Abstract
Whole-body optical molecular imaging of mouse models in preclinical research is rapidly developing in recent years. In this context, it is essential and necessary to develop novel simulation methods of light propagation for optical imaging, especially when a priori knowledge, large-volume domain and a wide-range of optical properties need to be considered in the reconstruction algorithm. In this paper, we propose a three dimensional parallel adaptive finite element method with simplified spherical harmonics (SPN) approximation to simulate optical photon propagation in large-volumes of heterogenous tissues. The simulation speed is significantly improved by a posteriori parallel adaptive mesh refinement and dynamic mesh repartitioning. Compared with the diffusion equation and the Monte Carlo methods, the SPN method shows improved performance and the necessity of high-order approximation in heterogeneous domains. Optimal solver selection and time-costing analysis in real mouse geometry further improve the performance of the proposed algorithm and show the superiority of the proposed parallel adaptive framework for whole-body optical molecular imaging in murine models.
Keywords: optical molecular imaging, photon migration, radiative transfer equation, SPN approximation, adaptive mesh refinement, parallel finite element
1. INTRODUCTION
As a strategy that reflects genomic and proteomic changes and interactions in vivo and at the macroscopic level, photonics-based molecular imaging has gained an indispensable position in the biological and medical research fields [1][2]. With very high sensitivity, low cost and easy operation, it is applied extensively in multiple biomedical research fields, such as oncology, gene therapy, and drug metabolism [3][4]. Typical tomographic optical molecular imaging includes bioluminescence tomography (BLT) [5] and fluorescence molecular tomography (FMT) [6]. Without tissue autofluorescence generated by external illumination, BLT is purported to have higher sensitivity than FMT. However, illumination provides many source-detector pairs for FMT, which make it become potentially easier and more accurate to reconstruct the internal sources compared with BLT [2].
Although optical tomography in molecular imaging is striving to reconstruct the information of the light source inside small animal models (the inverse problem), reflecting molecular and cellular changes, the essential problem in optical imaging is to develop fast, precise and efficient simulation algorithms of light propagation in biological tissues, that is the forward problem [7]. Through modeling individual photon trajectories, the Monte Carlo (MC) method and its variants can precisely depict light propagation information in tissues. The MC method is called as “gold standard” because of its accuracy. However, tracking a large number of photons results in slow simulation speeds, especially in complex heterogeneous geometries. A deterministic full description of light propagation can be obtained by the radiative transport equation (RTE) [8]. The RTE doesn’t consider statistical Poisson noise in photon propagation and therefore, it can reflect small changes of optical signals in the simulation. The diffusion equation (DE) is widely applied in optical imaging. In the DE, light propagation is assumed to be isotropic, which is not accurate when the light source is near surface and in anisotropic tissues with high absorption [7]. Complex living tissues and optical spectrum characteristics of existing optical probes aggravate the DE’s application range [9]. It is necessary to solve the RTE and its approximations for optical molecular imaging. Discrete ordinates (SN) and spherical harmonics (PN) methods, as two usual approximation methods, can obtain more precise numerical solution of the RTE with the increase of N depicting their approximation degree to the RTE [10]. Recently, several researchers have obtained desirable results using these approaches [11][12]. However, in order to generate precise solutions, one has to set N as big as possible and N(N + 2) and (N + 1)2 coupled equations corresponding to SN and PN methods need to be solved. This computational complexity results in long computational times with sequential execution even in high performance computers, restricting its application. In order to improve the simulation precision and speed, Cong et al propose an integral equation based on a generalized delta-Eddington phase function [13]. Based on a simplified spherical harmonics method, Klose et al deduce a set of coupled partial differential equations (PDEs) to reduce the computation time of PN methods [9].
Optical molecular imaging is an ill-posed problem by nature. Current research has shown that a priori knowledge is indispensable to reconstruct source information [14]. Source spectrum and small-animal anatomical information can significantly improve reconstruction results [15][16]. Numerical methods, especially the finite element method (FEM), become necessary when the heterogeneity of the small animal and the complex geometries of organs are considered. Using high order approximations to the RTE, multi- and hyper-spectral measurements further increase computation burden [17]. Simulation on a single PC is too costly, especially in the large-volume domain (eg whole-body mouse and fine mesh). Parallel computation strategy is an efficient method to perform simulations with large dimensionality. High-performance computer clusters dedicated to parallel computations are in general available at many research institutions. The adaptive finite element method is another approach. Not only has it improved simulation speed and precision of light propagation, but also has been successfully applied in source reconstruction [18][19]. Through local mesh refinement, the quality of reconstructed source distributions is remarkably improved. In fluorescence optical tomography, based on diffusion theory, Joshi et al consider multiple area illumination patterns to combine the parallel strategy and adaptive finite element method [20]. However, the parallel framework that each node of the cluster is responsible for one or multiple measurements is challenged when high order approximations to the RTE are applied. Therefore, a novel parallel framework with adaptive FEM needs to be developed for the simulation of light propagation in the large-volume domain with a complicated physical model.
In this paper, a novel parallel adaptive finite element framework is proposed to meet the current demand for whole-body small animal molecular imaging simulations. This framework efficiently tackles the computational burden through the combination of domain decomposition and local mesh refinement. To solve the computational load imbalance problem, dynamic repartitioning is implemented after the mesh is adaptively refined. Further, a set of coupled PDEs based on SPN method is used to realize the light propagation simulation. We also optimally select the preconditioner and iterative algorithm through comparing current popular methods in real heterogeneous mouse geometry to further accelerate the simulation speed. Regarding the Monte Carlo method and diffusion equation, comparison of the results illustrates the need for high order approximation in the heterogeneous phantom, further demonstrating the superiority of the parallel adaptive framework. The next section introduces the proposed simulation framework that incorporates parallel adaptive strategy and SPN approximation. Experimental setups, simulation results and relevant discussions are demonstrated in Section 3. Finally, the conclusion and discussion are provided in Section 4.
2. Algorithms
2.1. Radiative Transfer Equation and SPN Approximation
The radiative transfer equation (RTE) is an approximation to Maxwell’s equations. It comes from an energy conservation principle, that is the radiance ψ(r, ŝ, t) is equal to the sum of all factors affecting it (including absorption μa(r), scattering μs(r), and obtainable energy Q(r, t, ŝ) ) when light crosses a unit volume, where ψ(r, ŝ, t) denotes photons in unit volume traveling from point r in direction ŝ at time t [21]. In the RTE, some wave phenomena such as polarization and interference are ignored. When the signals are measured in optical imaging, light source intensity is generally assumed to be invariant. Therefore, the time-independent RTE in 3D is used [22]:
| (1) |
where p(ŝ, ŝ') is the scattering phase function and gives the probability of a photon anisotropically scattering from direction ŝ to direction ŝ'. Generally, the Henyey-Greenstein (HG) phase function is often used to characterize this probability [23]:
| (2) |
where g is the anisotropy parameter; cos θ denotes scattering angle and is equal to ŝ • ŝ' when we assume that the scattering probability only depends on the angle between the incoming and outgoing directions. The HG phase function is easily expanded by the Legendre polynomial. Therefore, it is convenient for numerical computation.
When photons reach the body surface of a mouse, that is r ∈ ∂Ω, some of them are reflected and can’t escape from the mouse body Ω because of the mismatch between the refractive indices nb for Ω and nm for the external medium. When the incidence angle θb from the mouse body is not beyond the critical angle θc (θc = arcsin(nm/nb) based on Snell’s law), the reflectivity R(cos θb) is given by [24]:
| (3) |
where θm is the transmission angle. Furthermore, we can get the exiting partial current J+(r) at each point r where the external source S(r) = 0 [9]:
| (4) |
After a series of deductions is performed in the planar geometry with PN method, the 3D SP7 approximation are obtained by replacing 1D diffusion operator with its 3D counterpart [9]:
| (5a) |
| (5b) |
| (5c) |
| (5d) |
where μan = μ s(1 − gn) + μ a(n = 1, 2, 3, 4, 5, 6, 7); φi(i = 1, 2, 3, 4) is the composite moments relevant to the Legendre moments. The Legendre moments can be obtained by expanding ψ with the PN approximation. The detailed deductions are described in [9]. We use
| (6) |
to depict the effect of reflectivity in different angular moments on SPN approximation, the corresponding boundaries are given [9]:
| (7a) |
| (7b) |
| (7c) |
| (7d) |
The coefficients A1,…,H1, …,A4, …,H4 can be found in [9]. Furthermore, the exiting partial current J+ is obtained at each point r where the source S(r) = 0:
| (8) |
where the coefficients J0, …, J7 can also be found in [9]. Note that SP5, SP3, and SP1 (diffusion equation) can be obtained correspondingly by setting φ4 = 0, φ4 = φ3 = 0, φ4 = φ3 = φ2 = 0. In order to describe the proposed algorithm, the general equation for Equs. 5a– 5d is followed:
| (9) |
2.2. Weak Formulation
When we select an arbitrary continuous test function υ, Equ. 9 is integrated over the whole domain Ω, the corresponding weighted residual formulation is as follows:
| (10) |
After applying the Gauss divergence theorem and regarding Robin boundary conditions (Equs. 7a–7d), we may get the general weak formulation for Equ. 9:
| (11) |
v· φi can be obtained through solving the boundary equations (7a–7d), and expressed by the linear combination of φ1, φ2, φ3 and φ4.
2.3. Finite Element Formulation
In the framework of adaptive finite element analysis, let {𝒯1, …𝒯l, …} be a sequence of nested triangulation of the given domain Ω based on the adaptive mesh evolution, where the sequence gradually changes as elements are refined and coarsened when l is increased. The spaces of linear finite elements 𝒱l are introduced on the discretized levels 𝒯l, satisfying ോ1 ⊂ … ⊂ 𝒱l ⊂ … ⊂ H1(Ω). We only consider the lth discretized level which includes N𝒯l elements and N𝒫l vertex nodes. is the nodal basis of the space 𝒱l. We use (i = 1, 2, 3, 4) to represent the finite dimensional approximations to φi on the lth level. When the standard Lagrange-type finite element basis is used, we have
| (12) |
Considering Equ. 11 and Equs. 5a–5d, we have
| (13) |
where
| (14) |
and
| (15) |
τe and ∂τe are the volumetric and boundary elements respectively. When all the matrices and vectors on each element are assembled, the FEM-based linear system on the lth level is as follows:
| (16) |
Here, the domain to be solved is decomposed into several subdomains depending on CPU numbers. Each processor of the cluster is responsible for matrix assembly in its subdomain.
2.4. Iterative Methods and Preconditioners
Compared with the diffusion equation, the linear equation 𝒜lΦl = bl has larger dimensionality in the high order SPN approximation. Fortunately, by observing the relevant coefficients in Equs. 5a–5d and Equs. 7a–7d, we may find that each is symmetric and each is equal to . This means that the mass matrix 𝒜l is sparse, symmetric, and positive definite. Iterative methods, especially the conjugate gradient (CG) method, are suitable for this type of linear equation [25]. However, with the increase of l, the size of 𝒜l becomes more and more larger and its condition number is worse. It is therefore difficult to improve simulation performance only using the CG method. Currently, with the development of computer hardware and computational techniques, scalability has become an important goal to develop numerical algorithms. Generally, if an algorithm is considered scalable, the time used to obtain the solution should be essentially invariable as the problem size and the computing resources increase [26]. Preconditioning is an important method to obtain scalable simulations. Several types of preconditioners have been developed with parallel mode. For the specified problem, it is important to decide which preconditioner is the best by relevant testings. Three main types of parallel preconditioners are as follows [27][28]:
Incomplete factorization methods [29] In nature, preconditioning is to find a matrix P which makes P−1A have better spectral characteristic than A. Therefore, some classical preconditioners are based on direct factorization methods, leading to incomplete factorization methods for saving computational time in fully factorization. Incomplete LU (ILU) factorization is the most popular method in this type. By decomposing A into L̃Ũ − R, we may define P = L̃Ũ, where R is the residucal of the factorization. Generally, an ILU factorization with less accuracy, such as ILU(0), takes lower preprocessing time, but requires more iterations to converge in preconditioned CG method. Therefore, it is necessary to find the suitable factorized degree of ILU, that is deciding the level k in ILU(k). For a sparse matrix, to efficiently implement the ILU method, reordering strategy and blocking technique are used to reduce fill-ins in L and U. The latter belongs to domain decomposition-type methods and is more suitable for the problems arising from discretization of PDEs over a given domain.
Sparse approximate inverse [30] The common idea of sparse approximate inverse is to obtain a sparse matrix P ≈ A−1. This method assumes that A is a sparse matrix. Although the inverse of a sparse matrix is generally dense, many entries of the inverse matrix are small. It is possible to get a sparse approximation of A−1. There are two types of approximate inverse methods which are classified by judging the expression of the inverse matrix. One classical method for finding an approximate inverse in one matrix is based on the minimization of the Frobenius norm of the residual matrix . The factored approximate inverse method is the other representative method which generates two or more matrices. Because sparse approximate inverse-based preconditioning operation is composed of matrix-vector products, it is attractive as a parallel preconditioner.
Multigrid/Multilevel methods [31] Multilevel methods, especially the multigrid method, have been very popular in PDE-based applications because of its desirable performance. It is easy to understand this method using the Fourier analysis of the residual in each iteration. The “high-frequency” components related to larger eigenvalues in the matrix A rapidly converge after several iterate steps, however, the convergence corresponding to “low-frequency” components is slow. Using several grids with different discretized degrees, that is with the multigrid method, the solution on the fine mesh is corrected by that on the coarse mesh. The establishment of geometric multigrid preconditioners depends to a large degree on the underlying mesh and is time-consuming. The algebraic multigrid (AMG) method exploring the characteristic of the underlying matrix and realizing convergence of the components with “different frequencies” is extensively applied. On the adaptively refined mesh, the AMG also shows the preferable performance in preprocessing. A parallel AMG preconditioner, that is BoomerAMG, is developed to meet the needs of parallel computation.
2.5. Adaptive Mesh Evolution and Loading Balance
2.5.1. Adaptive Mesh Evolution
Compared with the fixed or uniformly refined meshes, adaptively refined mesh needs to consider a posteriori error estimation and the processing of hanging nodes. A posteriori error estimators decide which element should be refined or coarsened based on current solution. In the parallel mode, local error indicators are suitable which reduce communications between processors. In this application, the gradient-jump error indicator is applied [32]. For the SPN, the indicator is as follows for element τe [33]
| (17) |
where wi is a weighted factor corresponding to φi, Ri,τe is the local residual for φi, which is defined by
| (18) |
vτe is the outward unit normal for element τep, and hτe is the diameter of τep. With ητe, a statistical strategy is used to decide the proportion of refined and coarsened elements [34]. In this method, the mean (m) and standard deviation (σ) of the distribution of the obtained indicator are computed. The refinement and coarsening ratios (γr and γc) are set to select elements. By judging whether the log-error of one element is higher than (m + γrσ), or lower than (m − γcσ), it is correspondingly refined or coarsened. Note that it is difficult to coarse a unstructured mesh beyond its initial level. Therefore, only the refined elements can be coarsened in the implementation.
The algebraic constraint method is selected to deal with hanging nodes [34]. The geometric method needs to consider the elements adjacent to the refined one. Generally, hanging nodes are dealt with by dividing the neighbor element into several subelements. Different types of elements need different constraint strategies. In the 3D simulation, the geometric method usually increases the processing complexity. Fox example, when a tetrahedron is locally refined by “red-green” method, four constraint cases need to be considered [35]. The algebraic constraint method deals with hanging nodes by constraining the corresponding degree of freedom at the matrix level. It is more flexible and easily realized.
2.5.2. Loading Balance
Parallelization of simulations with adaptive mesh evolution as a dynamic method bring new challenges. It is difficult to a priori know the element distribution in the final mesh. Imbalance loading deteriorates parallel performance and dynamic repartitioning of the mesh is indispensable to obtain loading balance. A k-way partitioning method is used to perform the partitioning after mesh refinement [36]. This method obtains superior performance by reducing the dimension of the mesh, partitioning it with smaller size, and refining it to the original.
3. Results and discussions
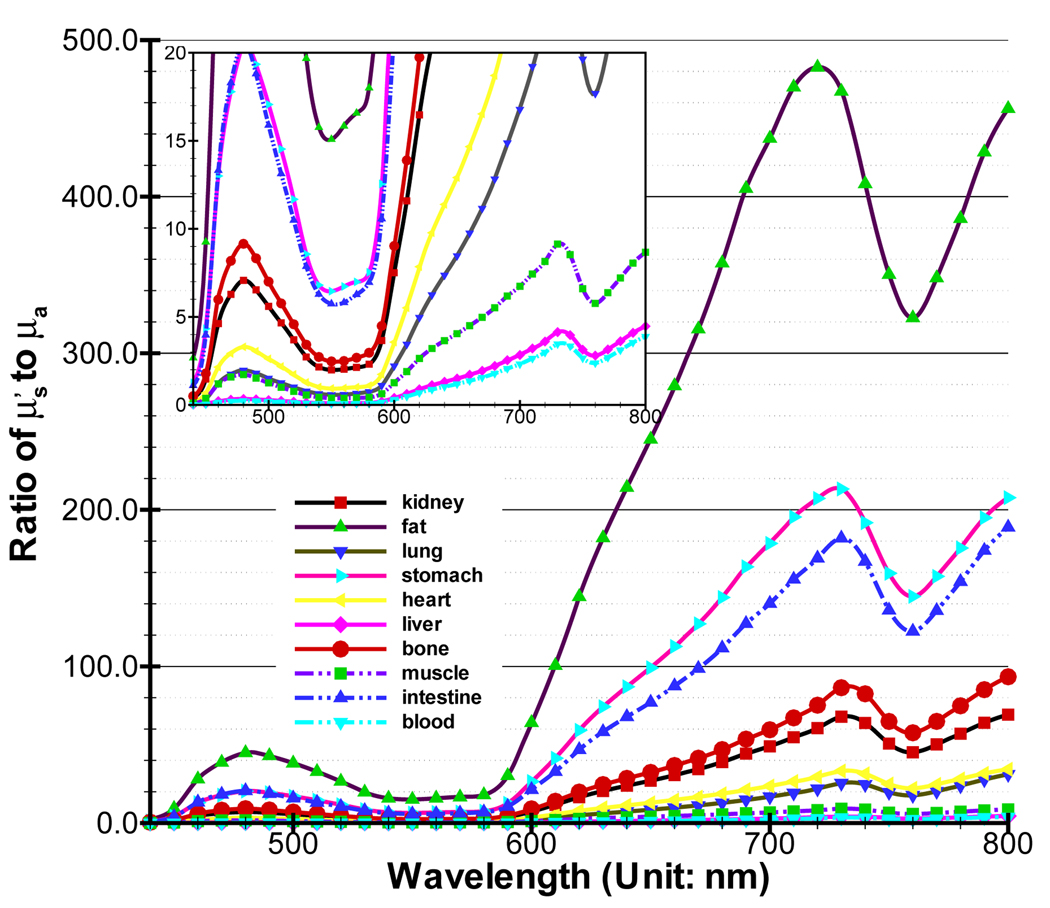
In biological tissues, blood and water play an important role in absorbing near-infrared (NIR) photons. Oxy-hemoglobin (HbO2) and deoxy-hemoglobin (Hb) are the main absorbers in blood [21]. The absorption coefficients of tissues are mainly decided by the concentration of three substances. With respect to the scattering coefficient, research has indicated that there is a small change in the NIR domain. Generally, it is an inverse power function of the wavelength [16]. Alexandrakis et al estimate the absorption (μa) and reduced scattering coefficients by compiling relevant data in [16]. The high scattering characteristic of tissue means . If is close to μa, diffusion theory is not accurate. Figure 3 is about the ratio for main organs in a mouse, which is calculated based on the formulas and data in [16]. At the shorter wavelength (440–650nm), almost all of the biological tissues have small ratios. Even though the spectrum of many applied bioluminescent and fluorescent probes is in this wavelength band, shorter wavelengths are heavily absorbed and therefore wavelengths less than 600 nm are not very important for in-vivo applications. However, the wavelength is between 650 – 800nm where NIR photons easily propagate, tissues with rich blood vessels, such as muscle and liver, also have small ratios, illustrating the necessity of developing new mathematical models.
Figure 3.
Comparison between diffusion equation, Monte Carlo method and SPN approximations in the homogeneous phantom when g = 0.0 and nb = 1.0. The ratios of (a), (b) and (c) are 100.0, 5.0 and 2.0 respectively. MC/SPi (i = 1, 3, 7) denote the ratios and “SPj to SP7” (j = 1, 3) are the relative errors.
Currently, many open-source, high-quality software packages are developed to meet the need of FEM-based simulation. This not only alleviates unnecessary effort and improves the efficiency, but also provides a platform to objectively evaluate various numerical algorithms. Here, we select libMesh as the basis development environment [37]. LibMesh provides almost all of the components used in parallel PDE-based simulation with unstructured discretization. Its design concept is also to use existing software packages as far as possible. PETSc developed by Argonne National Laboratory (ANL) is used to solve the linear systems in parallel mode [38]. By default, METIS realizing the k-way partitioning algorithm is used to dynamically partition the whole domain in libMesh.
In the following simulations, we first use the simple homogeneous and heterogeneous phantoms to verify the SPN method and evaluate its performance in three dimension. Subsequently, we use a whole-body digital mouse to further test the proposed algorithm [39]. We select the tetrahedron as the basis element of the mesh because of its flexibility in depicting complex geometries. We set the refinement and coarsening ratios (γr and γc) to 85% and 2%. Simulation for fluorescence imaging includes two phases, that is excitation and emission. Its emission process is similar to bioluminescence imaging. Therefore, we set the external source S to 0 for better comparison. Note that it is simple to realize photon propagation simulation for fluorescence imaging using the proposed algorithm. All the simulations are performed on a cluster of 27 nodes ( 2 CPUs of 3.2GHz and 4 GB RAM at each node) which is available at our lab.
3.1. Homogeneous Phantom Cases
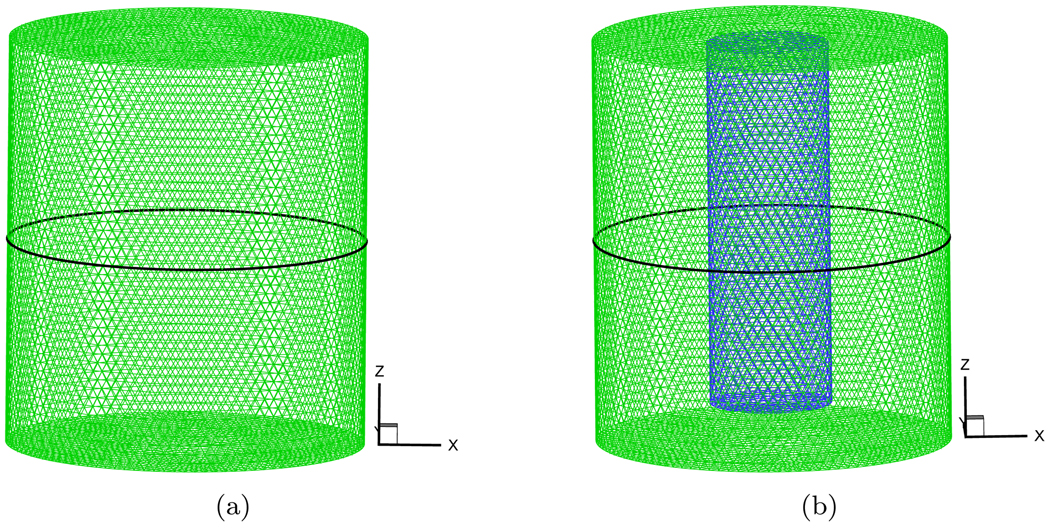
The maximal diameter of a normal mouse is about 35mm [39]. In order to better demonstrate the simulated results, we set the dimensions of the cylindrical homogeneous phantom to 17.5mm radius and 40.0mm height. Its center was at the coordinate origin. A solid spherical source was placed at half-radius position on the negative X-axis for observing the effect of different distances from source on the exiting partial current J+(r). The source radius was 0.01mm and was consider as a point source to reduce the effect of source shape. Because a posteriori error estimation depends on the variables φi, we select the fixed mesh for objective comparison. Figure 2(a) shows the discretized mesh which has the average element size of 1mm and includes 42524 nodes and 229019 elements. The Monte Carlo method can incorporate the Poisson noise into the simulation in a natural way. Its simulated results are very close to real measurements. An in-house MC-based simulator was coded to generate the exiting partial current based on the method in [40]. Its speed was fast because of the use of analytic geometry which was valid to this particular phantom geometry.
Figure 2.
The discretized meshes of the used homogeneous and heterogeneous phantoms. The black circles denote the detection position for comparison.
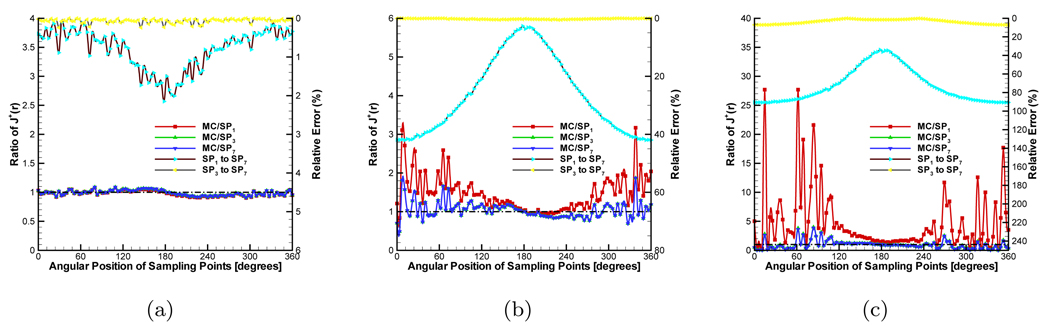
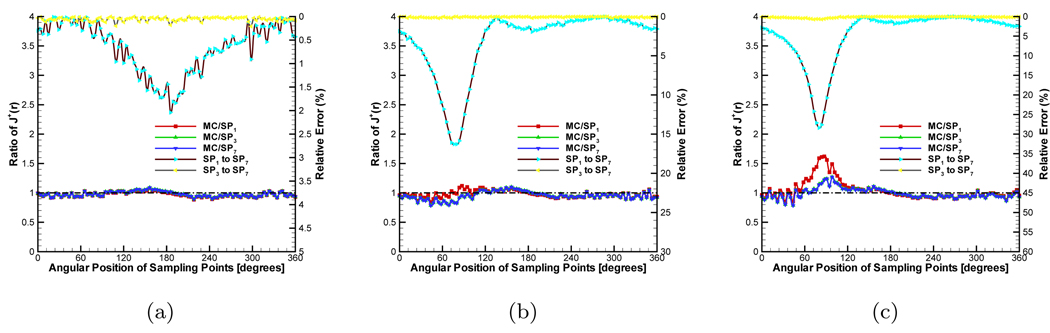
The ratio of to μa, the anisotropy g, and the refractive index nb have significant effect on J+(r). We consider these factors to observe their effect on the SPN approximation. In the MC simulation, 5 × 106 ~ 2 × 107 photons are tracked depending on the ratio to avoid the effect of a small amount of photons. The data from the MC simulation was normalized by the ratio of the average values obtained from SP7 approximation and the MC method, which ignores the effect of the source intensity. In the first group of experiments, we set g, nb and nm to 0.0, 1.0 and 1.0 respectively, which means that the phantom is isotropic and its refractive index matches that of the medium. The scattering coefficient μs was fixed to 0.5mm−1. The absorption coefficient μa was adjusted to obtain different ratios. The compared results are shown in Figure 3. The detected circle for J+ is shown in Figure 2(a). Figure 3(a) shows the compared results when μa is set to 0.005mm−1. The ratio is 100.0 and therefore the phantom had high scattering characteristic. Although there are some fluctuations in the compared results of MC/SPN(N = 1, 3, 7) because of the noise in the MC data, all the ratios are very close to the standard value “1.0”. The maximal value of the relative errors obtained by only is 2.15%. All of these results show the consistency of different SPN approximations in the 3D high scattering domain. Furthermore, we set the ratio to 5.0, Figure 3(b) shows the compared results. With the increase of μa, the values of J+ become smaller and the noise is comparatively large. Therefore, the fluctuations become larger. However, MC/SP3 and MC/SP7 always waver around “1.0” and MC/SP1 is far from “1.0” which shows the accuracy of the high order SPN approximation in the high absorption homogeneous medium. When μa is adjusted to 0.25mm−1, the much larger fluctuations of MC/SP1 shown in Figure 3(c) further demonstrate the inaccuracy of diffusion equation in the high absorption domain.
In order to observe the effect of the refractive indices mismatch on J+, all the settings were the same with the first group of experiments but nb was adjusted to 1.37. Figure 4 shows the compared results. The distinct difference from the previous case is that the errors between MC/SP1 and MC/SP7 become larger when the values of J+ are comparatively large, as shown in Figures 4(b) and 4(c). These errors cause a significant deviation on MC/SP1 from “1.0”. In the reconstruction, strong signals have much more effect than weak ones. Therefore, the accuracy of high order SPN approximation shows its ability to better tolerate the refractive indices mismatch.
Figure 4.
Comparison between diffusion equation, Monte Carlo method and SPN approximations in the homogeneous phantom when g = 0.0 and nb = 1.37. The ratios of (a), (b) and (c) are 100.0, 5.0 and 2.0 respectively. MC/SPi (i = 1, 3, 7) denote the ratios and “SPj to SP7” (j = 1, 3) are the relative errors.
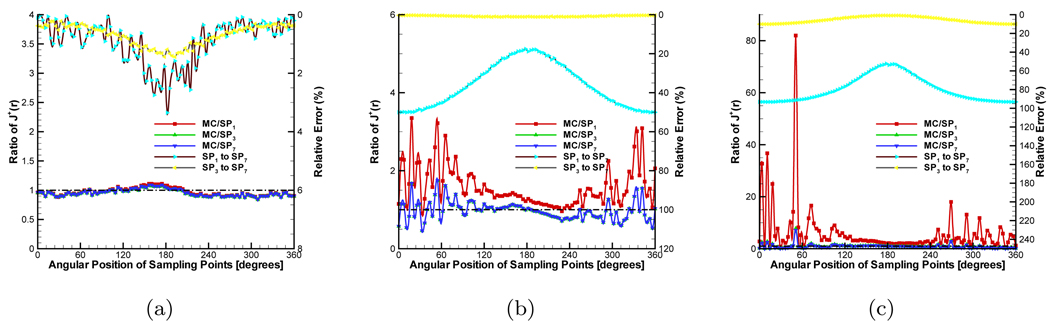
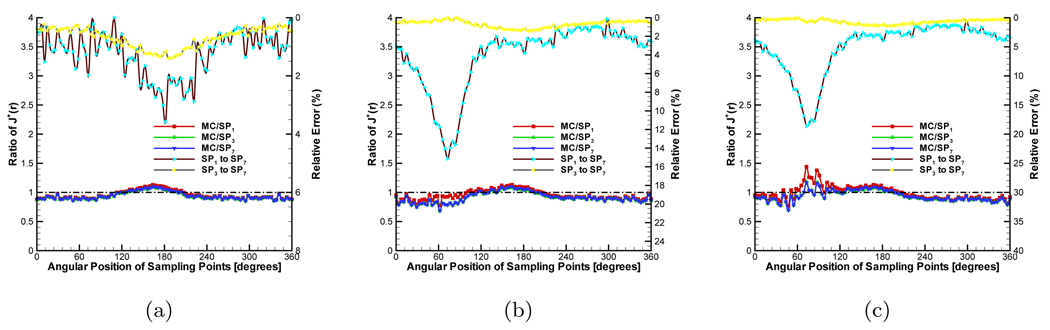
Finally, the anisotropy factor was considered in the simulation. For retaining the ratios, we set g to 0.9 and μs was correspondingly adjusted to 5.0mm−1. The compared results are shown in Figure 5. The high order SPN approximations obtain similar performance with in the pervious two groups of experiments. However, because diffusion equation can’t describe the anisotropic characteristic of the medium, the relative errors have comparatively large fluctuations. Note that though SP7 has better performance than SP3, the improvement is negligible relative to the MC-based statistical noise. Therefore, SP3 is an optimal choice for simulation and reconstruction on a 3D homogeneous phantom.
Figure 5.
Comparison between diffusion equation, Monte Carlo method and SPN approximations in the homogeneous phantom when g = 0.9 and nb = 1.37. The ratios of (a), (b) and (c) are 100.0, 5.0 and 2.0 respectively. MC/SPi (i = 1, 3, 7) denote the ratios and “SPj to SP7” (j = 1, 3) are the relative errors.
3.2. Heterogeneous Phantom Cases
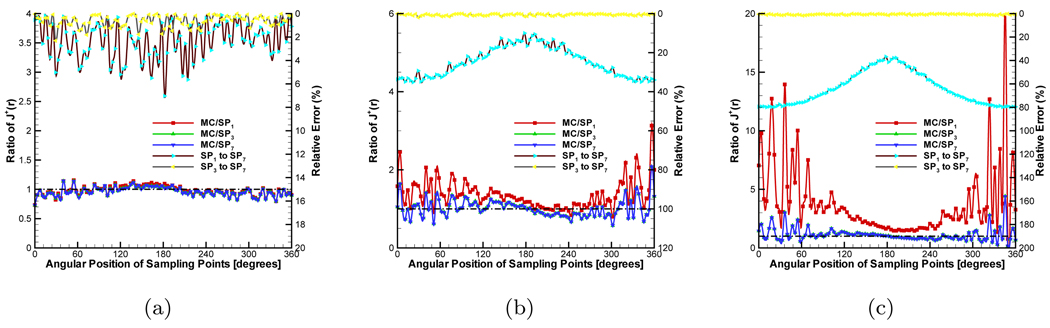
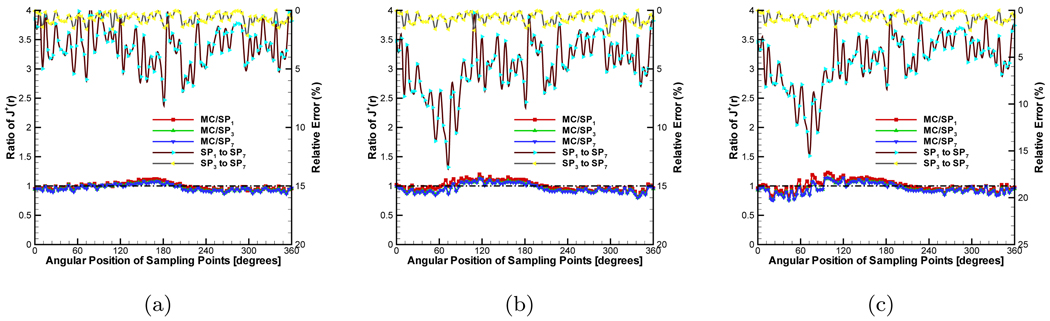
A heterogeneous phantom was considered for testing the SPN approximation, which is shown in Figure 2(b). This phantom was obtained by placing a cylinder with 6.0mm radius and 36.0mm height into the used homogeneous phantom. The center of the internal cylinder was at (0.0, 8.75, 0.0). The average element size of the discretized mesh was 1.0mm. The source settings were the same as that in the homogeneous phantom. In the simulations, the absorption coefficient μa of the external cylindrical component in the phantom was fixed to 0.005mm−1. μs was adjusted depending on g, guaranteeing that the ratio of to μa was 100.0. The refractive index was set to the same value with that of internal phantom. According to the homogeneous phantom cases, g and nb of the small cylindrical component of the phantom were adjusted. Figure 6, Figure 7 and Figure 8 show the compared results corresponding to Figure 3, Figure 4 and Figure 5. When the was set to 50.0, the similar performance was observed (Figure 6(a), Figure 7(a) and Figure 8(a)) with that in the homogeneous phantom indicating that the program can primely deal with the simulation with discontinuous coefficients. When we reduced the ratio of the small cylinder to 5.0 and 2.0, on the whole, it was a little difficult to observe the difference between diffusion equation and SP7 approximation regarding the MC-based statistical noise, especially when the relative errors were below 15%. The distinct difference between them can be observed from Figure 6(c) and Figure 7(c) though SP7 approximation becomes less accurate compared with MC method for depicting photon propagation. Because of the effect of the anisotropy factor, the performance of SP7 approximation is further deteriorated, as shown in Figure 8(c). It is necessary to use the higher order approximation for heterogeneous phantom especially regarding the weak measured signals on the surface.
Figure 6.
Comparison between diffusion equation, Monte Carlo method and SPN approximations in the heterogeneous phantom when g = 0.0 and nb = 1.0. μs and μa of the big cylindrical component of the phantom are fixed to 0.5mm−1 and 0.005mm−1 respectively. To the small cylindrical part, the ratios of (a), (b) and (c) are 50.0, 5.0 and 2.0 respectively. MC/SPi (i = 1, 3, 7) denote the ratios and “SPj to SP7” (j = 1, 3) are the relative errors.
Figure 7.
Comparison between diffusion equation, Monte Carlo method and SPN approximations in the heterogeneous phantom when g = 0.0 and nb = 1.37. μs and μa of the big cylindrical component of the phantom are fixed to 0.5mm−1 and 0.005mm−1 respectively. To the small cylindrical part, the ratios of (a), (b) and (c) are 50.0, 5.0 and 2.0 respectively. MC/SPi (i = 1, 3, 7) denote the ratios and “SPj to SP7” (j = 1, 3) are the relative errors.
Figure 8.
Comparison between diffusion equation, Monte Carlo method and SPN approximations in the heterogeneous phantom when g = 0.9 and nb = 1.37. μs and μa of the big cylindrical component of the phantom are fixed to 5.0mm−1 and 0.005mm−1 respectively. To the small cylindrical part, the ratios of (a), (b) and (c) are 50.0, 5.0 and 2.0 respectively. MC/SPi (i = 1, 3, 7) denote the ratios and “SPj to SP7” (j = 1, 3) are the relative errors.
3.3. Digital Mouse Cases
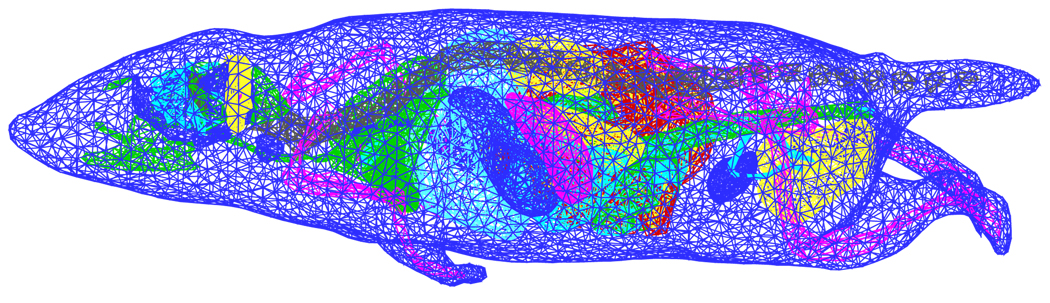
The whole-body mouse is the final test object of the proposed algorithm. To realize numerical simulation, a microMRI-basedmouse volume (MOBY) was applied. Furthermore, a commercial software Amira 3.0 (Mercury Computer Systems, Inc. Chelmsford, MA) was used to obtain the discretized mesh of MOBY. In the mesh generation, the simplified surface mesh was first obtained from the segmented mouse images. In order to retain the shape of organs, the surface mesh had 18708 points and 39007 triangles and a maximal element size of 1.5mm. Then, we obtained the volumetric mesh as coarse as possible. The final mesh initializing the simulation only had 21944 points. Almost all of the organs were depicted in the discretized mesh shown in Figure 9. Optical properties of organs were obtained by the formulas in [16].
Figure 9.
The discretized initial mesh of the MOBY mouse used in the proposed algorithm. Different mesh colors denote different organs.
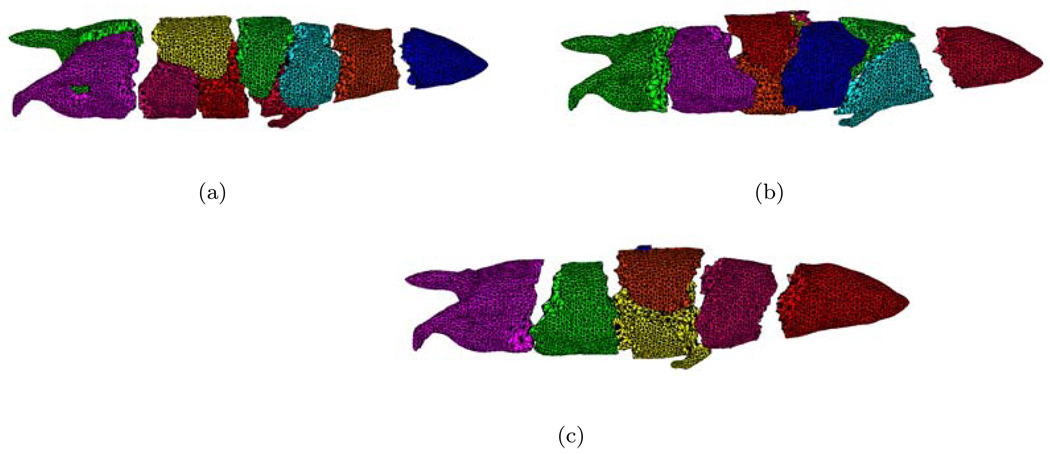
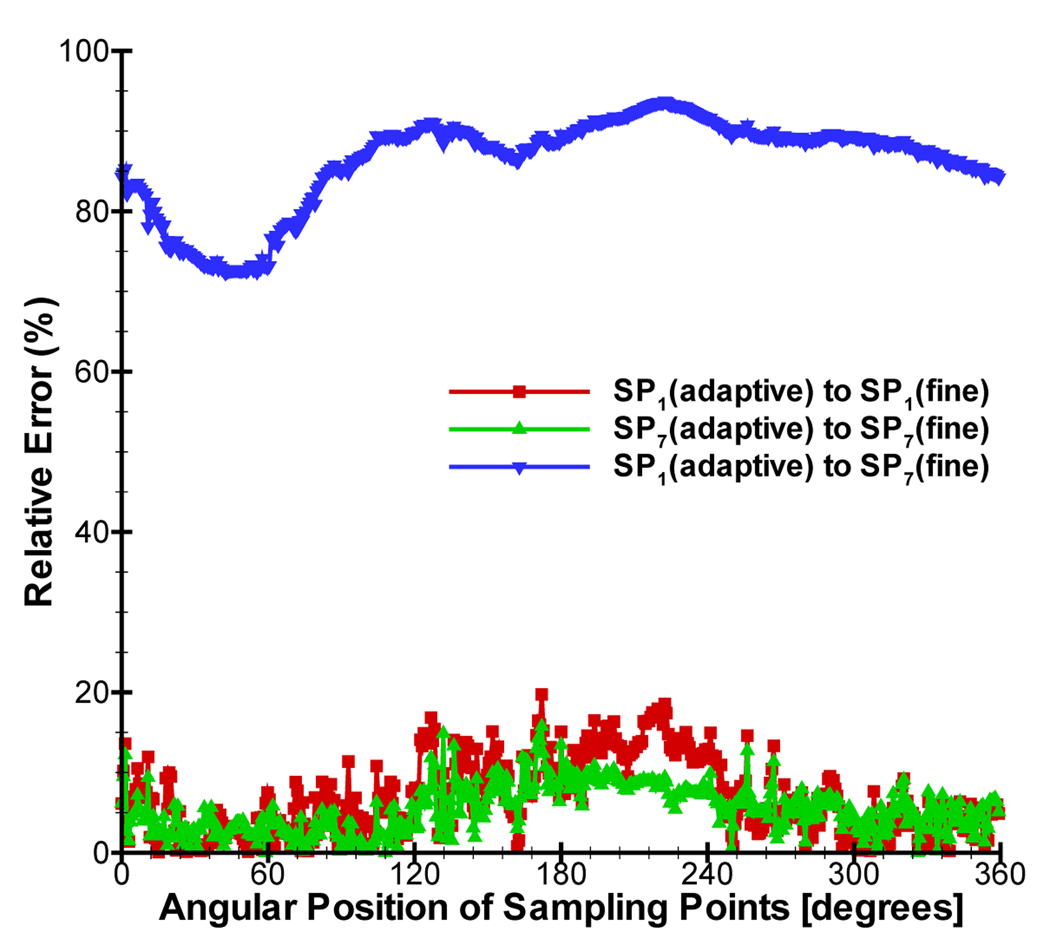
In the mesh generation, surface mesh simplification is liable to remove the small mesh domains. In order to preserve the shape of sources as far as possible, two cubic sources with 2.5mm width were set in the left kidney and liver. The optical properties of organs at 700nm were used for simulation. Figure 10 shows the mesh distribution using dynamic mesh repartitioning when 10 CPUs are used and five adaptive mesh refinements are performed. Based on a posteriori error estimation, the elements refined were centered on the vicinity of sources. Comparing Figure 10(a) with Figure 10(c), the parallel performance of the simulation will be significantly affected if only initial mesh partitioning is used in the whole simulation. The final meshes of SP1 and SP7 approximations had 49695 and 52047 points respectively. When 40 CPUs were used, the cost time of SP1 and SP7 was 275.83s and 698.13s only using CG method. Note that SP1 only required 1176 iterations after the final mesh refinement, whereas SP7 required 4210 iterations because of the ill-conditioned characteristic of the matrix. Therefore it is necessary to find good solvers for the high order SPN approximation. It is difficult to use the same geometries in the Monte Carlo method, therefore we didn’t obtain the simulation results based on the MC method. For comparison, a fine mesh of MOBY mouse was generated for obtaining the standard solution. The mesh had the average element size of 0.7mm and includes 188343 nodes and 2052092 tetrahedral elements. The solutions based on diffusion equation and SP7 were further obtained on the fine mesh. The relative errors of J+ on the adaptive refined and fine meshes were calculated by . Although the discretized points number of the adaptive refined mesh is about 1/4 of that of the fine mesh, the average relative errors of SP1(adaptive)-to-SP1(fine) and SP7(adaptive)-to-SP7(fine) only were 6.74% and 5.3%, showing the superiority of the adaptive strategy. However, the average relative errors of SP1(adaptive)-to-SP7(fine) reached 86.3%, indicating the necessity to use high order approximation in optical molecular imaging.
Figure 10.
Dynamic mesh repartitioning. (a) is the initial mesh partitioning. (b) and (c) denotes the third and fifth mesh partitionings.
3.3.1. Preconditioner Selection
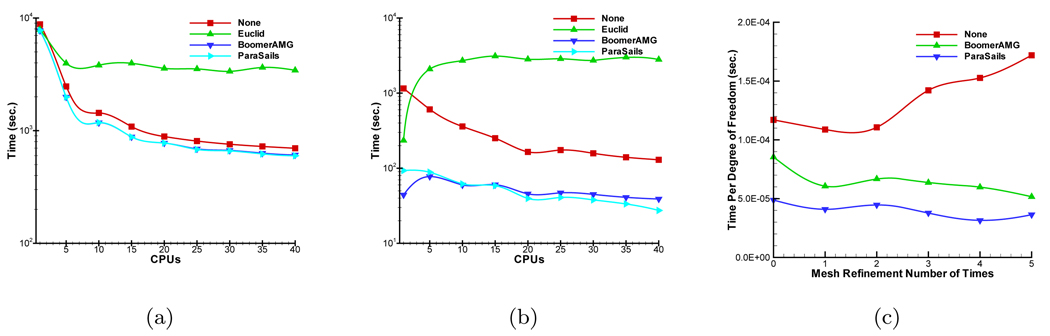
To improve the simulation speed, we anticipate to get the best preconditioner for the proposed algorithm by comparing current popular preconditioners. Hypre developed by Lawrence Livermore National Laboratory (LLNL) was used to help us select suitable preconditioner [41]. Hypre can provide several parallel preconditioners for realizing scalable computation, mainly including BoomerAMG [26](parallel algebraic multigrid), ParaSails [30](parallel sparse approximate inverse precondintioner), and Euclid [29](parallel ILU preconditioner). In this context, we tested multiple groups of parameters settings in preconditioners to obtain the best performance. For SP7 approximation, Figure 12(a) shows the comparison of total simulation time. BoomerAMG and ParaSails have almost similar performance. The parallel ILU preconditioner takes a longer time than only using CG method, which is not suitable for the proposed simulation. Furthermore, Figure 12(b) shows the running time of solver. When CPU number is less than 10, BoomerAMG takes shorter time than ParaSails. However, as the CPU number increases, ParaSails shows its superiority. In order to check the scalable characteristic of preconditioners, the cost time per degree of freedom is shown in Figure 12(c) when using 40 CPUs. The cost time of BoomerAMG and ParaSails is almost invariable with the increase of the matrix dimension, which shows that both of BoomerAMG and ParaSails are scalable. However, ParaSails takes less time. Therefore, Parasails is better selection when parallel simulation is performed in the proposed method.
Figure 12.
Comparison of time depending on CPU number between using different preconditioners. “None” denotes that only CG method is used in solver. (a) and (b) are the time of total simulation and solver. (c) denotes the cost time per degree of freedom depending on the mesh refinement number of times.
3.3.2. Time-costing Analysis
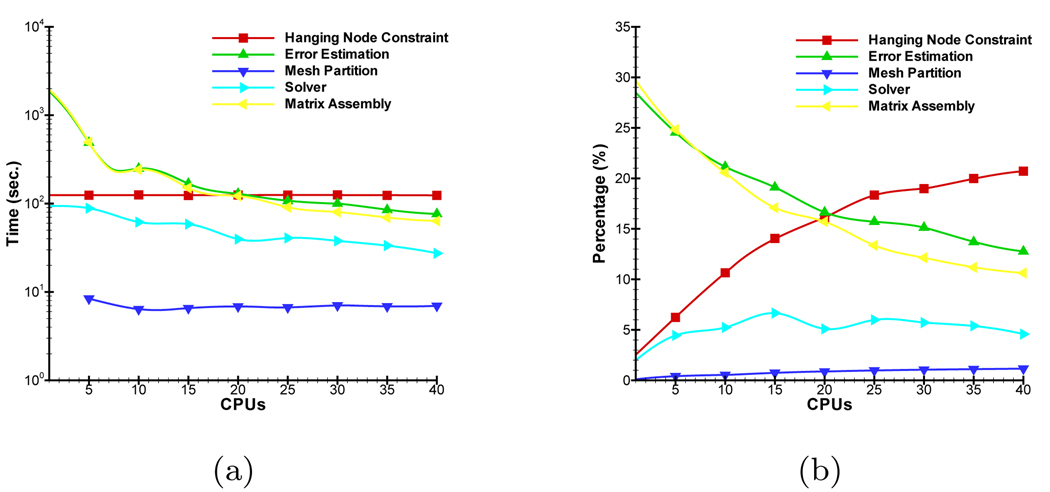
The proposed parallel adaptive element algorithm may be divided into five main modules, that is Matrix Assembly, Solver, Error Estimation, Hanging Node Constraint and Mesh Partition. Their cost time has a significant impact on the simulation performance. Figure 13 shows the change of their cost time and percentages of total simulation time depending on CPU number. The optimal selection of preconditioner reduces the time percentage of module Solver. Due to the use of the domain parallel and local a posteriori error estimator, the time of modules Matrix Assembly and Error Estimation is significantly reduced with the increase of CPU number. When a higher order approximation is used, there are much more variables at the discretized points of the mesh and this improvement becomes more significant. Module Mesh Partition realizes dynamic repartitioning, the time of which is almost negligible though it successfully tackles loading imbalance. Note that the time of Module Hanging Node Constraint is independent on CPU number. It is important to improve the parallel performance of this module in future research.
Figure 13.
Relationship between the cost time of different main modules and CPU number. (a) and (b) denote their absolute time and percentages of total time.
4. Conclusion
In this paper, we proposed a parallel adaptive finite element method to simulate photon propagation with SPN approximation. The combination of parallel mode, adaptive mesh evolution strategy and dynamic mesh repartitioning provides an elegant solution for the numerical simulation at the whole-body mouse level. Optimal solver selection improves numerical efficiency. The simulation results demonstrate that SP3 approximation is a good choice for the homogeneous domain especially with high absorption characteristics. Higher order approximation compared with SP7 is necessary for the heterogeneous high absorption domain. Time-costing analysis further shows the predominance of the proposed algorithm for such applications. Note that many approximations to RTE may be considered as a group of PDEs [10]. The proposed parallel adaptive algorithm is also a preferable framework for photon propagation simulation for these approximations. This work provides an efficient numerical simulation method and leads to high-performance significant investigations for optical molecular imaging.
Figure 1.
The ratio of main organs in a mouse. and μa are obtained from [16]. A small figure with different scale from the big figure is provided for better showing the data.
Figure 11.
Comparison between diffusion equation and SP7 approximation in the MOBY mouse.
ACKNOWLEDGEMENTS
We would like to thank Dr. Jianlin Xia for his comments on the manuscript. We would also like to thank Dr. Alexander D. Klose for the significant discussion about SPN approximation. We gratefully thank the development teams of libMesh, PETSc and hypre for discussions. This work is supported by the NIBIB R01-EB001458, NIH/NCI 2U24 CA092865 cooperative agreement, and DE-FC02-02ER63520 grants.
REFERENCES
- 1.Weissleder R. Scaling down imaging: Molecular mapping of cancer in mice. Nature Reviews Cancer. 2002;2:11–18. doi: 10.1038/nrc701. [DOI] [PubMed] [Google Scholar]
- 2.Ntziachristos V, Ripoll J, Wang LV, Weisslder R. Looking and listening to light: the evolution of whole body photonic imaging. Nature Biotechnology. 2005 March;23(3):313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- 3.Cherry SR. In vivo molecular and genomic imaging: new challenges for imaging physics. Phys. Med. Biol. 2004;49:R13–R48. doi: 10.1088/0031-9155/49/3/r01. [DOI] [PubMed] [Google Scholar]
- 4.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes and development. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 5.Wang G, Hoffman EA, McLennan G, Wang LV, Suter M, Meinel JF. Development of the first bioluminescence ct scanner. Radiology. 2003;566:229. [Google Scholar]
- 6.Schulz RB, Ripoll J, Ntziachristos V. Experimental fluorescence tomography of tissues with noncontact measurements. IEEE Transactions on Medical Imaging. 2004;23:492–500. doi: 10.1109/TMI.2004.825633. [DOI] [PubMed] [Google Scholar]
- 7.Gibson AP, Hebden JC, Arridge SR. Recent advances in diffuse optical imaging. Physics in Medicine and Biology. 2005;50:R1–R43. doi: 10.1088/0031-9155/50/4/r01. [DOI] [PubMed] [Google Scholar]
- 8.Arridge SR. Optical tomography in medical imaging. Inverse problems. 1999;15:R41–R93. [Google Scholar]
- 9.Klose AD, Larsen EW. Light transport in biological tissue based on the simplified spherical harmonics equations. Journal of Computational Physics. 2006;220(1):441–470. [Google Scholar]
- 10.Lewis EE, Warren F, Miller J. Computational Methods of Neutron Transport. New York: John Wiley & Sons; 1984. [Google Scholar]
- 11.Rasmussen JC, Joshi A, Pan T, Wareing T, McGhee J, Sevick-Muraca EM. Radiative transport in fluorescence-enhanced frequency domain photon migration. Medical Physics. 2006;33(12):4685–4700. doi: 10.1118/1.2388572. [DOI] [PubMed] [Google Scholar]
- 12.Aydin ED, de Oliveira CRE, Goddard AJH. A comparison between transport and diffusion calculations using a finite element-spherical harmonics radiation transport method. Medical Physics. 2002;29(9):2013–2023. doi: 10.1118/1.1500404. [DOI] [PubMed] [Google Scholar]
- 13.Cong W, Cong A, Shen H, Liu Y, Wang G. Flux vector formulation for photon propagation in the biological tissue. Optics Letters. 2007;32(19):2837–2839. doi: 10.1364/ol.32.002837. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, Li Y, Jiang M. Uniqueness theorems in bioluminescence tomography. Medical Physics. 2004 August;31(8):2289–2299. doi: 10.1118/1.1766420. [DOI] [PubMed] [Google Scholar]
- 15.Cong W, Wang G, Kumar D, Liu Y, Jiang M, Wang LV, Hoffman EA, McLennan G, McCray PB, Zabner J, et al. Practical reconstruction method for bioluminescence tomography. Optics Express. 2005 August;13(18):6756–6771. doi: 10.1364/opex.13.006756. [DOI] [PubMed] [Google Scholar]
- 16.Alexandrakis G, Rannou FR, Chatziioannou AF. Tomographic bioluminescence imaging by use of a combined optical-pet (opet) system: a computer simulation feasibility study. Physics in Medicine and Biology. 2005;50:4225–4241. doi: 10.1088/0031-9155/50/17/021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhari AJ, Darvas F, Bading JR, Moats RA, Conti PS, Smith DJ, Cherry SR, Leahy RM. Hyperspectral and multispectral bioluminescence optical tomography for small animal imaging. Physics in Medicine and Biology. 2005;50:5421–5441. doi: 10.1088/0031-9155/50/23/001. [DOI] [PubMed] [Google Scholar]
- 18.Joshi A, Bangerth W, Sevick-Muraca EM. Adaptive finite element based tomography for fluorescence optical imaging in tissue. Optics Express. 2004;12(22):5402–5417. doi: 10.1364/opex.12.005402. [DOI] [PubMed] [Google Scholar]
- 19.Lv Y, Tian J, Cong W, Wang G, Yang W, Qin C, Xu M. Spectrally resolved bioluminescence tomography with adaptive finite element analysis: methodology and simulation. Physics in Medicine and Biology. 2007;52:4497–4512. doi: 10.1088/0031-9155/52/15/009. [DOI] [PubMed] [Google Scholar]
- 20.Joshi A, Bangerth W, Sevick-Muraca EM. Non-contact fluorescence optical tomography with scanning patterned illumination. Optics Express. 2006;14(14):6516–6534. doi: 10.1364/oe.14.006516. [DOI] [PubMed] [Google Scholar]
- 21.Vo-Dinh T. Biomedical Photonics Handbook. CRC Press; 2002. [Google Scholar]
- 22.Klose AD, Ntziachristos V, Hielscher AH. The inverse source problem based on the radiative transfer equation in optical molecular imaging. Journal of Computational Physics. 2005;202(1):323–345. [Google Scholar]
- 23.Ishimaru A. Wave propagation and scattering in random media. IEEE Press; 1997. [Google Scholar]
- 24.Haskell RC, Svaasand LO, Tsay TT, Feng TC, McAdams MS, Tromberg BJ. Boundary conditions for the diffusion equation in radiative transfer. Journal of the Optical Society of America A. 1994;11(10):2727. doi: 10.1364/josaa.11.002727. [DOI] [PubMed] [Google Scholar]
- 25.Saad Y. Iterative Methods for Sparse Linear Systems. Society for Industrial and Applied Mathematics; 2003. [Google Scholar]
- 26.Henson VE, Yang UM. Boomeramg: a parallel algebraic multigrid solver and preconditioner. Applied Numerical Mathematics. 2002;41(1):155–177. [Google Scholar]
- 27.Benzi M. Preconditioning techniques for large linear systems: a survey. Journal of Computational Physics. 2002;182(2):418–477. [Google Scholar]
- 28.Saad Y, van der Vorst HA. Iterative solution of linear systems in the 20th century. Journal of Computational and Applied Mathematics. 2000;123:1–33. [Google Scholar]
- 29.Hysom D, Pothen A. A scalable parallel algorithm for incomplete factor preconditioning. SIAM Journal on Scientific Computing. 2000;22(6):2194–2215. [Google Scholar]
- 30.Chow E. A priori sparsity patterns for parallel sparse approximate inverse preconditioners. SIAM Journal on Scientific Computing. 2000;21(5):1804–1822. [Google Scholar]
- 31.Smith BF, Bjørstad PE, Gropp WD. Domain decomposition: parallel multilevel methods for elliptic partial differential equations. Cambridge University Press; 1996. [Google Scholar]
- 32.Kelly DW, Gago JPDSR, Zienkiewicz OC, Babuska I. A posteriori error analysis and adaptive processes in the finite element method: Part i - error analysis. International Journal for Numerical Methods in Engineering. 1983;19(11):1593–1619. [Google Scholar]
- 33.Kirk BS. PhD Thesis. The University of Texas at Austin; 2007. Adaptive finite element simulation of flow and transport applications on parallel computers. [Google Scholar]
- 34.Carey GF. Computational Grids : Generation, Adaptation, and Solution Strategies. Taylor & Francis; 1997. [Google Scholar]
- 35.Bey J. Tetrahedral grid refinement. Computing. 1995;55:355–378. [Google Scholar]
- 36.Karypis G, Kumar V. Multilevel k-way partitioning scheme for irregular graphs. Journal of Parallel and Distributed Computing. 1998;48(1):96–129. [Google Scholar]
- 37.Kirk B, Peterson JW, Stogner RH, Carey GF. libMesh: A C++ Library for Parallel Adaptive Mesh Refinement/Coarsening Simulations. Engineering with Computers. 2006;22:237–254. [Google Scholar]
- 38.Balay S, Buschelman K, Gropp WD, Kaushik D, Knepley MG, McInnes LC, Smith BF, Zhang H. PETSc Web page. 2001 Http://www.mcs.anl.gov/petsc.
- 39.Segars WP, Tsui BMW, Frey EC, Johnson GA, Berr SS. Development of a 4d digital mouse phantom for molecular imaging research. Molecular Imaging & Biology. 2004;6:149–159. doi: 10.1016/j.mibio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Jacques SL, Zheng L. Mcml - monte carlo modeling of photon transport in multi-layered tissues. Computer Methods and Programs in Biomedicine. 1995;47:131–146. doi: 10.1016/0169-2607(95)01640-f. [DOI] [PubMed] [Google Scholar]
- 41.Falgout RD, Yang UM. hypre: A library of high performance preconditioners. ICCS ’02: Proceedings of the International Conference on Computational Science-Part III; 2002. pp. 632–641. [Google Scholar]