Abstract
Acute lung injury and acute respiratory distress syndrome are the result of intense inflammation in the lungs leading to respiratory failure. The causes of acute lung injury/acute respiratory distress syndrome are numerous (e.g., pneumonia, sepsis and trauma) but the reasons why certain individuals develop lung injury in response to these stimuli and others do not are not well understood. There is ample evidence in the literature that gene–host and gene–environment interactions may play a large role in the morbidity and mortality associated with this syndrome. In this review, we initially discuss methods for identification of candidate acute lung injury/acute respiratory distress syndrome susceptibility genes using a number of model systems including in vitro cell systems and inbred mice. We then describe examples of polymorphisms in genes that have been associated with the pathogenesis of acute lung injury/acute respiratory distress syndrome in human case–control studies. Systematic bench to bedside approaches to understand the genetic contribution to acute lung injury/acute respiratory distress syndrome have provided important insight to this complex disease and continuation of these investigations could lead to the development of novel prevention or intervention strategies.
Keywords: acute respiratory distress syndrome, ARDS, association study, genetical genomics, genome-wide association studies, GWAS, haplotype, translational investigation
Acute lung injury (ALI) is a common and devastating illness in the intensive care unit, with mortality rates exceeding 30–50% [1]. The diagnosis of ALI/acute respiratory distress syndrome (ARDS) is by clinical criteria, established by the presence of new bilateral pulmonary infiltrates on chest radiography and severe hypoxia in the absence of the clinical diagnosis of congestive heart failure [2]. The degree of hypoxia dictates whether a patient has ALI (PaO2/FiO2 < 300) versus ARDS (PaO2/FiO2 < 200). ALI/ARDS occurs secondarily in a number of disease processes, most commonly sepsis, pneumonia, aspiration, trauma, pancreatitis, blood transfusions, smoke or toxic gas inhalation, and certain types of drug toxicity [3].
The pathogenesis of ALI/ARDS is not well understood. The disease process is characterized by diffuse damage to the alveoli resulting in disruption of the endothelium and epithelium. Fluid accumulates in the alveolar spaces, and is accompanied by severe inflammation and gas exchange abnormalities. These changes comprise the acute phase of ALI/ARDS. The subsequent fibrotic phase results in diffuse interstitial thickening, fibrosis, increased dead space and loss of lung compliance. In the recovery phase, which is not always present, hypoxemia resolves and lung compliance improves. After recovery, though radiographic infiltrates may improve and even resolve, residual microscopic fibrosis will still be present and clinical abnormalities in lung function remain [4]. Numerous attempts have been made to improve the management of ALI/ARDS with medical interventions, such as low tidal volume ventilation [5] and manipulations of the coagulation cascade [6], but mortality from ALI/ARDS remains unacceptably high.
The biochemical reasons why certain patients are more susceptible to ALI/ARDS are not understood. Considerable research has led to the identification of protein biomarkers that may be useful in predicting pathogenesis or outcome in ALI. Biomarkers include proinflammatory cytokines TNF-α [7,8] and IL-6 [7], VEGF [9,10], plasminogen activator inhibitor-1 [11], surfactant protein B [12], P-selectin [13], angiopoietin 2 [14] and peptidase inhibitor 3 (PI3; [15,16]). While the identification of biomarkers has provided important insight to the etiology of ALI, and may have some predictive value, biomarker-based novel intervention strategies have not been developed [17].
It has been suggested that genes involved in inflammatory and immune pathways may play a role in conferring susceptibility and morbidity in lung injury [18], in addition to gene–environment interactions. A genetic approach to assessing individual susceptibility is attractive since genotype can be easily determined from peripheral blood with minimal risk. Furthermore, in contrast to protein biomarkers that may be transiently expressed during disease pathogenesis, gene polymorphisms also do not vary in response to underlying illnesses, and may be predictive indicators of disease susceptibility.
Identification of candidate genes
The recent emergence of genomics and proteomics technologies has resulted in an enormous volume of sequence information to help understand human disease. Where genomics sequences and analyzes genes, proteomics concentrates on the analysis of complete sets mechanisms of disease can be determined. In the following section we briefly discuss genetic and genomic approaches that have been used in a variety of model systems to identify candidate genes that contribute to the pathogenesis of and/or susceptibility to ALI.
Linkage & association analyses
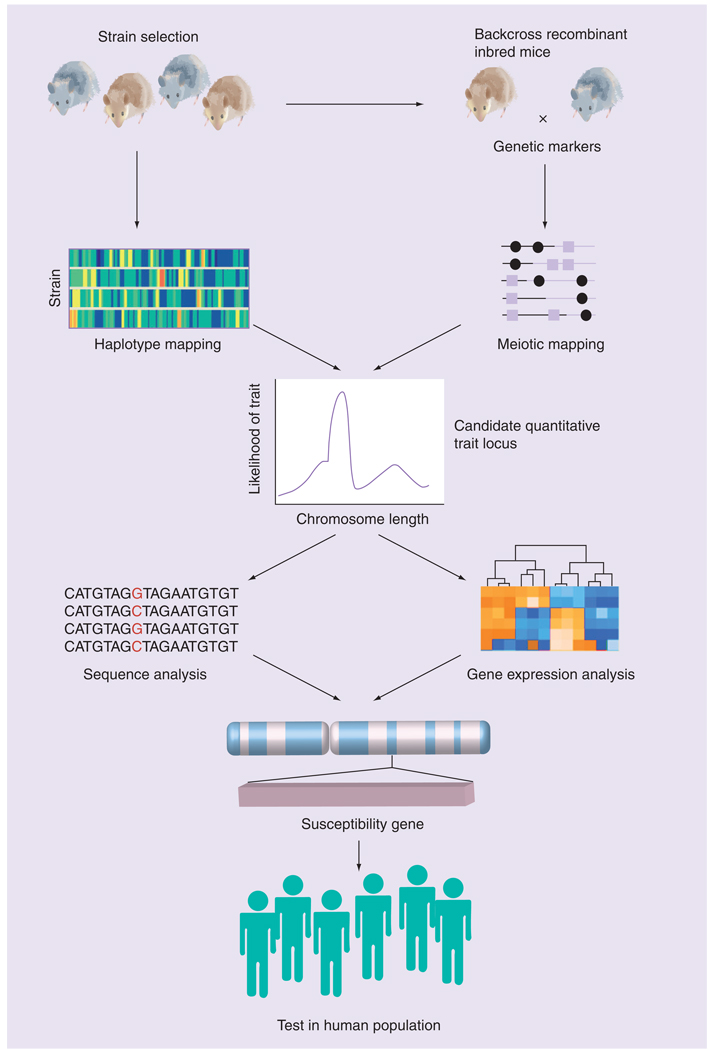
Two broad research strategies have been utilized to identify genes (or quantititative trait loci [QTLs]) that determine disease susceptibility. The first is meiotic (linkage) mapping and positional cloning (FIGURE 1). Linkage mapping exploits within-family associations between marker alleles and putative trait-influencing alleles that arise within families and may be followed by methods of co-segregation analyses. This approach is designed to identify association of a chromosomal interval(s) within the entire genome that may contain genes that are polymorphic and account for the differential response phenotype under study. That is, no a priori hypothesis regarding the role of a specific gene or genes is tested. Unfortunately, this approach is not feasible for human ALI/ARDS as the disease is sporadic, the result of extreme environmental insult(s), and ALI is rarely found to cluster in families. Furthermore, given the complex etiology and multifactorial nature of ALI/ARDS it is likely that few polymorphisms will confer a high degree of risk and fine-mapping these polymorphisms would be difficult with family-based designs. However, linkage analyses are ideally suited for genetically well-controlled models, particularly inbred mice. Furthermore, because of the highly significant homologies in gene order and chromosomal structure that exist between mouse and man, identification of the chromosomal location of a susceptibility gene in the mouse provides the basis for potentially localizing a homologous human gene.
Figure 1.
Positional cloning strategy using inbred mice to identify candidate susceptibility genes that may be tested for association with disease in human populations.
Adapted from [19].
A number of laboratories have used various exogenous and endogenous stimuli (e.g., hyperoxia, metals and endotoxin) to develop genetic models of ALI in mice. As is the case with all animal models of human disease, these models have limitations because, while simulating human lung injury phenotypes, the ALI phenotypes elicited by these challenges may not share the same pathogenetic mechanisms. However, models such as these have been successful in identification of candidate genes for testing in human populations (see below). For example, continuous exposure of mice to hyperoxia (>95%) induces inflammation and noncardiogenic edema in the lung, which are phenotypes of ALI/ARDS. The pathogenesis of this process involves the production of reactive oxygen species, which overwhelms the natural antioxidant system, leading to tissue injury [19]. Excessive production of reactive oxygen species occurs upon exposure to hyperoxia, leading to a profound activation of the inflammatory response. The release of inflammatory mediators and reactive oxygen species leads to endothelial cell injury, interstitial and perivascular edema, epithelial cell hypertrophy and proliferation, and denudation of the alveolar basement membrane. There is also evidence of both apoptotic and necrotic cell death of the endothelium and epithelium [20]. Clinically, prolonged exposure of normal subjects to high levels of oxygen (90–95% O2) results in tracheobronchitis [21,22], reduced tracheal mucous velocity [21] and cardiovascular effects (e.g., reduction in heart rate; increased mean arterial pressure, systemic vascular resistance and large artery stiffness) [23]. Hyperoxia may ultimately contribute to adverse outcomes in septic patients (e.g., [24]), but the overall role of hyperoxia in the pathogenesis of ARDS remains uncertain and controversial [25]. Nonetheless, understanding of the mechanisms of oxygen toxicity in rodent models may provide important insights to the pathogenesis of ALI/ARDS, as similar phenotypes are found in these diseases. Interestingly, interstrain variation in time course and magnitude of change in total lavageable protein concentration (a marker of lung permeability) and inflammatory cells (e.g., polymorphonuclear leukocytes) was significantly greater than intrastrain variation among the strains of mice in response to hyperoxia [26]. Using hyperoxia-susceptible (C57BL/6J, B6) and -resistant (C3H/HeJ, C3) mice, and intercross (F2) and recombinant inbred cohorts derived from them, genome-wide linkage analysis identified significant and suggestive QTLs on chromosomes 2 (hyperoxia susceptibility locus 1 [Hsl1]) and 3 (Hsl2), respectively. Fine and comparative mapping of Hsl1 identified a strong candidate gene, Nfe2l2 (nuclear factor, erythroid derived 2, like 2 or Nrf2) that encodes Nrf2, a cap’n’collar basic leucine zipper transcription factor that regulates antioxidant and phase 2 gene expression through binding of a Nrf2/small Maf protein heterodimer to promoter antioxidant response elements [27]. Proof of concept for Nrf2 as a candidate susceptibility gene for hyperoxia-induced ALI was provided by strain-specific variation in lung Nrf2 messenger RNA expression and a T→A substitution in the B6 Nrf2 promoter that cosegregated with susceptibility phenotypes in F2animals [27]. Furthermore, mice with a site-directed mutation of Nrf2 (Nrf2−/−) have significantly greater hyperoxia-induced ALI phenotypes compared with wild-type mice (Nrf2+/+) [28].
Prows and Leikauf developed a model of ALI by exposing mice to nickel sulfate aerosol, an occupational contaminant and respiratory irritant [29]. Continuous exposure of inbred mice to 150 µg/m3 nickel sulfate causes death in a strain-dependent manner; a QTL analysis with backcross mice derived from susceptible A/J and resistant B6 strains identified significant linkage to chromosome 6, and suggestive linkage to chromosomes 1 and 12. Interestingly, these QTLs are distinct from those identified for acute lung injury induced by hyperoxia, but are similar to those identified by these investigators for death induced by continuous exposure to high concentrations of the oxidant ozone [30]. Tgfa was identified as a chromosome 6 QTL candidate gene for susceptibility to nickel sulfate and subsequent investigations have confirmed a role for this gene in the pathogenesis of ALI phenotypes [31].
Ozone is a highly reactive oxidant that also occurs in air pollution that stimulates inflammation and epithelial injury in the lung. Investigations by Kleeberger et al. using inbred mice demonstrated that different sets of genes are responsible for injury due to acute or subacute ozone exposures [32]. Linkage analysis studies to identify a QTL for subacute ozone induced lung injury identified a region on chromosome 17, which included a number of candidate susceptibility genes including TNF [33]. Subsequent studies using anti-TNF-α antibodies [33] and TNF receptor knock out mice [34] confirmed a significant role for TNF-α in ozone-induced inflammation and injury.
These examples of linkage analyses of ALI susceptibility phenotypes illustrate the utility of this process to first identify, and then functionally test, candidate genes in mouse models. Further underscoring the usefulness of linkage analyses is that, because of the strong homology between genomes, functionally relevant murine candidate genes may be directly translatable to human disease in hypothesis-driven case–control investigations (e.g., NRF2, see below).
The emergence of whole-genome sequencing and high-density SNP information across multiple species has tremendously enhanced the power of association studies. One type of association study tests whether functional SNPs in candidate genes associate with risk of disease phenotypes. As mentioned above, these investigations rely on biological plausibility of the gene under investigation. Another category of association study asks whether all SNPs in a gene, irrespective of function, associate with disease singly or in haplotype blocks. The completion of the HapMap project and development of high-density genome-wide SNP arrays have enabled genome-wide association studies (GWAS) for many human complex diseases [35]. Unbiased GWAS have provided important insight to novel susceptibility genes for Type II diabetes [36], prostate cancer [37], pulmonary sarcoidosis [38] and asthma phenotypes [39]. However, it should be noted that GWAS studies are not without challenges and limitations [40]. A number of study design issues and requirements have been identified that must be considered including disease heterogeneity and phenotype definition, population substructure, epistatic interactions and replication across independent populations [41–43]. No GWAS studies have yet been published for ALI/ARDS, but the experience with other complex diseases suggests that, with the appropriate study design, the approach may lead to novel insights into this disease. In mouse models, recently developed emergent haplotype mapping algorithms based on high-density SNP mapping across multiple inbred strains have provided additional tools for investigators to identify disease genes (see e.g., [44,45]). The developing collaborative cross which will create 1000 recombinant inbred strains derived from eight parental strains should also greatly advance our ability to determine the genetic basis of disease phenotypes [46].
Genomic approaches
High-density gene-expression array technologies and their application to primary cell culture systems, cell lines and animal models under similar stress conditions have also provided novel insight to susceptibility genes and gene patterns that correlate with responsivity and/or susceptibility to the stress. For example, Grigoryev et al. have used whole-genome expression profiling in multiple species (rat, mouse, dog and human) to identify candidate genes that were differentially expressed in response to ventilator-associated ALI [18,47]. Identification of those differentially expressed genes that were conserved across species enabled prioritization of gene candidates to be validated (i.e., proof of concept) in the models. Priority genes include IL6, macrophage migration inhibitory factor (MIF), myosin light chain kinase (MLCK), VEGF and heat shock protein 70.
Perkowski and colleagues sought to identify genes that were differentially expressed during the early response to hyperoxia [48]. Using B6 mice, genome-wide gene expression was analyzed after 0, 8, 24 and 48 h after exposure. A total of 385 genes were found to be differentially expressed, 175 of which were upregulated and 210 were downregulated in response to hyperoxia. They found that many antioxidants such as catalase and superoxide dismutase (both manganese and copper-zinc forms) showed no change in expression, while antioxidant enzymes glutathione peroxidase and heme-oxygenase 1 expression increased. Expression of proinflammatory genes was largely unchanged after 24 and 48 h exposures. Gene-expression changes also indicated an overall inhibition of cell cycle progression. Interestingly, thrombomudulin expression was decreased significantly, suggesting a role for the coagulation and inflammatory pathways in the pathogenesis of hyperoxia-induced lung injury.
Gene-expression arrays may also be used to evaluate downstream effector genes or pathways altered by targeted disruption of genes known to be functionally relevant to ALI/ARDS. For example, Cho et al. sought to identify the genes that were differentially expressed in mice lacking Nrf2 compared with wild-type mice after hyperoxia exposure [49]. In particular, antioxidant response element-containing antioxidant/redox-cycle enzyme genes were differentially expressed, including NAD(P)H:quinone oxidoreductase (NQO1), GST – Ya and -Yc subunits, UDP glycosyl transferase, glutathione peroxidase 2, and heme oxygenase 1. Genes involved in cell growth, signal transduction, inflammation and immunity, and transcription were also differentially expressed in Nrf2 knockout mice compared with controls after hyperoxia. These results suggested that modification of expression of antioxidant genes and antioxidant defenses via Nrf2 may play a role in ALI, but other potentially important biological pathways that may affect differential responsiveness to hyperoxia were also implicated in this study, and have provided novel mechanistic insight.
Genetical genomics
The genetic and genomic approaches applied to animal and cell models have clearly proved to be useful in the identification of candidate susceptibility genes for testing in human populations. Relatively recently, investigations have integrated genetics and genomics to draw upon the properties of both to provide additional insight to the genetic contribution to disease susceptibility and pathogenesis. ‘Genetical genomics’ seeks to combine QTLs for protein level and gene expression with traditional disease phenotype QTL approaches to help identify and prioritize candidate genes for further investigation (for recent reviews, see [50,51]). Genetical genomics has identified important gene networks for complex diseases and physiological traits such as hematopoietic stem cell function [52], obesity [53], neurobehavioral phenotypes [54] and cardiovascular disease [50]. While genetical genomics has not yet been published for ALI/ARDS models, studies are ongoing in our laboratory.
Genetic polymorphisms associated with acute lung injury
Using the methods described above, various candidate susceptibility genes have been identified in cell and animals models. Many of the studies have implicated genes involved in inflammation and immune modulation, as well as antioxidant/cell cycle related processes. To determine whether any of these genes have relevance to susceptibility and severity associated with ALI/ARDS, a number of investigations have evaluated the role of functional polymorphisms in case–control investigations. Reviewed below are some of the genes that have been investigated for association with development of lung injury. This list is not exhaustive, but rather illustrates a subset of genes and biological processes that have been tested for relevance to ALI/ARDS pathogenesis. Additional candidate genes that are distributed on 11 chromosomes throughout the human genome have been investigated for association with susceptibility to ALI/ARDS and are included with their chromosomal location in Table 1.
Table 1.
Summary of candidate genes investigate in case–control investigations of acute lung injury.
| Candidate gene | Symbol | Chromosome (location) | Ref. |
|---|---|---|---|
| Angiotensin converting enzyme | ACE | 17 (q23.3) | [55,60–62] |
| Epidermal growth factor | EGF | 4 (q25) | [106] |
| Glutathione S-transferase M1 | GSTM1 | 1 (p13.3) | [107] |
| Inhibitor κB-α | NFKBIA | 14 (q13) | [108] |
| Interleukin-6 | IL6 | 7 (p21) | [109] |
| Interleukin-8 | IL8 | 4 (q13-q21) | [110] |
| Interleukin-10 | IL10 | 1 (q31-q32) | [67,70] |
| Macrophage migration inhibitory factor | MIF | 22 (q11.23) | [111] |
| Mannose binding lectin | MBL2 | 10 (q11.2-q21) | [73] |
| Myosin light chain kinase | MYLK | 3 (q21) | [77,78] |
| NF-E2 related factor 2 | NRF2 | 2 (q31) | [81] |
| NAD(P)H:quinone oxidoreductase 1 | NQO1 | 16 (q22.1) | [86] |
| Nuclear factor κB | NFKB1 | 4 (q24) | [112] |
| Plasminogen activator inhibitor-1 | PAI1 | 7 (q21.3-q22) | [113] |
| Pre-B cell colony enhancing factor | PBEF | 7 (q22.2) | [90,91] |
| Superoxide dismutase 3 | SOD3 | 4 (p15.3-p15.1) | [64] |
| Surfactant protein B | SFTPB | 2 (p12-p11.2) | [85–87] |
| Toll-like receptor 1 | TLR1 | 4 (p14) | [114] |
| Tumor necrosis factor-α | TNF | 6 (p21.3) | [89] |
| Urokinase | PLAU | 10 (q24) | [115] |
| Vascular endothelial growth factor A | VEGFA | 6 (p12) | [10,101] |
Angiotensin-converting enzyme
Activation of the pulmonary renin–angiotensin system has been speculated to influence the pathogenesis of ARDS by altering vascular permeability, vascular tone, fibroblast activity and alveolar epithelial cell survival [55]. Serum angiotensin converting enzyme (ACE) levels have been shown to be decreased in patients with ARDS [56], while ACE levels in bronchoalveolar lavage fluid is elevated [57].
Studies have shown that Ace knockout mice are protected from severe ALI induced by acid aspiration or sepsis [58]. Furthermore, pretreatment of mice with a systemic ACE inhibitor, enalapril, significantly attenuated endotoxin-induced acute lung inflammation [59]. These results also suggest that ACE promotes ALI through edema formation and decreased elastance.
The human ACE gene (ACE) contains a RFLP consisting of an insertion or deletion (I/D) of a 287 base pair Alu repeat sequence in intron 16. Presence of a DD genotype has been associated with increased plasma ACE concentrations [60]. Marshall et al. genotyped patients with ARDS, patients with non-ARDS respiratory failure, patients undergoing coronary artery bypass grafting and a general population group [55]. They found that the DD genotype frequency was increased in patients with ARDS compared with other non-ARDS patients, coronary artery bypass grafting patients and the general population, as well as being associated with a higher mortality in the ARDS group (p < 0.02).
Jerng et al. studied Chinese patients with ARDS, ‘at risk’ intensive care unit patients with acute respiratory failure (non-ARDS), and not-at-risk individuals [61]. Patients with the II genotype had a significantly increased chance of survival compared with the DD genotype, but no increased risk for ARDS for patients with the D allele was identified in this study. Interestingly, Villar et al. found that the ACE gene I/D polymorphism did not associate with susceptibility or mortality in a Spanish cohort [62]. The contradictory findings between studies and populations illustrate the necessity for replication of association studies as well as careful consideration of study design (see Future Perspective).
Extracellular superoxide dismutase
Extracellular superoxide dismutase (SOD3) is one of three human SODs, and is a potent extracellular antioxidant enzyme. SOD3 is found in the extracellular spaces of many tissues including the lung. Experiments with SOD3 knockout mice have shown that deletion of Sod3 enhanced susceptibility to lipopolysaccharide (LPS)-induced lung injury and inflammation, whereas overexpression of Sod3 reduced inflammation induced by LPS thus confirming an important role for this gene in protection against lung injury [63]. Arcaroli et al. resequenced SOD3 and found a GCCT haplotype that reduced risk on the ventilator and mortality in two patient populations with infection-associated ALI [64]. Although the protective mechanism conferred by the SOD3 haplotype is not yet known, the strong association of the haplotype with protection against ALI suggests that SOD3 is an important determinant of disease susceptibility.
IL-10
IL-10 suppresses the expression of proinflammatory cytokines and has been shown to be elevated in trauma and is associated with multiorgan dysfunction [65], and it has been reported that 50–75% of the variation of IL-10 production is genetically controlled. The 5′ flanking region of IL10 contains numerous polymorphisms that are completely or strongly linked and three haplotypes have been described (GCC, ACC and ATA [-1082, -819 and -592 positions respectively]) and are associated with varying levels of IL-10 production with GCC/GCC individuals with the highest IL-10 levels [66].
Schroder et al. genotyped 119 severely injured trauma patients for each IL10 polymorphism [67]. Though the polymorphisms were not associated with specific IL-10 levels, the -592AC polymorphism was associated with a 3.3-fold increase in relative risk in developing multiorgan dysfunction.
Data from studies measuring IL-10 levels in ARDS have varied. Patients with ARDS have been shown to have lower levels of IL-10 compared with critically ill non-ARDS patients [68]. But in ARDS patients, low bronchoalveolar lavage concentrations of IL-10 and high plasma IL-10 correlated with increased mortality [69]. Gong et al. have shown, in a nested case–control study of patients at risk for ARDS, that IL10 polymorphism -1082GG (high IL-10 producing) is associated with lower severity of illness, lower mortality and lower organ failure amongst patients with ARDS depending on age [70].
Mannose binding lectin
Mannose binding lectin (MBL) activates the complement system in an antibody-dependent manner and binds to mannose and N-acetyl glucosamine residues on microorganisms. Point mutations in MBL within exon 1 leading to amino acid substitutions result in decreased serum levels of MBL. Promoter polymorphisms also exist that influence MBL levels and have been associated with increased susceptibility to infections [71].
Fidler et al. sequenced MBL2 in DNA from 100 children admitted to a pediatric intensive care unit [72]. MBL2 variant alleles were associated with increased risk (sevenfold) and severity of systemic response to infection, which also correlated with lower MBL levels. Gong et al. genotyped codons -221(X), 52(D), 54(B) and 57(C) for variant MBL2 alleles in patients with ARDS and healthy individuals [73]. Variant codon 54BB was associated with increased severity of illness and increased odds of ARDS (OR: 6.7), especially in patients with septic shock.
Myosin light-chain kinase
Dysfunction of the endothelial cell layer in ALI can result in cytokine release and increased endothelial permeability. This barrier is thought to be regulated by MLCK through the phosphorylation of myosin light chains and subsequent effects on the actin–myosin interaction and cell contraction [74].
Myosin light-chain kinase knockout mice have been shown to be protected from LPS-induced lung injury [75]. Another study utilized a MLCK inhibitor, 5-iodonaphthalene-1-sulphonylhomopiperazine (ML-7), in mice treated with intratracheal instillation of LPS. Pretreatment with ML-7 inhibited LPS induced airway epithelial permeability and inflammation [76].
Gao et al. sequenced the MLCK gene in sepsis-associated ALI patients, sepsis patients and healthy individuals [77]. A total of 51 SNPs were identified, and numerous polymorphisms were associated with risk for sepsis and ALI, as well as ALI alone. Christie et al. also found that variation in MYLK associates with development of ALI in a major trauma cohort [78].
NF-E2 related factor 2
In response to a number of stressors including oxidants, NF-E2 related factor 2 (NRF2) dissociates from the cytoplasmic inhibitor, Kelchlike ECH-associated protein (Keap1), and translocates to the nucleus where it induces the transcription of antioxidant response element bearing detoxifying enzymes [79,80]. Prior positional cloning studies performed in our laboratory have suggested that Nrf2 is a candidate susceptibility gene in hyperoxia-induced lung injury ([27], see above).
To determine whether this gene is also important in human disease, Marzec et al. resequenced NRF2 and identified three novel, potentially functional promoter SNPs at positions -617 (C/A), -651 (G/A) and -653 (A/G) [81]. Luciferase reporter and transcription factor binding assays confirmed that the -617 SNP conferred loss of NRF2 function. Further, in a nested case–control study of major trauma patients, those with the -617 A SNP had a significantly higher risk for developing ALI after major trauma (OR: 6.44; 95% CI: 1.34, 30.8; p = 0.021) relative to patients with the wild-type (-617 CC).
NQO1
NAD(P)H:quinone oxidoreductase 1 (NQO1) is a phase II/antioxidant enzyme that catalyzes the two electron reduction of a variety of quinine compounds, which prevents the generation of free radicals and reactive oxygen species. In some instances, NQO1 metabolism creates a more active product where redox-labile hydroquinones can react with molecular oxygen to form semiquinones that can generate reactive oxygen species and cause DNA alkylation [82]. Furthermore, NQO1 is highly inducible [83–85] and NQO1 has been associated with oxidant stress, which is thought to be an important component of ALI/ARDS. These observations led Reddy et al. to test whether functional NQO1 promoter SNPs associate with risk of ALI [86]. These investigators found a SNP (A-1221C) that decreased transcription of NQO1 basally and after oxidant stress in BEAS-2B cells. They also found that, in a prospective cohort of major trauma patients, the -1221 C allele conferred protection (OR: 0.46; 95% CI: 0.23, 0.90; p = 0.024) against ALI compared with patients homozygous for the wild-type allele.
Pre-B cell colony-enhancing factor
Pre-B cell colony-enhancing factor/nicotinamide phosphoribosyl transferase (PBEF) is a cytokine induced by mechanical distension/force [87] and other inflammatory cytokines [88]. PBEF was significantly expressed in the neutrophils of septic patients [89]. Ye et al. genotyped two PBEF polymorphisms (-1001TG and -1543CT) in Caucasian patients with sepsis-induced ALI, patients with severe sepsis and healthy individuals [90]. They found that the haplotype GC had a higher risk of developing ALI (7.7-fold), while the TT haplotype was protective against ALI. Bajwa et al. also genotyped these polymorphisms in patients with ARDS and at-risk controls and reported that patients with SNP -1001G had a significantly increased odds of developing ARDS (OR: 1.35) and increased intensive care unit mortality, and SNP -1543T was associated with a decreased odds of developing ARDS (OR: 0.66) and decreased 28-day and 60-day mortality and shorter duration of mechanical ventilation [91].
Surfactant protein
Surfactant protein (SP) reduces the surface tension in the alveoli to allow for inflation of the lungs. During ALI, epithelial injury results in increased serum levels and decreased bronchoalveolar lavage levels of surfactant and has been used as a marker of lung injury and increased permeability [12]. Increased plasma levels of surfactant have been associated with worse clinical outcomes in ALI patients [92,93].
Floros et al. have reported an insertion/deletion polymorphism in SP-B (SFTPB) intron 4 is associated with neonatal respiratory distress syndrome [94]. Furthermore, this variant polymorphism of the SFTPB was associated with ARDS and with direct pulmonary injury in women, but not in men [95]. The SFTPB +1580CT polymorphism, which results in decreased activity of SP-B, was also significantly associated with ARDS in patients with community acquired pneumonia [96]. Interestingly, Currier et al. found that a variable tandem repeat polymorphism in intron 4 of SFTPB associated with increased 60-day mortality in ARDS while the +1580CT polymorphism was not significantly associated with disease in the same cohort [97]. Therefore, while a consistent association of SFTPB with risk of ARDS appears to be emerging, the specific mechanism(s) of SFTPB involvement with disease pathogenesis is not clear.
TNF
TNF-α is a proinflammatory cytokine that has a role in the development of ALI by increasing endothelial permeability [98]. Increased serum TNF-α levels have been correlated to increased severity and mortality in ARDS in some studies, but this finding has not been consistent [7,8]. TNFSNP -308GA has been shown to be associated with protection from ARDS and improved mortality in patients with direct pulmonary injury [99].
VEGF
VEGF is involved in angiogenesis, endothelial cell proliferation and cell permeability, and therefore possibly involved in the development of pulmonary edema. Furthermore, increased levels of VEGF protected against hyperoxic acute lung injury in mice [100]. Plasma VEGF was elevated in patients with ARDS with intrapulmonary depletion of VEGF [9]. Medford et al. genotyped ARDS patients and at risk individuals for VEGFA SNP +936CT and found that this polymorphism was associated with higher APACHE III scores and increased risk of developing ARDS (OR: 1.77), but there were no differences in mortality rates [101]. Zhai et al. found that the VEGF +936TT and +936CT+TT genotypes and the TCT haplotype were associated with increased mortality in patients with ARDS [10]. They also found these genotypes were associated with decreased plasma VEGF levels, suggesting a protective role of VEGF in the severity of ARDS. These studies suggest VEGF may have an important role in the development of, and mortality associated with, ARDS but more studies are necessary to fully understand the role of VEGF in this disease.
Future perspective
Continued development of predictive/informative animal and cell models will provide additional candidate genes and gene networks that increase our understanding of susceptibility to ALI/ARDS. These studies should leverage the continued emergence of genetic and genomic data from multiple species (e.g., human cell lines, inbred mice and rats) and approaches (e.g., collaborative cross), as well as the development of more sophisticated bioinformatics tools, to identify candidate susceptibility genes for ALI/ARDS phenotypes. However, due to the multiple etiologies and phenotypes of complex diseases such as ALI/ARDS, careful consideration of study design is essential to avoid false-positive or false-negative findings (e.g., [102]). Potential pitfalls in case–control investigations include selection of cases and controls, population stratification, observation bias, linkage disequilibrium and sample size (see [103–105] for excellent reviews of case–control study design and interpretation). Well-characterized, replicated case–control populations will be critical for hypothesis-based candidate gene SNP association studies. Coupled with well-designed genome-wide association studies that have no a priori hypotheses, these genetic analyses should provide tremendous insight to the mechanisms of susceptibility to ALI/ARDS and may provide novel therapeutic intervention strategies.
Executive summary
Acute lung injury (ALI)/acute respiratory distress syndrome (ARDS) is a complex disease with multiple causes and diverse etiologies
The contribution of genetic background is becoming increasingly clear, but only recently have investigations begun to identify genes that may be involved in ALI/ARDS susceptibility. A number of these genes have been tested for association with disease.
Furthermore, gene–environment and gene–gene interactions will almost certainly be important in disease susceptibility.
-
Traditional family-based studies are not feasible, so alternative approaches have been applied to identify candidate genes:
Positional cloning (genetic) investigations in animal models;
Gene-expression (genomic) investigations in cell and animal models, with cross-species comparisons;
Genetical genomics, an integration of genetic and genomics approaches to identify candidate genes.
-
Case–control association studies have begun to clarify the importance of some genes and gene categories that are likely to have an impact on susceptibility to disease incidence and progression/severity, including:
Inflammation/immunity (e.g., TNF, IL10, MBL2 and TLR1);
Antioxidant defense (e.g., NRF2, NQO1 and SOD3);
Cell integrity (e.g., MLCK).
The complexity of ALI/ARDS has indicated the necessity for well-designed case–control and genome-wide association study investigations and replication in independent study populations to minimize false-positive and false-negative results.
Increased understanding of the role of genetic background to ALI/ARDS should lead to development of novel prevention and intervention strategies.
Acknowledgments
The authors thank Drs Donald Cook and Michael Fessler for critical review of the manuscript, and Dr Susan Edelstein for her creation of Figure 1.
Footnotes
Financial & competing interests disclosure
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Goss CH, Brower RG, Hudson LD, Rubenfeld GD. ARDS Network: Incidence of acute lung injury in the United States. Crit. Care Med. 2003;31:1607–1611. doi: 10.1097/01.CCM.0000063475.65751.1D. [DOI] [PubMed] [Google Scholar]
- 2.Bernard GR, Artigas A, Brigham KL, et al. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am. J. Respir. Crit. Care Med. 1994;149:818–824. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- 3.Hudson LD, Milberg JA, Anardi D, Maunder RJ. Clinical risks for development of the acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 1995;151:293–301. doi: 10.1164/ajrccm.151.2.7842182. [DOI] [PubMed] [Google Scholar]
- 4. Piantadosi CA, Schwartz DA. The acute respiratory distress syndrome. Ann. Intern. Med. 2004;141:460–470. doi: 10.7326/0003-4819-141-6-200409210-00012. ▪▪ Very good review of the etiology, pathogenesis and susceptibility factors for acute respiratory distress syndrome.
- 5.ARDS Net. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N. Engl. J. Med. 2000;342:1301–1308. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- 6.Bernard GR PROWESS study group. Efficacy and safety of recombinant human activated protein C for severe sepsis. N. Engl. J. Med. 2001;344:699–709. doi: 10.1056/NEJM200103083441001. [DOI] [PubMed] [Google Scholar]
- 7.Meduri GU, Headley S, Kohler G, et al. Persistent elevation of inflammatory cytokines predicts a poor outcome in ARDS. Plasma IL-1β and IL-6 levels are consistent and efficient predictors of outcome over time. Chest. 1995;107:1062–1073. doi: 10.1378/chest.107.4.1062. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly TJ, Meade P, Jagels M, et al. Cytokine, complement, and endotoxin profiles associated with the development of the adult respiratory distress syndrome after severe injury. Crit. Care Med. 1994;22:768–776. doi: 10.1097/00003246-199405000-00010. [DOI] [PubMed] [Google Scholar]
- 9.Thickett DR, Armstrong L, Millar AB. A role for vascular endothelial growth factor in acute and resolving lung injury. Am. J. Respir. Crit. Care Med. 2002;166:1332–1337. doi: 10.1164/rccm.2105057. [DOI] [PubMed] [Google Scholar]
- 10.Zhai R, Gong MN, Zhou W, et al. Genotypes and haplotypes of the VEGF gene are associated with higher mortality and lower VEGF plasma levels in patients with ARDS. Thorax. 2007;62:718–722. doi: 10.1136/thx.2006.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prabhakaran P, Ware LB, White KE, et al. Elevated levels of plasminogen activator inhibitor-1 in pulmonary edema fluid are associated with mortality in acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;285:L20–L28. doi: 10.1152/ajplung.00312.2002. [DOI] [PubMed] [Google Scholar]
- 12.Pan T, Nielsen LD, Allen MJ, et al. Serum SP-D is a marker of lung injury in rats. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;282:L284–L832. doi: 10.1152/ajplung.00421.2000. [DOI] [PubMed] [Google Scholar]
- 13.Kawut SM, Okun J, Shimbo D, et al. Soluble P-selectin and the risk of primary graft dysfunction after lung transplantation. Chest. 2009 doi: 10.1378/chest.08-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhandari V, Choo-Wing R, Lee CG, et al. Hyperoxia causes angiopoietin 2-mediated acute lung injury and necrotic cell death. Nat. Med. 2006;12:1286–1293. doi: 10.1038/nm1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Beach D, Su L, Zhai R, Christiani DC. A genome-wide expression analysis in blood identifies pre-elafin as a biomarker in ARDS. Am. J. Respir. Cell Mol. Biol. 2008;38:724–732. doi: 10.1165/rcmb.2007-0354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Chen F, Zhai, et al. Plasma neutrophil elastase and elafin imbalance is associated with acute respiratory distress syndrome (ARDS) development. PLoS ONE. 2009;4:1–10. doi: 10.1371/journal.pone.0004380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong MN. Genetic epidemiology of acute respiratory distress syndrome: implications for future prevention and treatment. Clin. Chest Med. 2006;27:705–724. doi: 10.1016/j.ccm.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grigoryev DN, Finigan JH, Hassoun P, Garcia JG. Science review: Searching for gene candidates in acute lung injury. Crit. Care. 2004;8:440–447. doi: 10.1186/cc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho H-Y, Kleeberger SR. Genetic mechanisms of susceptibility to oxidative lung injury in mice. Free Radic. Biol. Med. 2007;42:433–445. doi: 10.1016/j.freeradbiomed.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 20.Fracica PJ, Knapp MJ, Piantadosi CA, et al. Responses of baboons to prolonged hyperoxia: physiology and qualitative pathology. J. Appl. Physiol. 1991;71:2352–2362. doi: 10.1152/jappl.1991.71.6.2352. [DOI] [PubMed] [Google Scholar]
- 21.Sackner MA, Landa J, Hirsch J, Zapata A. Pulmonary effects of oxygen breathing. A 6-hour study in normal men. Ann. Intern. Med. 1975;82:40–43. doi: 10.7326/0003-4819-82-1-40. [DOI] [PubMed] [Google Scholar]
- 22.Erzurum SC, Danel C, Gillissen A, Chu C-S, Trapnell BC, Crystal RG. In vivo antioxidant gene expression in human airway epithelium of normal individuals exposed to 100% O2. J. Appl. Physiol. 1993;75:1256–1262. doi: 10.1152/jappl.1993.75.3.1256. [DOI] [PubMed] [Google Scholar]
- 23.Waring WS, Thomson AJ, Adwani SH, et al. Cardiovascular effects of acute oxygen administration in healthy adults. J. Cardiovasc. Pharmacol. 2003;42:245–250. doi: 10.1097/00005344-200308000-00014. [DOI] [PubMed] [Google Scholar]
- 24.Rossi P, Tauzin L, Weiss M, Rostain J-C, Sainty J-M, Boussuges A. Could hyperoxic ventilation impair oxygen delivery in septic patients? Clin. Physiol. Funct. Imaging. 2007;27:180–184. doi: 10.1111/j.1475-097X.2007.00732.x. [DOI] [PubMed] [Google Scholar]
- 25.Matute-Bello G, Frevert CW, Martin TR. Animal models of acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;295:L379–L399. doi: 10.1152/ajplung.00010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudak BB, Zhang LY, Kleeberger SR. Inter-strain variation in susceptibility to hyperoxic injury of the murine lung. Pharmacogenetics. 1993;3:135–143. doi: 10.1097/00008571-199306000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Cho HY, Jedlicka AE, Reddy SP, Zhang LY, Kensler TW, Kleeberger SR. Linkage analysis of susceptibility to hyperoxia: Nrf2 is a candidate gene. Am. J. Respir. Cell Mol. Biol. 2002;26:42–51. doi: 10.1165/ajrcmb.26.1.4536. [DOI] [PubMed] [Google Scholar]
- 28.Cho HY, Jedlicka AE, Reddy SPM, et al. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 29.Prows DR, Leikauf GD. Quantitative trait analysis of nickel-induced acute lung injury in mice. Am. J. Respir. Cell Mol. Biol. 2001;24:740–746. doi: 10.1165/ajrcmb.24.6.4303. [DOI] [PubMed] [Google Scholar]
- 30.Prows DR, Shertzer HG, Daly MJ, Sidman CL, Leikauf GD. Genetic analysis of ozone-induced acute lung injury in sensitive and resistant strains of mice. Nat. Genet. 1997;17:471–474. doi: 10.1038/ng1297-471. [DOI] [PubMed] [Google Scholar]
- 31.Bein K, Wesselkamper SC, Liu X, et al. Surfactant associated protein B is critical to survival in nickel-induced injury in mice. Am. J. Respir. Cell Mol. Biol. 2009;41(2):226–236. doi: 10.1165/rcmb.2008-0317OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleeberger SR, Levitt RC, Zhang LY. Susceptibility to ozone induced inflammation: II. Separate loci control responses to acute and subacute exposures. Am. J. Physiol. 1993;264:L21–L26. doi: 10.1152/ajplung.1993.264.1.L21. [DOI] [PubMed] [Google Scholar]
- 33.Kleeberger SR, Levitt RC, Zhang LY, et al. Linkage analysis of susceptibility to ozone-induced lung inflammation in inbred mice. Nat. Genet. 1997;17:475–478. doi: 10.1038/ng1297-475. [DOI] [PubMed] [Google Scholar]
- 34.Cho HY, Morgan DL, Bauer AK, Kleeberger SR, et al. Signal transduction pathways of tumor necrosis factor-mediated lung injury induced by ozone in mice. Am. J. Respir. Crit. Care Med. 2007;175:829–839. doi: 10.1164/rccm.200509-1527OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson AD, O’Donnell CJ. An open access database of genome-wide association results. BMC Med. Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 37.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 38.Hofmann S, Franke A, Fischer A, et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat. Genet. 2008;40:1103–1106. doi: 10.1038/ng.198. [DOI] [PubMed] [Google Scholar]
- 39.Weidinger S, Gieger C, Rodriguez E, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:8. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. ▪ Very good overview of the advantages and challenges associated with genome-wide association study investigations.
- 41.Psychiatric GWAS Consortium Coordinating Committee. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am. J. Psychiatry. 2009;166:540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elbers CC, van Eijk KR, Franke L, et al. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet. Epidemiol. 2009;33(5):419–431. doi: 10.1002/gepi.20395. [DOI] [PubMed] [Google Scholar]
- 43. Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. ▪ Good overview of GWAS investigations and their potential application.
- 44. Liao G, Wang J, Guo J, et al. In silico genetics: identification of a functional element regulating H2-Eα gene expression. Science. 2004;306:690–695. doi: 10.1126/science.1100636. ▪ Example of the application of genome-wide mouse haplotypes to identify candidate genes.
- 45.Pletcher MT, McClurg P, Batalov S, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:2159–2169. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Complex Trait Consortium. The collaborative cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. ▪ Very good description of the development and potential application of this new resource for identifying disease susceptibility genes.
- 47.Grigoryev DN, Ma SF, Irizarry RA, Ye SQ, Quackenbush J, Garcia JG. Orthologous gene-expression profiling in multi-species models: search for candidate genes. Genome Biol. 2004;5:R34. doi: 10.1186/gb-2004-5-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perkowski S, Sun J, Singhal S, et al. Gene expression profiling of the early pulmonary response to hyperoxia in mice. Am. J. Respir. Cell Mol. Biol. 2003;28:682–696. doi: 10.1165/rcmb.4692. [DOI] [PubMed] [Google Scholar]
- 49.Cho HY, Reddy SP, Debiase A, Yamamoto M, Kleeberger SR. Gene expression profiling of NRF2-mediated protection against oxidative injury. Free Radic. Biol. Med. 2005;38:325–343. doi: 10.1016/j.freeradbiomed.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 50. Drake TA, Schadt EE, Lusis AJ. Integrating genetic and gene expression data: application to cardiovascular and metabolic traits in mice. Mammal. Genome. 2006;17:466–479. doi: 10.1007/s00335-005-0175-z. ▪ Description and examples of genetical genomics and how this approach may be used to investigate complex traits.
- 51.Li J, Burmeister M. Genetical genomics: combining genetics with gene expression analysis. Hum. Mol. Genet. 2005;14:R163–R169. doi: 10.1093/hmg/ddi267. [DOI] [PubMed] [Google Scholar]
- 52.Bystrykh L, Weersing E, Dontje B, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat. Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- 53.Mehrabian M, Allayee H, Stockton J, et al. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat. Genet. 2005;37:1224–1233. doi: 10.1038/ng1619. [DOI] [PubMed] [Google Scholar]
- 54.Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet. 2008;4(11):E1000260. doi: 10.1371/journal.pgen.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marshall RP, Webb S, Bellingan GJ, et al. Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 2002;166:646–650. doi: 10.1164/rccm.2108086. [DOI] [PubMed] [Google Scholar]
- 56.Casey L, Krieger B, Kohler J, Rice C, Oparil S, Szidon P. Decreased serum angiotensin converting enzyme in adult respiratory distress syndrome associated with sepsis: a preliminary report. Crit. Care Med. 1981;9:651–654. doi: 10.1097/00003246-198109000-00008. [DOI] [PubMed] [Google Scholar]
- 57.Fourrier F, Chopin C, Wallaert B, et al. Compared evolution of plasma fibronectin and angiotensin-converting enzyme levels in septic ARDS. Chest. 1985;87:191–195. doi: 10.1378/chest.87.2.191. [DOI] [PubMed] [Google Scholar]
- 58.Imai Y, Kuba K, Rao S, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arndt PG, Young SK, Poch KR, et al. Systemic inhibition of the angiotensin-converting enzyme limits lipopolysaccharide-induced lung neutrophils recruitment through both bradykinin and angiotensin II-regulated pathways. J. Immunol. 2006;177:7233–7241. doi: 10.4049/jimmunol.177.10.7233. [DOI] [PubMed] [Google Scholar]
- 60.Rigat B, Hubert C, Alhenc-Gelas F, et al. An insertion/deletion polymorphism in the angiotensin-1-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jerng JS, Yu CJ, Wang HC, et al. Polymorphism of the angiotensin-converting enzyme gene affects the outcome of acute respiratory distress syndrome. Crit. Care Med. 2006;34:1001–1006. doi: 10.1097/01.CCM.0000206107.92476.39. [DOI] [PubMed] [Google Scholar]
- 62.Villar J, Flores C, Perez-Mendez L, et al. Angiotensin-converting enzyme insertion/deletion polymorphism is not associated with susceptibility and outcome in sepsis and acute respiratory distress syndrome. Intensive Care Med. 2008;34:488–495. doi: 10.1007/s00134-007-0937-z. [DOI] [PubMed] [Google Scholar]
- 63.Bowler RP, Nicks M, Tran K, et al. Extracellular superoxide dismutase attenuates lipopolysaccharide-induced neutrophilic inflammation. Am. J. Respir. Cell Mol. Biol. 2004;31:432–439. doi: 10.1165/rcmb.2004-0057OC. [DOI] [PubMed] [Google Scholar]
- 64.Arcaroli JJ, Hokanson JE, Abraham E, et al. Extracellular superoxide dismutase haplotypes are associated with acute lung injury and mortality. Am. J. Respir. Crit. Care Med. 2009;179:105–112. doi: 10.1164/rccm.200710-1566OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neidhardt R, Keel M, Steckholzer U, et al. Relationship of interleukin-10 plasma levels to severity of injury and clinical outcome in injured patients. J. Trauma. 1997;42:863–871. doi: 10.1097/00005373-199705000-00017. [DOI] [PubMed] [Google Scholar]
- 66.Crawley E, Kay R, Sillibourne J, et al. Polymorphic haplotypes of the interleukin-10 5′ flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile arthritis. Arthritis Rheum. 1999;42:1101–1108. doi: 10.1002/1529-0131(199906)42:6<1101::AID-ANR6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 67.Schroder O, Laun RA, Held B, Ekkernkamp A, Schulte KM. Association of interleukin-10 promoter polymorphism with the incidence of multiple organ dysfunction following major trauma: results of a prospective pilot study. Shock. 2004;21:306–310. doi: 10.1097/00024382-200404000-00003. [DOI] [PubMed] [Google Scholar]
- 68.Armstrong L, Millar AB. Relative production of tumor necrosis factor α and interleukin-10 in adult respiratory distress syndrome. Thorax. 1997;52:442–446. doi: 10.1136/thx.52.5.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parson PE, Moss M, Vannice JL, et al. Circulating IL-1RA and IL-10 levels are increased but do not predict the development of acute respiratory distress syndrome in at-risk patients. Am. J. Resp. Crit. Care Med. 1997;155:1469–1473. doi: 10.1164/ajrccm.155.4.9105096. [DOI] [PubMed] [Google Scholar]
- 70.Gong MN, Thompson BT, Williams PL, et al. Interleukin-10 polymorphism in position -1082 and acute respiratory distress syndrome. Eur. Respir. J. 2006;27:674–681. doi: 10.1183/09031936.06.00046405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Summerfield JA, Sumiya M, Levin M, Turner MW. Association of mutations in mannose binding protein gene with childhood infection in consecutive hospital series. BMJ. 1997;314:1229–1232. doi: 10.1136/bmj.314.7089.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fidler KJ, Wilson P, Davies JC, et al. Increased incidence and severity of the systemic inflammatory response syndrome in patients deficient in mannose-binding lectin. Intensive Care Med. 2004;30:1438–1445. doi: 10.1007/s00134-004-2303-8. [DOI] [PubMed] [Google Scholar]
- 73.Gong MN, Zhou W, Williams PL, et al. Polymorphisms in the mannose binding lectin-2 gene and acute respiratory distress syndrome. Crit. Care Med. 2007;35:48–56. doi: 10.1097/01.CCM.0000251132.10689.F3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goeckeler ZM, Wysolmerski RB. Myosin light chain kinase-regulated endothelial cell contraction: the relationship between isometric tension, actin polymerization, and myosin phosphorylation. J. Cell Biol. 1995;130:613–627. doi: 10.1083/jcb.130.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wainwright MS, Rossi J, Schavocky J, et al. Protein kinase involved in lung injury susceptibility: evidence from enzyme isoform genetic knockout and in vivo inhibitor treatment. Proc. Natl Acad. Sci. USA. 2003;100:6233–6238. doi: 10.1073/pnas.1031595100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eutamene H, Theodorou V, Schmidlin F, et al. LPS-induced lung inflammation is linked to increased epithelial permeability: role of MLCK. Eur. Resp. J. 2005;25:789–796. doi: 10.1183/09031936.05.00064704. [DOI] [PubMed] [Google Scholar]
- 77.Gao L, Grant A, Halder I, et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am. J. Respir. Cell Mol. Biol. 2006;34:487–495. doi: 10.1165/rcmb.2005-0404OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christie JD, Ma SF, Aplenc R, et al. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit. Care Med. 2008;36:2794–2800. doi: 10.1097/ccm.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- 79.Itoh K, Tong KI, Yamamoto M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 2004;36:1208–1213. doi: 10.1016/j.freeradbiomed.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 80. Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. ▪ Very good review of the role of the Nrf2 signaling system in cellular responses to environmental stimuli.
- 81.Marzec JM, Christie JD, Reddy SP, et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 2007;21:2237–2246. doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- 82.Ross D, Beall H, Traver RD, Siegel D, Phillips RM, Gibson NW. Bioactivation of quinines by DT-diaphorase: molecular, biochemical, and chemical studies. Oncol. Res. 1994;6:493–500. [PubMed] [Google Scholar]
- 83.Lim JH, Kim K-M, Kim SW, Hwang O, Choi HJ. Bromocriptine activates NQO1 via Nrf2-PI3K/Akt signaling: novel cytoprotective mechanism against oxidative damage. Pharmacol. Res. 2008;57:325–331. doi: 10.1016/j.phrs.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 84.Jaiswal AK. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic. Biol. Med. 2000;29:254–262. doi: 10.1016/s0891-5849(00)00306-3. [DOI] [PubMed] [Google Scholar]
- 85.Korashy HM, Brocks DR, El-Kadi AOS. Induction of the NAD(P)H:quinone oxidoreductase 1 by ketoconazole and itraconazole; a mechanism of cancer chemoprotection. Cancer Lett. 2007;258:135–143. doi: 10.1016/j.canlet.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 86.Reddy AJ, Christie JD, Aplenc R, Fuchs B, Lanken PN, Kleeberger SR. Association of human NAD(P)H:quinone oxidoreductase 1 (NQO1) polymorphism with development of acute lung injury. J. Cell. Mol. Med. 2009 doi: 10.1111/j.1582-4934.2008.00581.x. (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nemeth E, Millar LK, Bryant-Greenwood G. Fetal membrane distension:II. Differentially expressed genes regulated by acute distension in vitro. Am. J. Obstet. Gynecol. 2007;182:60–67. doi: 10.1016/s0002-9378(00)70491-1. [DOI] [PubMed] [Google Scholar]
- 88.Ognjanovic S, Bao S, Yamamoto SY, et al. Genomic organization of the gene coding for human pre-B-cell colony enhancing factor and expression in human fetal membranes. J. Mol. Endocrinol. 2001;26:107–117. doi: 10.1677/jme.0.0260107. [DOI] [PubMed] [Google Scholar]
- 89.Jia SH, Li Y, Parodo J, et al. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J. Clin. Invest. 2004;113:1318–1327. doi: 10.1172/JCI19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ye SQ, Simon BA, Maloney JP, et al. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am. J. Respir. Crit. Care Med. 2005;171:361–370. doi: 10.1164/rccm.200404-563OC. [DOI] [PubMed] [Google Scholar]
- 91.Bajwa EK, Yu CL, Gong MN, et al. Pre-B-cell colony-enhancing factor gene polymorphisms and risk of acute respiratory distress syndrome. Crit. Care Med. 2007;35:1290–1295. doi: 10.1097/01.CCM.0000260243.22758.4F. [DOI] [PubMed] [Google Scholar]
- 92.Eisner MD, Parsons PE, Matthay MA, et al. Plasma surfactant protein levels and clinical outcomes in patients with acute lung injury. Thorax. 2003;58:983–988. doi: 10.1136/thorax.58.11.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Doyle IR, Bersten AD, Nicholas TE. Surfactant proteins-A and -B are elevated in plasma of patients with acute respiratory failure. Am. J. Respir. Crit. Care Med. 1997;156:1217–1229. doi: 10.1164/ajrccm.156.4.9603061. [DOI] [PubMed] [Google Scholar]
- 94.Floros J, Veletza SV, Kotikalapudi P, et al. Dinucleotide repeats in the human surfactant protein-B gene and respiratory-distress syndrome. Biochem. J. 1995;305:583–590. doi: 10.1042/bj3050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gong MN, Wei Z, Xu L-L, et al. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest. 2004;125:203–211. doi: 10.1378/chest.125.1.203. [DOI] [PubMed] [Google Scholar]
- 96.Quasney MW, Waterer GW, Dahmer MK, et al. Association between surfactant protein-B +1580 polymorphism and the risk of respiratory failure in adults with community-acquired pneumonia. Crit. Care Med. 2004;32:1115–1119. doi: 10.1097/01.ccm.0000124872.55243.5a. [DOI] [PubMed] [Google Scholar]
- 97.Currier PF, Gong MN, Zhai R, et al. Surfactant protein-B polymorphisms and mortality in the acute respiratory distress syndrome. Crit. Care Med. 2008;36:2511–2516. doi: 10.1097/CCM.0b013e318183f608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seybold J, Thomas D, Witzenrath M, et al. Tumor necrosis factor-α-dependent expression of phosphodiesterase 2: role in endothelial hyperpermeability. Blood. 2005;105:3569–3576. doi: 10.1182/blood-2004-07-2729. [DOI] [PubMed] [Google Scholar]
- 99.Gong MN, Zhou W, Williams PL, et al. -308GA and TNFB polymorphisms in acute respiratory distress syndrome. Eur. Resp. J. 2005;26 doi: 10.1183/09031936.05.00000505. 382-289. [DOI] [PubMed] [Google Scholar]
- 100.Corne J, Chupp G, Lee CG, et al. IL-13 stimulates vascular endothelial growth factor and protects against hyperoxic lung injury. J. Clin. Invest. 2000;106:783–791. doi: 10.1172/JCI9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Medford AR, Keen LJ, Bidwell JL, Millar AB. Vascular endothelial growth factor gene polymorphism and acute respiratory distress syndrome. Thorax. 2005;60:244–248. doi: 10.1136/thx.2004.034785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morgan TM, Krumholz HM, Lifton RP, Spertus JA. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007;297:1551–1561. doi: 10.1001/jama.297.14.1551. [DOI] [PubMed] [Google Scholar]
- 103.Daly AK. Candidate gene case-control studies. Pharmacogenomics. 2003;4:127–139. doi: 10.1517/phgs.4.2.127.22629. [DOI] [PubMed] [Google Scholar]
- 104.Keating BJ, Tischfield S, Murray SS, et al. Concept, design and implementation of a cardiovascular gene-centric 50 K SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:1–9. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rao DC. An overview of the genetic dissection of complex traits. Adv. Genet. 2008;60:3–34. doi: 10.1016/S0065-2660(07)00401-4. [DOI] [PubMed] [Google Scholar]
- 106.Sheu CC, Zhai R, Su L, et al. Sex-specific association of epidermal growth factor gene polymorphisms with acute respiratory distress syndrome. Eur. Respir. J. 2009;33:543–550. doi: 10.1183/09031936.00091308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moradi M, Mojtahedzadeh M, Mandegari A, et al. The role of glutathione-S-transferase polymorphisms on clinical outcome of ALI/ARDS patients treated with N-acetylcysteine. Respir. Med. 2009;103:434–441. doi: 10.1016/j.rmed.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 108.Zhai R, Zhou W, Gong MN, et al. Inhibitor κβ-α haplotype GTC is associated with susceptibility to acute respiratory distress syndrome in Caucasians. Crit. Care Med. 2007;35:893–898. doi: 10.1097/01.CCM.0000256845.92640.38. [DOI] [PubMed] [Google Scholar]
- 109.Flores C, Ma SF, Maresso K, Wade MS, Villar J, Garcia JG. IL6 gene-wide haplotype is associated with susceptibility to acute lung injury. Transl. Res. 2008;152:11–17. doi: 10.1016/j.trsl.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 110.Hildebrand F, Stuhrmann M, van Giensven M, et al. Association of IL-8-251A/T polymorphism with incidence of acute respiratory distress syndrome (ARDS) and IL-8 synthesis after multiple trauma. Cytokine. 2007;37:192–199. doi: 10.1016/j.cyto.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 111.Gao L, Flores C, Fan-Ma S, et al. Macrophage migration inhibitory factor in acute lung injury: expression, biomarker, and associations. Transl. Res. 2007;150:18–29. doi: 10.1016/j.trsl.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Adamzik M, Frey UH, Rieman K, et al. Insertion/deletion in the promoter of NFKB1 influences severity but not mortality of acute respiratory distress syndrome. Intensive Care Med. 2007;33:1199–1203. doi: 10.1007/s00134-007-0649-4. [DOI] [PubMed] [Google Scholar]
- 113.Sapru A, Hansen H, Ajayi T, et al. 4G/5G polymorphism of plasminogen activator inhibitor-1 gene is associated with mortality in intensive care unit patients with severe pneumonia. Anesthesiology. 2008;110:1086–1091. doi: 10.1097/ALN.0b013e3181a1081d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wurfel MM, Gordon AC, Holden TD, et al. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am. J. Respir. Crit. Care Med. 2008;178:710–720. doi: 10.1164/rccm.200803-462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arcaroli J, Sankoff J, Liu N, Allison DB, Maloney J, Abraham E. Association between urokinase haplotypes and outcome from infection-associated acute lung injury. Intensive Care Med. 2008;34:300–307. doi: 10.1007/s00134-007-0930-6. [DOI] [PubMed] [Google Scholar]