]
- •6.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. This commentary highlights recent shifts in our view of biological membranes, from a random two-dimensional fluid to a mosaic of segregated functional structures. The precise organizing functionality of modern membranes could have played a significant role in chemical evolution. [DOI] [PubMed] [Google Scholar]
- 7.Pereto J, Lopez-Garcia P, Moreira D. Ancestral lipid biosynthesis and early membrane evolution. Trends BiochemSci. 2004;29:469–477. doi: 10.1016/j.tibs.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Mulkidjanian AY, Galperin MY, Koonin EV. Co-evolution of primordial membranes and membrane proteins. Trends BiochemSci. 2009;34:206–215. doi: 10.1016/j.tibs.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deamer DW. Boundary structures are formed by organic-components of the murchison carbonaceous chondrite. Nature. 1985;317:792–794. [Google Scholar]
- 10.Namani T, Deamer DW. Stability of model membranes in extreme environments. Orig Life Evol Biosph. 2008;38:329–341. doi: 10.1007/s11084-008-9131-8. [DOI] [PubMed] [Google Scholar]
- ••11.Mansy SS, Szostak JW. Thermostability of model protocell membranes. Proc Natl Acad Sci U S A. 2008;105:13351–13355. doi: 10.1073/pnas.0805086105. A minimal protocell composed of simple fatty acids,fatty alcohols, and fatty-acid glycerol esters was designed. At elevated temperatures, the prebiotic protocell allows uptake of charged nucleotides. Duplex DNA can be denatured while encapsulated inside protocell to permit the eventual replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apel CL, Deamer DW, Mautner MN. Self-assembled vesicles of monocarboxylic acids and alcohols: Conditions for stability and for the encapsulation of biopolymers. Biochim Biophys Acta-Biomembr. 2002;1559:1–9. doi: 10.1016/s0005-2736(01)00400-x. [DOI] [PubMed] [Google Scholar]
- 13.Monnard PA, Luptak A, Deamer DW. Models of primitive cellular life: Polymerases and templates in liposomes. Philos Trans R Soc B-Biol Sci. 2007;362:1741–1750. doi: 10.1098/rstb.2007.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansy SS, Schrum JP, Krishnamurthy M, Tobe S, Treco DA, Szostak JW. Template-directed synthesis of a genetic polymer in a model protocell. Nature. 2008;454:122–125. doi: 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansy SS. Model protocells from single-chain lipids. Int J Mol Sci. 2009;10:835–843. doi: 10.3390/ijms10030835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••16.Luisi PL. The emergence of life: From chemical origins to synthetic biology. edn 1. Cambridge University Press; Cambridge: 2006. A basic textbook that describes the concepts of the chemical origins of life for undergraduate and graduate students. [Google Scholar]
- 17.Miller SL. A production of amino acids under possible primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- •18.Johnson AP, Cleaves HJ, Dworkin JP, Glavin DP, Lazcano A, Bada JL. The miller volcanic spark discharge experiment. Science. 2008;322:404–404. doi: 10.1126/science.1161527. The authors re-evaluate Miller's volcanic discharge experiment to simulate the spark discharge synthesis by lightning in a steam-rich volcanic eruption. A total of 22 different amino acids and 5 amines were identified via HPLC and LC-TOF mass spectrometry. [DOI] [PubMed] [Google Scholar]
- 19.Ehrenfreund P, Glavin DP, Botta O, Cooper G, Bada JL. Extraterrestrial amino acids in Orgueil and Ivuna: Tracing the parent body of cl type carbonaceous chondrites. Proc Natl Acad Sci U S A. 2001;98:2138–2141. doi: 10.1073/pnas.051502898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagawa H, Kojima K, Ito M, Handa N. Synthesis of polypeptides by microwave-heating .1. Formation of polypeptides during repeated hydration-dehydration cycles and their characterization. J Mol Evol. 1990;31:180–186. doi: 10.1007/BF02109494. [DOI] [PubMed] [Google Scholar]
- 21.Imai E, Honda H, Hatori K, Brack A, Matsuno K. Elongation of oligopeptides in a simulated submarine hydrothermal system. Science. 1999;283:831–833. doi: 10.1126/science.283.5403.831. [DOI] [PubMed] [Google Scholar]
- 22.Plankensteiner K, Reiner H, Rode BM. Catalytically increased prebiotic peptide formation: Ditryptophan, dilysine, and diserine. Origins of Life and Evolution of the Biosphere. 2005;35:411–419. doi: 10.1007/s11084-005-1971-x. [DOI] [PubMed] [Google Scholar]
- 23.Rimola A, Ugliengo P, Sodupe M. Formation versus hydrolysis of the peptide bond from a quantum-mechanical viewpoint: The role of mineral surfaces and implications for the origin of life. Int J Mol Sci. 2009;10:746–760. doi: 10.3390/ijms10030746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brack A, Barbier B, Bertrand M, Chabin A, Westall F. Polymerization of amino acid thioesters on mineral surfaces in dilute solution. In: Lacoste H, editor. 2nd European Workshop on Exo/Astrobiology Sep 16-19. Esa Publications Division C/O Estec; Graz, Austria: 2002. pp. 435–436. [Google Scholar]
- 25.Fox SW, Harada K. The thermal copolymerization of amino acids common to protein1. J Am Chem Soc. 1960;82:3745–3751. [Google Scholar]
- 26.Brack A, Orgel LE. Beta-structures of alternating polypeptides and their possible prebiotic significance. Nature. 1975;256:383–387. doi: 10.1038/256383a0. [DOI] [PubMed] [Google Scholar]
- 27.Brack A. From amino-acids to prebiotic active peptides - a chemical reconstitution. 18th International Symp on the Chemistry of Natural Products; Blackwell Science Ltd; Strasbourg, France. Aug 30-Sep 04.1992. pp. 1143–1151. [Google Scholar]
- ••28.Mehta AK, Lu K, Childers WS, Liang Y, Dublin SN, Dong J, Snyder JP, Pingali SV, Thiyagarajan P, Lynn DG. Facial symmetry in protein self-assembly. J Am Chem Soc. 2008;130:9829–9835. doi: 10.1021/ja801511n. A combination of solid-state NMR, diffraction and computational simulations to compare and distinguish two distinct self-assembled morphologies formed from the same peptide under different conditions. This article details an approach to characterize these self-assembly structures. [DOI] [PubMed] [Google Scholar]
- 29.de la Paz ML, Serrano L. Sequence determinants of amyloid fibril formation. Proc Natl Acad Sci U S A. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vauthey S, Santoso S, Gong HY, Watson N, Zhang SG. Molecular self-assembly of surfactant-like peptides to form nanotubes and nanovesicles. Proc Natl Acad Sci U S A. 2002;99:5355–5360. doi: 10.1073/pnas.072089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu K, Jacob J, Thiyagarajan P, Conticello VP, Lynn DG. Exploiting amyloid fibril lamination for nanotube self-assembly. J Am Chem Soc. 2003;125:6391–6393. doi: 10.1021/ja0341642. [DOI] [PubMed] [Google Scholar]
- 32.Reches M, Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300:625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 33.Santoso S, Hwang W, Hartman H, Zhang SG. Self-assembly of surfactant-like peptides with variable glycine tails to form nanotubes and nanovesicles. Nano Letters. 2002;2:687–691. [Google Scholar]
- •34.Dong J, Shokes JE, Scott RA, Lynn DG. Modulating amyloid self-assembly and fibril morphology with Zn(II) J Am Chem Soc. 2006;128:3540–3542. doi: 10.1021/ja055973j. Zn (II) was shown to modulate the assembly of HHQALVFFA through His-Zn-His ligation, resulting in extended beta-sheet stacking. The resulting hollow bilayer nanotube surfaces are arrayed with a 2D lattice of transition metal binding sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan X, He Q, Wang K, Duan L, Cui Y, Li J. Transition of cationic dipeptide nanotubes into vesicles and oligonucleotide delivery. Angew Chem Int Ed Engl. 2007;46:2431–2434. doi: 10.1002/anie.200603387. [DOI] [PubMed] [Google Scholar]
- ••36.Matsumoto K, Vaughn M, Bruce BD, Koutsopoulos S, Zhang SG. Designer peptide surfactants stabilize functional photosystem-I membrane complex in aqueous solution for extended time. J Phys Chem B. 2009;113:75–83. doi: 10.1021/jp8021425. This study reveals the potential of peptide surfactants to serve as an alternative medium for the stabalization and characterization of membrane bound proteins. The peptides localize functional multi-enzyme complexes of Photosystem I and several parameters including: N-terminal acetylation, C-terminal amidation, the number of charges on the hydrophilic head, and the bulkiness of the amino acid side chains are systematically explored. [DOI] [PubMed] [Google Scholar]
- 37.Yang SJ, Zhang SG. Self-assembling behavior of designer lipid-like peptides. Supramol Chem. 2006;18:389–396. [Google Scholar]
- 38.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim Biophys Acta-Rev Biomembr. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marrink SJ, de Vries AH, Mark AE. Coarse grained model for semiquantitative lipid simulations. J Phys Chem B. 2004;108:750–760. [Google Scholar]
- 40.Schnur JM. Lipid tubules - a paradigm for molecularly engineered structures. Science. 1993;262:1669–1676. doi: 10.1126/science.262.5140.1669. [DOI] [PubMed] [Google Scholar]
- 41.Sackmann E. Dynamic molecular-organization in vesicles and membranes. Ber Bunsen-Ges Phys Chem Chem Phys. 1978;82:891–909. [Google Scholar]
- 42.Yeh JI, Du SC, Tortajada A, Paulo J, Zhang SG. Peptergents: Peptide detergents that improve stability and functionality of a membrane protein, glycerol-3-phosphate dehydrogenase. Biochemistry. 2005;44:16912–16919. doi: 10.1021/bi051357o. [DOI] [PubMed] [Google Scholar]
- 43.Zhao XJ, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang SG. Designer short peptide surfactants stabilize G protein-coupled receptor bovine rhodopsin. Proc Natl Acad Sci U S A. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson CM. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM. Amyloid fibril formation by an SH3 domain. Proc Natl Acad Sci U S A. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su JY, Hodges RS, Kay CM. Effect of chain length on the formation and stability of synthetic Alpha.-helical coiled coils. Biochemistry. 1994;33:15501–15510. doi: 10.1021/bi00255a032. [DOI] [PubMed] [Google Scholar]
- 47.Dong J, Lu K, Lakdawala A, Mehta AK, Lynn DG. Controlling amyloid growth in multiple dimensions. Amyloid. 2006;13:206–215. doi: 10.1080/13506120600960809. [DOI] [PubMed] [Google Scholar]
- 48.Liang Y, Pingali SV, Jogalekar AS, Snyder JP, Thiyagarajan P, Lynn DG. Cross-strand pairing and amyloid assembly. Biochemistry. 2008;47:10018–10026. doi: 10.1021/bi801081c. [DOI] [PubMed] [Google Scholar]
- 49.Merkel JS, Regan L. Aromatic rescue of glycine in beta sheets. Fold Des. 1998;3:449–455. doi: 10.1016/s1359-0278(98)00062-5. [DOI] [PubMed] [Google Scholar]
- 50.Gazit E. A possible role for -stacking in the self-assembly of amyloid fibrils. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- 51.Waters ML. Aromatic interactions in peptides: Impact on structure and function. Biopolymers. 2004;76:435–445. doi: 10.1002/bip.20144. [DOI] [PubMed] [Google Scholar]
- 52.Waters ML. Aromatic interactions in model systems. Curr Opin Chem Biol. 2002;6:736–741. doi: 10.1016/s1367-5931(02)00359-9. [DOI] [PubMed] [Google Scholar]
- 53.Morgan DM, Dong JJ, Jacob J, Lu K, Apkarian RP, Thiyagarajan P, Lynn DG. Metal switch for amyloid formation: Insight into the structure of the nucleus. J Am Chem Soc. 2002;124:12644–12645. doi: 10.1021/ja0273086. [DOI] [PubMed] [Google Scholar]
- 54.Dong J, Canfield JM, Mehta AK, Shokes JE, Tian B, Childers WS, Simmons JA, Mao Z, Scott RA, Warncke K, et al. Engineering metal ion coordination to regulate amyloid fibril assembly and toxicity. Proc Natl Acad Sci. 2007;104:13313–13318. doi: 10.1073/pnas.0702669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••55.Liu P, Ni R, Mehta AK, Childers WS, Lakdawala A, Pingali SV, Thiyagarajan P, Lynn DG. Nucleobase-directed amyloid nanotube assembly. J Am Chem Soc. 2008;130:16867–16869. doi: 10.1021/ja807425h. Incorporation of the cytosine base dyad within the peptide backbone results in extensive β-sheet lamination and the formation of hollow nanotubes. The parallel β-strand organization results in distinct inner and outer surfaces that could be utilized to organize and segregate components inside or outside the tubes. [DOI] [PubMed] [Google Scholar]
- 56.Jin L-W, Claborn KA, Kurimoto M, Geday MA, Maezawa I, Sohraby F, Estrada M, Kaminksy W, Kahr B. Imaging linear birefringence and dichroism in cerebral amyloid pathologies. Proc Natl Acad Sci U S A. 2003;100:15294–15298. doi: 10.1073/pnas.2534647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••57.Childers WS, Mehta AK, Lu K, Lynn DG. Templating molecular arrays in amyloid's cross-beta grooves. J Am Chem Soc. 2009;131:10165–10172. doi: 10.1021/ja902332s. The ability of KLVFFAL peptide nanotubes to form small molecule binding sites was characterized. Each nanotube is composed of numerous binding sites along the β-termini packed in well defined 2D arrays. [DOI] [PubMed] [Google Scholar]
- 58.Krebs MRH, Bromley EHC, Donald AM. The binding of Thioflavin-T to amyloid fibrils: Localisation and implications. J Struct Biol. 2005;149:30–37. doi: 10.1016/j.jsb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- ••59.Biancalana M, Makabe K, Koide A, Koide S. Molecular mechanism of Thioflavin-T binding to the surface of β-rich peptide self-assemblies. J Mol Biol. 2009;385:1052–1063. doi: 10.1016/j.jmb.2008.11.006. The authors designed a “peptide self-assembly mimic” model system to understand the interaction of Thioflavin T with β-sheet rich structures. Introduction of a single tyrosine created a binding interface for Thioflavin T. This work illustrates that the β-sheet pleat surface could specifically harbor a variety of small molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu K, Guo L, Mehta AK, Childers WS, Dublin SN, Skanthakumar S, Conticello VP, Thiyagarajan P, Apkarian RP, Lynn DG. Macroscale assembly of peptide nanotubes. Chem Commun (Camb) 2007:2729–2731. doi: 10.1039/b701029j. [DOI] [PubMed] [Google Scholar]
- •61.Liang Y, Guo P, Pingali SV, Pabit S, Thiyagarajan P, Berland KM, Lynn DG. Light harvesting antenna on an amyloid scaffold. Chem Commun. 2008:6522–6524. doi: 10.1039/b814262a. Amyloid nanotubes were used as a scaffold to organize a FRET pair via covalent attachment of Rhodamine and non-covalent binding of Alexa 555. The resulting ordered pigment array provides the preliminary steps towards design of a functional light-harvesting antenna. [DOI] [PubMed] [Google Scholar]
- 62.Channon KJ, Devlin GL, Magennis SW, Finlayson CE, Tickler AK, Silva C, MacPhee CE. Modification of fluorophore photophysics through peptide-driven self-assembly. J Am Chem Soc. 2008;130:5487–5491. doi: 10.1021/ja710310c. [DOI] [PubMed] [Google Scholar]
- 63.Channon KJ, Devlin GL, MacPhee CE. Efficient energy transfer within self-assembling peptide fibers: A route to light-harvesting nanomaterials. J Am Chem Soc. 2009;131:12520–12521. doi: 10.1021/ja902825j. [DOI] [PubMed] [Google Scholar]
- 64.Channon K, MacPhee CE. Possibilities for ‘smart’ materials exploiting the self-assembly of polypeptides into fibrils. Soft Matter. 2008;4:647–652. doi: 10.1039/b713013a. [DOI] [PubMed] [Google Scholar]
- 65.Serio TR, Cashikar AG, Moslehi JJ, Kowal AS, Lindquist SL, Ronald W. Methods in enzymology. Volume 309. Academic Press; 1999. [41] yeast prion [[psi]+] and its determinant, sup35p; pp. 649–673. [DOI] [PubMed] [Google Scholar]
- 66.Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the saccharomyces cerevisiae ure2 protein. Proc Natl Acad Sci U S A. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •67.Guglielmi F, Monti DM, Arciello A, Torrassa S, Cozzolino F, Pucci P, Relini A, Piccoli R. Enzymatically active fibrils generated by the self-assembly of the ApoA-I fibrillogenic domain functionalized with a catalytic moiety. Biomaterials. 2009;30:829–835. doi: 10.1016/j.biomaterials.2008.10.036. A bifunctional chimeric protein containing an amyloidgenic domain coupled with a glutathione S-transferase (GST) catalytic domain was demonstrated to assemble into amyloid fibers and retain 100% catalytic function for at least 2 weeks. This work illustrates that amyloid can function as an active matrix to localize functional proteins. [DOI] [PubMed] [Google Scholar]
- 68.Baldwin AJ, Bader R, Christodoulou J, MacPhee CE, Dobson CM, Barker PD. Cytochrome display on amyloid fibrils. J Am Chem Soc. 2006;128:2162–2163. doi: 10.1021/ja0565673. [DOI] [PubMed] [Google Scholar]
- ••69.Blackmond DG. An examination of the role of autocatalytic cycles in the chemistry of proposed primordial reactions. Angew Chem-Int Edit. 2009;48:386–390. doi: 10.1002/anie.200804565. The authors identify key catalytic features that distinguish between auto-inductive and autocatalytic processes. [DOI] [PubMed] [Google Scholar]
- 70.Lee DH, Granja JR, Martinez JA, Severin K, Ghadiri MR. A self-replicating peptide. Nature. 1996;382:525–528. doi: 10.1038/382525a0. [DOI] [PubMed] [Google Scholar]
- 71.Yao S, Ghosh I, Zutshi R, Chmielewski J. A ph-modulated, self-replicating peptide. J Am Chem Soc. 1997;119:10559–10560. [Google Scholar]
- 72.Lee DH, Severin K, Yokobayashi Y, Ghadiri MR. Emergence of symbiosis in peptide self-replication through a hypercyclic network. Nature. 1997;390:591–594. doi: 10.1038/37569. [DOI] [PubMed] [Google Scholar]
- 73.Saghatelian A, Yokobayashi Y, Soltani K, Ghadiri MR. A chiroselective peptide replicator. Nature. 2001;409:797–801. doi: 10.1038/35057238. [DOI] [PubMed] [Google Scholar]
- ••74.Ryadnov MG, Woolfson DN. Self-assembled templates for polypeptide synthesis. J Am Chem Soc. 2007;129:14074–14081. doi: 10.1021/ja072960s. As an extension of self-replicating coiled-coil peptides [70-73], the authors exploit the organization of sticky-end peptide self-assembled fibers and proximity-driven chemical ligation of N- and C-terminal residues to catalyze the formation of polypeptides > 3MDa. [DOI] [PubMed] [Google Scholar]
- 75.Takahashi Y, Mihara H. Construction of a chemically and conformationally self-replicating system of amyloid-like fibrils. Bioorganic & Medicinal Chemistry. 2004;12:693–699. doi: 10.1016/j.bmc.2003.11.022. [DOI] [PubMed] [Google Scholar]
- •76.Rubinov B, Wagner N, Rapaport H, Ashkenasy G. Self-replicating amphiphilic beta-sheet peptides. Angew Chem Int Ed Engl. 2009;48:6683–6686. doi: 10.1002/anie.200902790. Using (Phe-Glu)n peptides as a beta-sheet forming template, the authors demonstrate the beta-sheet template can promote the ligation of two fragments to make a new beta-sheet replicator. [DOI] [PubMed] [Google Scholar]
- 77.Dawkins R. Man vs. God. The Wall Street Journal. 2009 [Google Scholar]
- 78.Cech TR. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987;236:1532–1539. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]
- 79.Joyce GF. RNA evolution and the origins of life. Nature. 1989;338:217–224. doi: 10.1038/338217a0. [DOI] [PubMed] [Google Scholar]
- 80.Gilbert W. Origin of life - the RNA world. Nature. 1986;319:618–618. [Google Scholar]
- 81.Robertson MP, Scott WG. The structural basis of ribozyme-catalyzed RNA assembly. Science. 2007;315:1549–1553. doi: 10.1126/science.1136231. [DOI] [PubMed] [Google Scholar]
- 82.McClain WH, Guerriertakada C, Altman S. Model substrates for an RNA enzyme. Science. 1987;238:527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]