Abstract
Tissue Engineering is a rapidly evolving field in terms of cell source and scaffold fabrication. As the template for three dimensional tissue growth, the scaffold should emulate the native extracellular matrix, which is nano-fibrous. Currently, there are three basic techniques capable of generating nano-fibrous scaffolding: electrospinning, molecular self-assembly, and thermally induced phase separation. These scaffolds can then be further modified by various three dimensional surface modification techniques if necessary to more precisely emulate the native extracellular matrix. However, even without further modification, nano-fibrous scaffolds have been shown to have advantageous effects on cellular behavior and tissue formation when compared to more traditional types of scaffolding. This review focuses on the current state of tissue engineering with nano-fibrous scaffolding with particular emphasis on bone tissue engineering.
Introduction
Organ failure (heart, kidney, liver, lungs, pancreas, etc) or tissue loss (bone, ligaments, corneas, arteries, veins, skin, etc) accounts for around half of the medical spending in the U.S. leading to roughly 8 million surgical procedures and 40–90 million hospital days per year required for treatment of these ailments1. In 2005, there were 27,527 organ transplantation procedures in the United States, while approximately 90,000 patients remained on waiting lists waiting for organs2. Many of those left on the waiting list will die before an organ becomes available. In addition to organ transplants, there were approximately 1.5 million transplantations of human tissue in 2004. This number has doubled over the past 10 years3, 4. As the need for organs and tissue continues to increase and surpass the supply, the interdisciplinary field of tissue engineering has emerged to help meet these needs. Tissue engineering aims to develop biological substitutes which restore, maintain or improve tissue function through the application of engineering principles and the life sciences5, 6.
There are three basic approaches to tissue engineering5, 6: use of isolated cells or cell substitutes to replace the cells that supply a needed function; delivery of tissue-inducing substances such as growth factors to a targeted location; and growing cells in a three-dimensional scaffold. For small, well-contained defects the first two approaches may be suitable. However, to produce larger blocks of tissue with predesigned shapes only the third approach, using a scaffold to direct cell growth, is sufficient. As such, both cells and materials play an important role in de novo tissue development.
Traditionally, there are several important factors to consider when designing a scaffold for tissue engineering applications, including scaffold morphologies (porosity, pore size and interpore connectivity), mechanical properties and degradation7. Recent development has focused on designing biomimetic scaffolds to elicit favourable biological effects. It has been indicated that the architecture of natural extracellular matrix (ECM) plays an important role in regulating cellular behaviour8–10. For example, type I collagen has a nano-fibrous structure11 and is the base attachment structure for cells in many tissues12. As ECM matures, other proteins and bio-molecules are either adsorbed from the serum or secreted from the cells joining type I collagen to form the tissues native ECM. In bone, type I collagen composes 95% of the organic ECM which is strengthened by the cellular deposition of hydroxyapatite13to form the mature ECM of bone. It is this maturation process which nano-fibrous and surface modified tissue engineering scaffolds attempt to mimic in order to create replacement tissues.
As you progress through this review, cell sourcing for tissue engineering then methods of nano-fibrous scaffold fabrication and methods of surface modification will be addressed followed by the biological effects of nano-fibrous architecture on bone formation.
Cell sources for tissue engineering
Since its inception, a variety of cells have been used in tissue engineering. Currently due to their ability to produce multiple cell types and self renew14, stem cells have gained popularity as a cell source for tissue engineering. Stem cells can be isolated from multiple sources including adult tissue15–19, umbilical cord blood20, amniotic fluid21 and embryos22. Of the adult tissue stem cells, haematopoietic stem cells have been used clinically for years to restore the haematopoietic system23 and mesenchymal stem cells are currently being investigated in clinical trials for the treatment of multiple conditions24–28 due to their multiple lineage potentials15, 29 and ability to illicit a reduced immune response30–33. However, mesenchymal stem cells are not immortal and their ability to proliferate and differentiate are affected by donor age and culture time34, 35.
In contrast, embryonic stem cells, isolated from the inner cell mass of blastocysts22, are immortal providing a potentially unlimited source of cells for tissue engineering and are capable of differentiating into all the cell types in the human body. Recent evidence indicates that embryonic stem cells and embryonic stem cell derived cells may be less immunogenic than adult cells36 and with the use of somatic nuclear transfer embryonic stem cells may become autologous. However, embryonic stem cells cultured with animal products have been found to carry immunogenic non-human surface markers37. This contamination from animal products, the tumorgenicity of the undifferentiated cells, and the heterogeneous cell population generated by current differentiation protocols all must be resolved before embryonic stem cells are used clinically as a cell source for tissue engineering. However, it is important to note that the study of these cells is still in its infancy and these obstacles will most likely be overcome in time with further studies.
Nano-fibrous scaffold fabrication
While a large number of scaffolding fabrication methods have been developed, the techniques of controlling the architecture of scaffold at nano-scale level, which is required to emulate the size scale of collagen, is still limited. Only three techniques have been developed in the fabrication of nano-fibrous scaffolds for use in tissue engineering: electrospinning, self-assembly, and phase separation.
Electrospinning
The electrospinning process has long been utilized to fabricate industrial products before it was recently applied to produce nano-fibrous structures for use in tissue engineering 38, 39. The principle of electrospinning is to use an electric field to draw a polymer solution from an orifice to a collector, producing polymer fibers with diameters in the range of nanometers to micrometers38, 40. Due to the simplicity and the ability to produce nano-fibers from various materials of this method, electrospinning has attracted considerable attention for use in tissue engineering. A variety of synthetic and natural biomaterials, including poly(l-lactic acid) (PLLA), poly(lactic-co-glycolic acid) (PLGA), poly(caprolactone) (PCL), polyethylene terephthalate (PET), poly(ethylene oxide) (PEO), poly(vinyl alcohol) (PVA), collagen, gelatin, chitosan, silk protein and fibrinogen have been used to form nano-fibrous scaffolds for tissue engineering39, 41–52. The fiber diameters can simply be controlled by altering the concentration of the polymer solution, that is, solutions made of higher concentrations produce larger diameter fibers. In addition, fiber alignment can be controlled by rotating the grounded target. While the simplicity of electrospinning makes it a very active research field, significant challenges include the difficulties to create three-dimensional (3D) scaffolds with well-defined pore architecture and complex geometries.
Molecular self-assembly
Molecular self-assembly is a useful approach for fabricating supramolecular architectures53 and can be defined as a spontaneous process to form structurally ordered and stable arrangement through a number of non-covalent interactions, such as hydrogen bonds, van der Waals interactions, electrostatic interactions, and hydrophobic interactions54, 55. Found throughout biology, self-assembly of biomolecules, such as peptides and proteins, to well-defined architectures perform a variety of functions55. A good example is the self-assembly of collagen molecules to collagen fibrils, and the collagen fibrils are further packed side-by-side in parallel bundles to form collagen fibers with diameter ranged from 50 to 500 nm56. Inspired by nature, several groups have designed and synthesized polypeptides or oligopeptides molecules to self-assemble into nano-fibrous structures under suitable conditions53, 57–59. The formation of nano-fibers by molecular self-assembly is a “bottom-up” strategy and usually the fiber diameter is much smaller than those produced by using electrospinning57. While molecular self-assembly is a fairly new technique for the formation of nano-scale scaffold, it has not been demonstrated how to control the pore size and pore structures, which are important to allow for cell incorporation, migration and proliferation. The mechanical strength of self-assembled scaffolds also has to be addressed before they can be truly used in tissue engineering applications.
Thermally induced phase separation
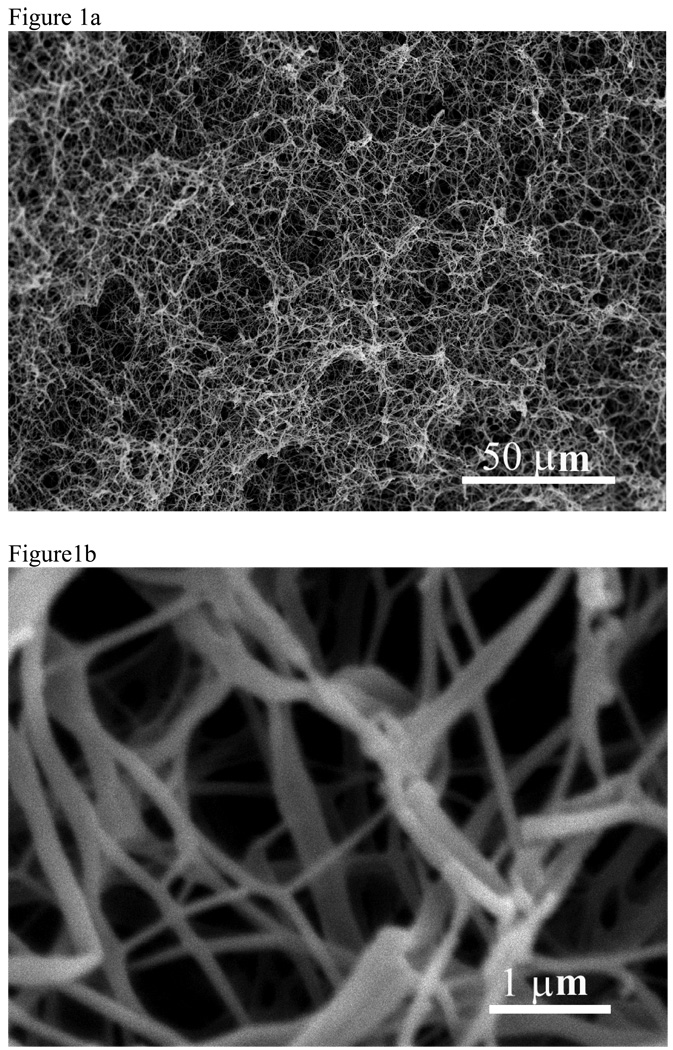
Thermally induced phase separation (TIPS) has been used for several years to fabricate synthetic porous scaffolds for tissue engineering60–62. In this process, the temperature of a polymer solution is controlled to induce a phase separation into two phases, a polymer-rich phase and a polymer-lean phase. After removal of the solvent by extraction, evaporation, or sublimation, the polymer-rich phase solidifies and forms polymer foam. By varying the types of polymer and solvent, polymer concentration, and phase-separation temperature, different pore morphology and structures can be achieved. To mimic the fibrous structure of natural type I collagen, a novel TIPS technique has recently been developed to fabricate nano-fibrous matrices by using synthetic biodegradable polymers63. For example, a solution of PLLA dissolved in THF is thermally induced following a series of processes to phase separate. The solvent is exchanged with water and then freeze-dried to yield nano-fibrous PLLA matrices. The fibers formed in this manner have diameters ranging from 50–500 nm, and have a porosity as high as 98% (Figure 1).
Figure 1.
SEM micrographs of a PLLA nano-fibrous matrix prepared from 2.5% (wt/v) PLLA/THF solution at a phase separation temperature of 8°C. A) 500x; B) 20,000x. From Ma and Zhang63, Copyright ©John Wiley & Sons. Reprinted by permission of John Wiley & Sons.
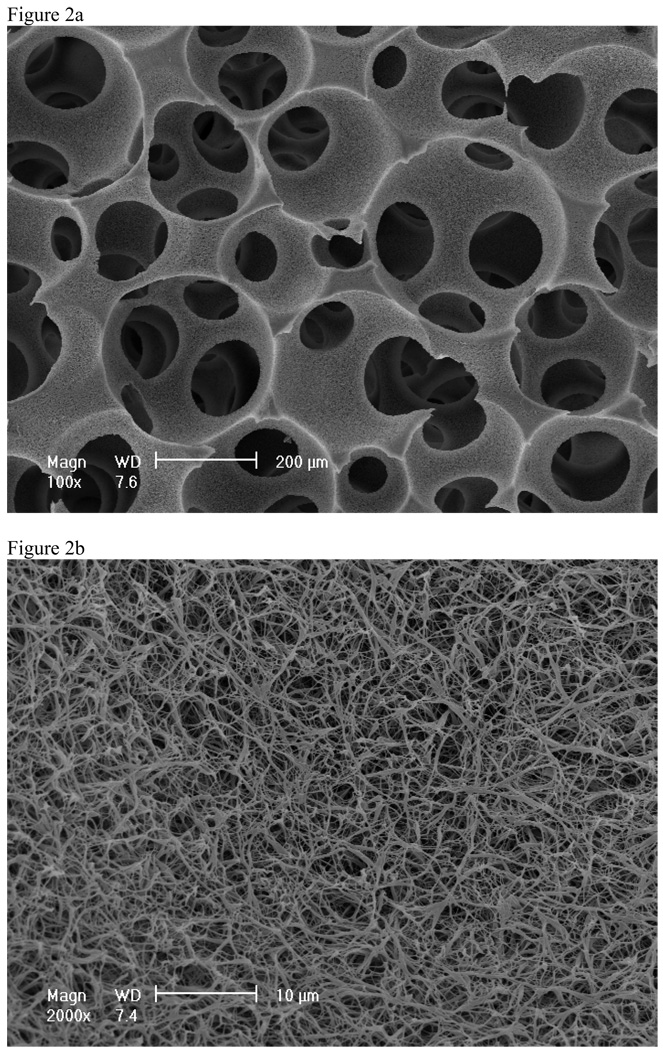
By combining this TIPS with other processing techniques (such as particulate leaching or solid free-form fabrication), scaffolds with complex 3D structures and well-defined pore morphologies can be produced64, 65. For example, a solution of PLLA in THF was dripped onto sugar fiber assembly in a mold and then cooled to a preset gelation temperature. After phase separation, the gel-sugar composite was immersed in distilled water to extract the solvent and leach the sugar from the composite. The sample was freeze-dried, resulting in a 3D nano-fibrous matrix with macropores left behind from the leached sugar fibers 64. To further control the interconnectivity between pores in the scaffold, a novel technique to generate interconnected spherical pore network has been combined with TIPS to fabricate nano-fibrous scaffolds with interconnected spherical macropores (Figure 2)66–68. The combined technique advantageously controls macropore shape and size by sugar spheres, interpore opening size by assembly conditions (time and temperature of heat treatment), and pore wall morphology by phase-separation parameters.
Figure 2.
SEM micrographs of PLLA nano-fibrous scaffolds prepared from 10% (wt/v) PLLA/THF solutions at a phase separation temperature of −20°C. A) 100x; B) 2000x. From Liu et al.66, Copyright © American Scientific Publishers. Reprinted by permission of American Scientific Publishers.
Surface modification of nano-fibrous scaffolds
Although a variety of synthetic biodegradable polymers have been used as tissue engineering scaffolding materials, one disadvantage of these materials is their lack of biological recognition. The surface of the scaffold fabricated with these synthetic biomaterials can be modified to obtain more desirable characteristics to positively enhance cell-scaffold interactions 69.
Several approaches have been developed to modify the scaffold surface 70–73. For example, low pressure ammonia plasma treatment has been used for the modification of poly(3-hydroxybutyrate) (PHB) thin film 70. The introduction of amine functions was used in order to permit subsequent protein immobilization. The plasma treatment of PHB induced a durable conversion of hydrophobic material into hydrophilic but did not cause significant changes in the morphology of the analyzed thin films. Such surface modification work, however, is limited to 2D film surfaces or very thin 3D constructs. Our lab has recently developed several techniques to effectively modify the surface of 3D scaffolds 66, 74, 75. For example, an electrostatic layer-by-layer self-assembly technique has been used to modify nano-fibrous PLLA (NF-PLLA) scaffolds with gelatin 66. The NF-PLLA scaffold was first fabricated and activated in an aqueous poly(diallyldimethylammonium chloride) (PDAC) solution to obtain positive charges on the scaffold surface. The scaffold was then subsequently immersed in a solution of negatively charged gelatin, so that gelatin molecules were self-assembled onto the activated scaffold surface. By alternately immersing the scaffold into the solutions of positively charged PDAC and negatively charged gelatin, polyelectrolyte multilayers containing gelatin molecules were deposited on the NF-PLLA surfaces. This technique provides a means to create polycation-polyanion polyelectrolyte complexes one molecular layer at a time, thereby allowing for an unprecedented level of control over the composition and surface functionality of materials. The layer-by-layer self-assembly method can be used for any complex 3D geometry as long as the pores are interconnected.
Effect of nano-fibrous scaffold on cellular behaviour and tissue development
Although limited data is available, the effects on cellular behaviour and tissue formation due to nano-fibrous scaffolds have been observed with numerous cell types76. For example, nano-fibrous scaffolding has recently been shown to facilitate recovery from spinal cord injury in mice77. However, due to space limitations we will principally focus our discussion on the cellular effects of nano-fibrous scaffolding in bone tissue engineering.
Attachment and proliferation
Several cells types, including osteoblasts78, 79, fibroblasts80, 81, rat kidney cells80, smooth muscle cells82, neural stem cells83 and embryonic stem cells84, have shown increased attachment on various nano-fibers compared to their corresponding control materials. Additionally, a recent study found that branched nano-fibers improve fibroblast attachment over linear nano-fibers85.
Integrins are a large family of heterodimeric transmembrane proteins which mediate cell attachment through ECM binding. Several studies have noted differences in integrin expression on nano-fibrous materials compared to control78, 80, 86, 87. Increased expression of α2, αV, β1, and β3 integrins has been seen in osteoblasts on nano-fibrous scaffolds compared to comparable solid-walled scaffolds78. A previous study with these same scaffolds found that several integrin-binding protein components of the ECM (fibronectin, victronectin and laminin) were adsorbed selectively at a higher level on nano-fibrous scaffolds compared to the solid-walled scaffolds79, which may be contributing to the increased integrin expression on nano-fibrous scaffolds compared to solid-walled scaffolds. Notably, the upregulation of α2 and β1 integrins, which are associated with type I collagen binding, were maintained on nano-fibrous scaffolds compared to solid-walled scaffolds after cellular formation of collagen fibrils was blocked, indicating that the cells on the nano-fibrous scaffold may be interacting directly with the synthetic scaffold78, 88.
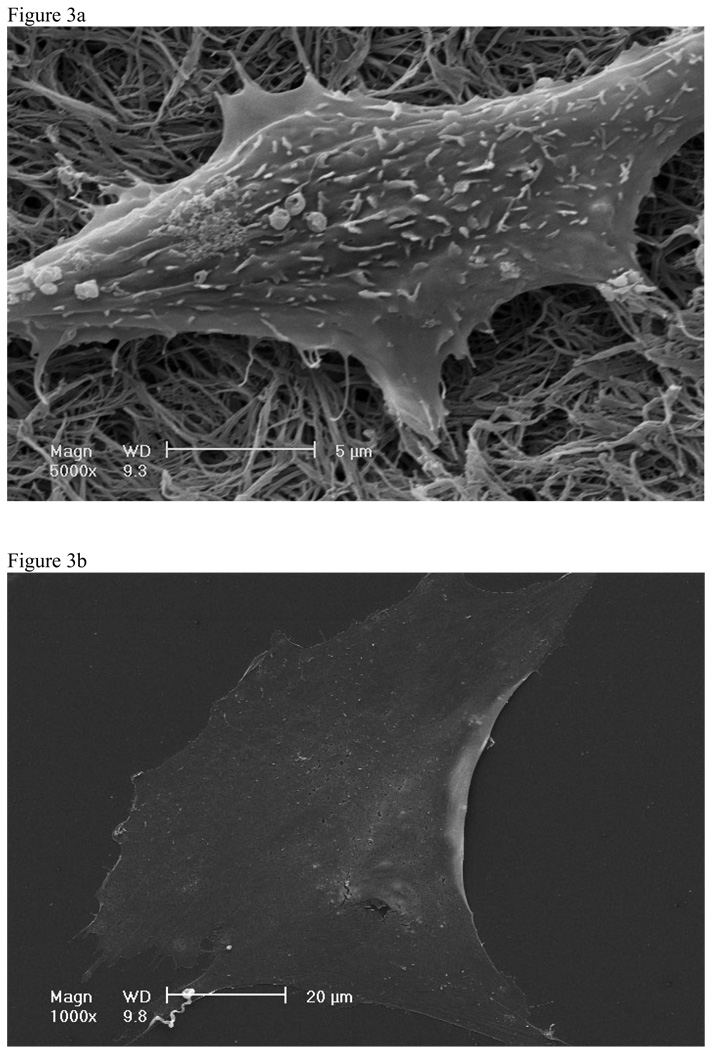
Cells cultured on nano-fibrous material also have a cellular morphology more similar to in vivo compared to cells cultured on control materials 78, 80, 89. Mouse pre-osteoblasts cultured on nano-fibrous matrices exhibited processes interacting with nano-fibers while cells on the flat films were flat and spread over large areas (Figure 3)88. Mouse pre-osteoblasts cultured on nano-fibrous matrices also exhibited fewer stress fibers than cells on the flat films88. In several cell types these morphological characteristics have been linked to increased Rac expression90, a regulator of actin cytoskeleton assembly known to affect attachment and other cellular functions.
Figure 3.
SEM of MC-4 cells after 24 hours of culture on A) nano-fibrous matrices and B) flat films. From Hu et al.88, Copyright © 2008 by Elsevier.
After cells attach and spread on the scaffolds, they must then proliferate in order to fully populate the scaffold and form tissue. Nano-fibrous materials have enhanced the proliferation of several cell types compared to various control materials that do not have nano-fibrous features 65, 80–82, 86, 87, 89, 91. Specifically, after 7 days of growth nearly 3 times more osteoblasts were present on the nano-fibrous scaffold compared to the solid walled scaffold65.
Differentiation and tissue formation
Various cells types, including osteoblasts65, 78, chondrocytes86, 92, neural progenitors83, 93, and hepatocytes94, 95 have shown enhanced differentiation on a few types of nano-fibrous materials compared to their corresponding control materials. Considering the space limitation, we will focus on osteoblast differentiation as an example.
In addition to their role in cellular attachment, integrins also activate signalling pathways which stimulate cellular differentiation. After 24hrs of culture increased paxillin and focal adhesion kinase phosphorylation, components in integrin activated differentiation pathways96, were observed in osteoblasts on nano-fibrous scaffolds compared to solid-walled scaffolds78. This indicates that the increased integrin expression during the cellular attachment translates into increased cellular differentiation and underscores how the selective adsorption of serum proteins onto nano-fibrous materials creates a different micro-environment from the solid-walled materials, which then alters the pathways which contribute to differentiation.
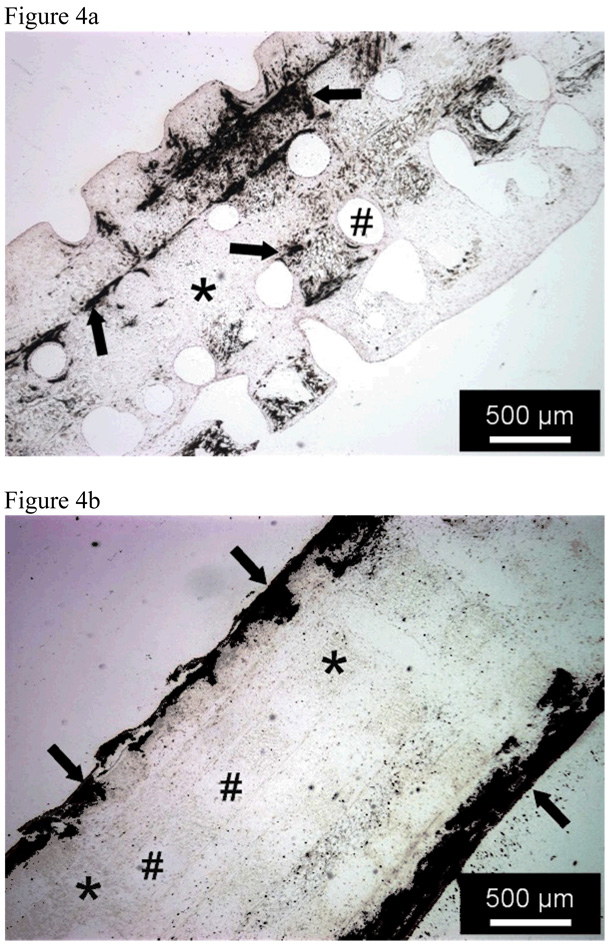
As the extracellular matrix matures, the osteoblasts and their progenitor cells express osteogenic markers. Increased expression of alkaline phosphatase, an early marker of osteogenic differentiation, has been seen on nano-fibrous matrices compared to solid-walled scaffolds after 3 or more days of culture78, 97, while increased expression of bone sialoprotein and osteocalcin, later markers of osteogenic differentiation, have been seen on nano-fibrous matrices compared to solid-walled scaffolds after 1 week of culture65, 78, 88. The expression of these later bone markers is typically accompanied by mineralization of the extracellular matrix. Upon quantification, the nano-fibrous scaffolds have been found to contain up to 13 times more calcium content compared to solid-walled scaffolds65, 78. The mineral on the nano-fibrous matrix spread more evenly throughout the scaffolds, while the mineral on the solid-walled scaffold is principally in its exterior (Figure 4). Overall, this data suggests that nano-fibrous scaffolds better promote cellular differentiation and tissue formation over more traditional scaffolds.
Figure 4.
Von Kossa’s silver nitrate staining of histological sections after 6 weeks of MC3T3-E1 osteoblasts cultured under A) nano-fibrous scaffolds and B) solid-walled scaffold Scale bars is 500 µm. * denotes the PLLA scaffold, # a scaffold pore. Arrows denote mineralization. From Chen et al.65, Copyright © 2006 by Elsevier.
Conclusions
Tissue engineering is a rapidly evolving field in which scaffolding plays a pivotal role. As our understanding of tissue development expands, the complexity of the scaffolding has increased to mimic the native ECM. Currently, there are three techniques capable of producing nano-fibrous scaffolds. Of these, phase separation has shown high potential to meet the needs of three dimensional tissue regeneration due to its ability to incorporate any pore shape and size or any overall 3-D geometry. Through mimicking the natural extracellular matrix, cellular interactions have been enhanced over previous scaffolding strategies. Increased cellular attachment, proliferation, and differentiation have all been observed on nano-fibrous scaffolding compared to more traditional scaffolds. However, completely duplicating the ECM may not be the best strategy since mature ECM often does not contain highly interconnected pores to allow for the quick even cell dispersion that tissue engineering strives. Additionally, tissue engineering seeks to accelerate the natural development and wound healing processes which may render mimicking some aspects of the ECM unnecessary. As the field continues to mature, the scaffolds will most likely become more complex and bring us closer to the goal of functional tissue regeneration.
Notes and references
- 1.Karp JM, Langer R. Current Opinion in Biotechnology. 2007;18:454–459. doi: 10.1016/j.copbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 2.2006 Annual Report of the U.S. Organ Procurement and Transplantation Network and the Scientific Registry of Transplant Recipients: Transplant Data 1996–2005. Rockville, MD: Health Resources and Services Administration, Healthcare Systems Bureau, Division of Transplantation; 2006. [Google Scholar]
- 3.Statistical Abstract of the United States: 2004–2005 Section 3 Health and Nutrition, No 161 Organ Transplants and Grafts: 1990 to 2001. Washington DC: U.S. Census Bureau; 2004. [Google Scholar]
- 4.Wang S, Zinderman C, Wise R, Braun M. Cell and tissue Banking. 2007;8:211–219. doi: 10.1007/s10561-007-9034-3. [DOI] [PubMed] [Google Scholar]
- 5.Langer R, Vacanti J. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 6.Ma PX. Materials Today. 2004;7:30–40. [Google Scholar]
- 7.Ma PX. Adv Drug Deliv Rev. 2008;60:184–189. doi: 10.1016/j.addr.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbott A. Nature. 2003;424:870–872. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 9.Cukierman E, Pankov R, Stevens D, Yamada K. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 10.Schmeichel KL, Bissell MJ. J Cell Sci. 2003;116:2377–2388. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsdale T, Bard J. J cell biology. 1972;54:626–637. doi: 10.1083/jcb.54.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadler K. Birth Defects Res C Embryo Today. 2004;72:1–11. doi: 10.1002/bdrc.20002. [DOI] [PubMed] [Google Scholar]
- 13.Marks S, Odgren P. In: Principles of Bone Biology. 2nd edn. Bilezikian J, Raisz L, Rodan G, editors. vol. 1. San Diego, CA: Academic Press; 2002. pp. 3–16. [Google Scholar]
- 14.Smith A. Nature. 2006;441:1060. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 15.Pittenger MF, Mackay AM, Deck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 16.Alison M. Curr Opin Cell Biol. 1998;10:710–715. doi: 10.1016/s0955-0674(98)80111-7. [DOI] [PubMed] [Google Scholar]
- 17.Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, LI S, Hatch H, Ahrens K, Cornelius JG, Petersen BE, Peck AB. Proc Natl Acad Sci U S A. 2002;99:8078–8083. doi: 10.1073/pnas.122210699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke DL, Johansson CB, Wilbertz J, Veress B, Nilsson E, Karlstrom H, Lendahl U, Frisen J. Science. 2000;288:1660–1663. doi: 10.1126/science.288.5471.1660. [DOI] [PubMed] [Google Scholar]
- 20.Lee MW, Choi J, Yang MS, Moon YJ, Park JS, Kim HC, Kim YJ. Biochem Biophys Res Commun. 2004;320:273–278. doi: 10.1016/j.bbrc.2004.04.206. [DOI] [PubMed] [Google Scholar]
- 21.De Coppi P, Bartsh G, Jr, Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ, Furth ME, Soker S, Atala A. Nat Biotechnol. 2007;25:100–106. doi: 10.1038/nbt1274. [DOI] [PubMed] [Google Scholar]
- 22.Thomson JA, Itskovitiz-Eldor J, Shapiro SS, Waknitz MS, Swiergiel JJ, Marshall VS, Jones JM. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 23.Mathe G, Amiel JL, Schwarzenberg L, C A, Schneider M. Br Med J. 1963;2:1633–1635. doi: 10.1136/bmj.2.5373.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen SL, Fang WW, Ye F, Lui YH, Qian J, Shan SJ, Zhang JJ, Chunhua RZ, Liao LM, Lin S, Sun JP. Am J Cardiol. 2004;94:92–95. doi: 10.1016/j.amjcard.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 25.Bang OY, Lee JS, Lee PH, Lee G. Ann Neurol. 2005;57:874–882. doi: 10.1002/ana.20501. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz EM, Gordon PL, Koo WK, Marx JC, Neel MD, McNall RY, Muul L, Hofmann T. Proc Natl Acad Sci U S A. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koc ON, Day J, Nieder M, Gerson SL, Lazarus HM, Krivit W. Bone Marrow Transplant. 2002;30:215–222. doi: 10.1038/sj.bmt.1703650. [DOI] [PubMed] [Google Scholar]
- 28.Karitisis DG, Sotiropoulou PA, Karvouni E, Karabinos I, Korovesis S, Perez SA, Voridis EM, Papamichail M. Catheter Cardiovas Interv. 2005;65:321–329. doi: 10.1002/ccd.20406. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, Du J, Alsrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 30.Aggarwal S, Pittenger MF. Blood. 2005;105:1815–1822. doi: 10.1182/blood-2004-04-1559. [DOI] [PubMed] [Google Scholar]
- 31.Di Nicola M, Carlo-Stella C, Magni M, Milanesi M, Longoni P, Matteucci P, S, Gianni A. Blood. 2002;99:3838–3843. doi: 10.1182/blood.v99.10.3838. [DOI] [PubMed] [Google Scholar]
- 32.Corcione A, Benvenuto F, Ferretti E, Giunti D, Cappiello V, Cazzanti F, Risso M, Gualandi F, Mancardi G, Pistoia V, Uccelli A. Blood. 2006;107:367–372. doi: 10.1182/blood-2005-07-2657. [DOI] [PubMed] [Google Scholar]
- 33.Beyth S, Borovsky Z, Mevorach D, Liebergall M, Gazit Z, Aslan H, Galun E, Rachmilewitz J. 2005;105 doi: 10.1182/blood-2004-07-2921. [DOI] [PubMed] [Google Scholar]
- 34.Fehrer C, Lepperdinger G. Exp Gerontol. 2005;40:926–930. doi: 10.1016/j.exger.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Muraglia A, Cancedda R, Quarto R. J Cell Sci. 2000;113:1161–1166. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- 36.Drukker M, Katchman H, Katz G, Friedman SE, Shezen E, Hornstein E, Mandelboim O, Reisner Y, Benvenisty N. Stem Cells. 2006;24:221–229. doi: 10.1634/stemcells.2005-0188. [DOI] [PubMed] [Google Scholar]
- 37.Martin MJ, Muotri A, Gage R, Varki A. Nat Med. 2005;11:228–232. doi: 10.1038/nm1181. [DOI] [PubMed] [Google Scholar]
- 38.US Pat., 1,975,504. 1934
- 39.Li WJ, Laurencin CT, Caterson EJ, Tuan RS, Ko FK. Journal of Biomedical Materials Research. 2002;60:613–621. doi: 10.1002/jbm.10167. [DOI] [PubMed] [Google Scholar]
- 40.Reneker DH, Chun I. Nanotechnology. 1996;7:216–223. [Google Scholar]
- 41.Yoshimoto H, Shin YM, Terai H, Vacanti JP. Biomaterials. 2003;24:2077–2082. doi: 10.1016/s0142-9612(02)00635-x. [DOI] [PubMed] [Google Scholar]
- 42.Li MY, Mondrinos MJ, Gandhi MR, Ko FK, Weiss AS, Lelkes PI. Biomaterials. 2005;26:5999–6008. doi: 10.1016/j.biomaterials.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 43.Smith LA, Ma PX. Colloids and Surfaces B-Biointerfaces. 2004;39:125–131. doi: 10.1016/j.colsurfb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Min BM, Lee G, Kim SH, Nam YS, Lee TS, Park WH. Biomaterials. 2004;25:1289–1297. doi: 10.1016/j.biomaterials.2003.08.045. [DOI] [PubMed] [Google Scholar]
- 45.Jin HJ, Chen JS, Karageorgiou V, Altman GH, Kaplan DL. Biomaterials. 2004;25:1039–1047. doi: 10.1016/s0142-9612(03)00609-4. [DOI] [PubMed] [Google Scholar]
- 46.Luu YK, Kim K, Hsiao BS, Chu B, Hadjiargyrou M. Journal of Controlled Release. 2003;89:341–353. doi: 10.1016/s0168-3659(03)00097-x. [DOI] [PubMed] [Google Scholar]
- 47.Wnek GE, Carr ME, Simpson DG, Bowlin GL. Nano Letters. 2003;3:213–216. [Google Scholar]
- 48.Kenawy ER, Layman JM, Watkins JR, Bowlin GL, Matthews JA, Simpson DG, Wnek GE. Biomaterials. 2003;24:907–913. doi: 10.1016/s0142-9612(02)00422-2. [DOI] [PubMed] [Google Scholar]
- 49.Matthews JA, Wnek GE, Simpson DG, Bowlin GL. Biomacromolecules. 2002;3:232–238. doi: 10.1021/bm015533u. [DOI] [PubMed] [Google Scholar]
- 50.Deitzel JM, Kleinmeyer JD, Hirvonen JK, Tan NCB. Polymer. 2001;42:8163–8170. [Google Scholar]
- 51.Bhattarai N, Edmondson D, Veiseh O, Matsen FA, Zhang MQ. Biomaterials. 2005;26:6176–6184. doi: 10.1016/j.biomaterials.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 52.Ma ZW, Kotaki M, Yong T, He W, Ramakrishna S. Biomaterials. 2005;26:2527–2536. doi: 10.1016/j.biomaterials.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 53.Whitesides GM, Mathias JP, Seto CT. Science. 1991;254:1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 54.Decher G. Science. 1997;277:1232–1237. [Google Scholar]
- 55.Philp D, Stoddart JF. Angewandte Chemie-International Edition in English. 1996;35:1155–1196. [Google Scholar]
- 56.Kadler KE, Holmes DF, Trotter JA, Chapman JA. Biochemical Journal. 1996;316:1–11. doi: 10.1042/bj3160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 58.Zhang SG. Nature Biotechnology. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 59.Yu YC, Tirrell M, Fields GB. Journal of the American Chemical Society. 1998;120:9979–9987. [Google Scholar]
- 60.Nam YS, Park TG. Journal of Biomedical Materials Research. 1999;47:8–17. doi: 10.1002/(sici)1097-4636(199910)47:1<8::aid-jbm2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 61.Zhang RY, Ma PX. Journal of Biomedical Materials Research. 1999;44:446–455. doi: 10.1002/(sici)1097-4636(19990315)44:4<446::aid-jbm11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 62.Lee SH, Kim BS, Kim SH, Kang SW, Kim YH. Macromolecular Bioscience. 2004;4:802–810. doi: 10.1002/mabi.200400021. [DOI] [PubMed] [Google Scholar]
- 63.Ma PX, Zhang RY. Journal of Biomedical Materials Research. 1999;46:60–72. doi: 10.1002/(sici)1097-4636(199907)46:1<60::aid-jbm7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 64.Zhang RY, Ma PX. Journal of Biomedical Materials Research. 2000;52:430–438. doi: 10.1002/1097-4636(200011)52:2<430::aid-jbm25>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 65.Chen VJ, Smith LA, Ma PX. Biomaterials. 2006;27:3973–3979. doi: 10.1016/j.biomaterials.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 66.Liu XH, Smith LA, Wei G, Won YJ, Ma PX. Journal of biomedical nanotechnology. 2005;1:54–60. [Google Scholar]
- 67.Chen VJ, Ma PX. Biomaterials. 2004;25:2065–2073. doi: 10.1016/j.biomaterials.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 68.Wei GB, Ma PX. Journal of Biomedical Materials Research Part A. 2006;78A:306–315. doi: 10.1002/jbm.a.30704. [DOI] [PubMed] [Google Scholar]
- 69.Liu XH, Ma PX. Annals of Biomedical Engineering. 2004;32:477–486. doi: 10.1023/b:abme.0000017544.36001.8e. [DOI] [PubMed] [Google Scholar]
- 70.Nitschke M, Schmack G, Janke A, Simon F, Pleul D, Werner C. Journal of Biomedical Materials Research. 2002;59:632–638. doi: 10.1002/jbm.1274. [DOI] [PubMed] [Google Scholar]
- 71.Cai KY, Yao KD, Cui YL, Lin SB, Yang ZM, Li XQ, Xie HQ, Qing TW, Luo J. J. Biomed. Mater. Res. 2002;60:398–404. doi: 10.1002/jbm.10008. [DOI] [PubMed] [Google Scholar]
- 72.Gao JM, Niklason L, Langer R. Journal of Biomedical Materials Research. 1998;42:417–424. doi: 10.1002/(sici)1097-4636(19981205)42:3<417::aid-jbm11>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 73.Hu YH, Winn SR, Krajbich I, Hollinger JO. J. Biomed. Mater. Res. A. 2003;64A:583–590. doi: 10.1002/jbm.a.10438. [DOI] [PubMed] [Google Scholar]
- 74.Liu XH, Won YJ, Ma PX. Biomaterials. 2006;27:3980–3987. doi: 10.1016/j.biomaterials.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 75.Liu XH, Won YJ, Ma PX. Journal of Biomedical Materials Research Part A. 2005;74A:84–91. doi: 10.1002/jbm.a.30367. [DOI] [PubMed] [Google Scholar]
- 76.Smith LA, Beck J, Ma PX. In: Nanotechnologies for Tissue, Cell and Organ Engineering. Kumar C, editor. Weinheim, Germany: Wiley-VCH; 2007. [Google Scholar]
- 77.Tysseling-Mattiace V, Sahni V, Niece KL, Birch D, Czeisler C, Fehling M, Stupp S, Kessler J. J Neurosci. 2008;28:3814–3814. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woo KM, Jun JH, Chen VJ, Seo J, Baek JH, Ryoo HM, Kim GS, Somerman MJ, Ma PX. Biomaterials. 2007;28:335–343. doi: 10.1016/j.biomaterials.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 79.Woo KM, Chen VJ, Ma PX. J Biomed Mater Res A. 2003;67:531–537. doi: 10.1002/jbm.a.10098. [DOI] [PubMed] [Google Scholar]
- 80.Schindler M, Ahmed I, Kamal J, Nur-E-Kamal A, Grafe TH, Chung HY, Meiners S. Biomaterials. 2005;26:5624–5631. doi: 10.1016/j.biomaterials.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 81.Chen M, Prabir K, Warner SB, Bhowmick S. Tissue Eng. 2007;13:579–587. doi: 10.1089/ten.2006.0205. [DOI] [PubMed] [Google Scholar]
- 82.Xu CY, Inai R, Kotaki M, Ramakrishna S. Biomaterials. 2004;25:877–886. doi: 10.1016/s0142-9612(03)00593-3. [DOI] [PubMed] [Google Scholar]
- 83.Yang F, Xu CY, Kotaki M, Wang S, Ramakrishna S. J Biomater Scit Polymer Edn. 2004;15:1483–1497. doi: 10.1163/1568562042459733. [DOI] [PubMed] [Google Scholar]
- 84.Nur-E-Kamal A, Ahmed I, Kamal J, Schindler M, Meiners S. Stem Cells. 2006;24:426–433. doi: 10.1634/stemcells.2005-0170. [DOI] [PubMed] [Google Scholar]
- 85.Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B, Stupp SI. Biomaterials. 2007;28:4608–4618. doi: 10.1016/j.biomaterials.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 86.Li WJ, Danielson KG, Alexander PG, Tuan RS. J Biomed Mater Res. 2003;67A:1105–1114. doi: 10.1002/jbm.a.10101. [DOI] [PubMed] [Google Scholar]
- 87.Bondar B, Fuchs S, Motta A, Migliaresi C, Kirkpatrick CJ. Biomaterials. 2008;29:561–572. doi: 10.1016/j.biomaterials.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 88.Hu J, Liu X, Ma PX. Biomaterials. In Press. [Google Scholar]
- 89.Shih YV, Chen CN, Tsai SW, Wang YJ, Lee OK. Stem Cells. 2006;24:2391–2397. doi: 10.1634/stemcells.2006-0253. [DOI] [PubMed] [Google Scholar]
- 90.Nur-E-Kamal A, Ahmed I, Kamal J, Schindler M, Meiners S. Biochem Biophys Res Commun. 2005;331:428–434. doi: 10.1016/j.bbrc.2005.03.195. [DOI] [PubMed] [Google Scholar]
- 91.Lee CH, Shin HO, Cho IH, Kang YM, Kim IA, Park KD, Shin JW. Biomaterials. 2005;26:1261–1270. doi: 10.1016/j.biomaterials.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 92.Li WJ, Jiang YJ, Tuan RS. Tissue Eng. 2006;12:1775–1785. doi: 10.1089/ten.2006.12.1775. [DOI] [PubMed] [Google Scholar]
- 93.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 94.Chua KN, Lim WS, Zhang P, Lu H, Wen J, Ramakrishna S, Leong KW, Mao HQ. Biomaterials. 2005;26:2537–2547. doi: 10.1016/j.biomaterials.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 95.Semino CE, Merok JR, Crane GG, Panagiotakos G, Zhang S. Differentiation. 2003;71:262–270. doi: 10.1046/j.1432-0436.2003.7104503.x. [DOI] [PubMed] [Google Scholar]
- 96.Parson JT. J Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- 97.Li W, Tuli R, Huang X, Laquerriere P, Tuan R. Biomaterials. 2005;26:5158–5166. doi: 10.1016/j.biomaterials.2005.01.002. [DOI] [PubMed] [Google Scholar]