Abstract
Phycocyanobilin:ferredoxin oxidoreductase (PcyA) is the best characterized member of the ferredoxin-dependent bilin reductase family. Unlike other ferredoxin-dependent bilin reductases that catalyze a two-electron reduction, PcyA sequentially reduces D-ring (exo) and A-ring (endo) vinyl groups of biliverdin IXα (BV) to yield phycocyanobilin, a key pigment precursor of the light-harvesting antennae complexes of red algae, cyanobacteria, and cryptophytes. To address the structural basis for the reduction regiospecificity of PcyA, we report new high resolution crystal structures of bilin substrate complexes of PcyA from Synechocystis sp. PCC6803, all of which lack exo-vinyl reduction activity. These include the BV complex of the E76Q mutant as well as substrate-bound complexes of wild-type PcyA with the reaction intermediate 181,182-dihydrobiliverdin IXα (18EtBV) and with biliverdin XIIIα (BV13), a synthetic substrate that lacks an exo-vinyl group. Although the overall folds and the binding sites of the U-shaped substrates of all three complexes were similar with wild-type PcyA-BV, the orientation of the Glu-76 side chain, which was in close contact with the exo-vinyl group in PcyA-BV, was rotated away from the bilin D-ring. The local structures around the A-rings in the three complexes, which all retain the ability to reduce the A-ring of their bound pigments, were nearly identical with that of wild-type PcyA-BV. Consistent with the proposed proton-donating role of the carboxylic acid side chain of Glu-76 for exo-vinyl reduction, these structures reveal new insight into the reduction regiospecificity of PcyA.
Introduction
Bilins are linear tetrapyrrole pigments that are utilized for light sensing and/or light harvesting in plants, red algae, cyanobacteria, and cryptophytes (1–3). The red/far red light-sensitive phytochromes are bilin-containing photoreceptors involved in plant photomorphogenesis, photoperiodic induction of flowering, chloroplast development, and leaf abscission (4, 5). Photoisomerization of the phytochromobilin (PΦB)2 of phytochrome chromophore triggers these physiological reactions (6). In oxygenic photosynthetic organisms including red algae, cyanobacteria, and cryptophytes, bilins comprise the major pigment components of thylakoid-associated antennae complexes known as phycobilisomes (7, 8).
Bilin biosynthesis is initiated by the oxygen-dependent cleavage of the porphyrin ring of heme catalyzed by ferredoxin-dependent heme oxygenases (9–13). This conversion produces biliverdin IXα (BV) that is subsequently converted to a variety of bilins by ferredoxin-dependent bilin reductases (FDBRs) (14). Members of the FDBR family include PΦB synthase (HY2), phycocyanobilin:ferredoxin oxidoreductase (PcyA), phycoerythrobilin:ferredoxin oxidoreductase (PebA), 15,16-dihydrobiliverdin:ferredoxin oxidoreductase (PebB), and phycoerythrobilin synthase (PebS), each of which reduces distinct double bonds of their bilin substrates (14–16).
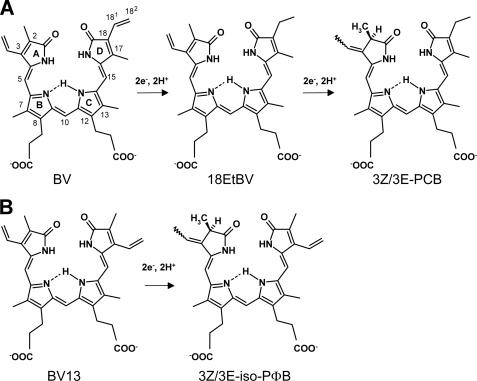
Previous biochemical studies have established that FDBRs contain neither organic nor metal cofactors and have revealed that these ferredoxin-dependent oxidoreductases yield bilin radical intermediates during catalysis (17). PcyA is the best characterized FDBR that catalyzes an overall four-electron reduction of BV to yield 3Z/3E-phycocyanobilin (PCB) (Fig. 1A), the major pigment in most phycobilisomes (18). Via sequential reduction of the D-ring exo-vinyl group of BV to generate 181,182-dihydrobiliverdin IXα (18EtBV) followed by reduction of the A-ring endo-vinyl group, PcyA rigorously controls the regiospecificity of BV vinyl group reduction. All FDBRs share ferredoxin (Fd) as an electron donor; they show, however, only weak sequence homology that likely reflects their ability to recognize widely divergent bilin substrates. Recent studies show that red chlorophyll catabolite reductase, an FDBR-related enzyme involved in chlorophyll catabolism (19), also adopts a characteristic three-layer α/β/α sandwich fold of the FDBR family (20). In view of their unusual chemistry and their distinct substrate and product specificities, a mechanistic understanding of the FDBR family is of fundamental interest in the design and control of these radical catalysts.
FIGURE 1.
Reactions catalyzed by PcyA. A, PcyA reduces BV to 3Z/3E-PCB via an intermediate of 18EtBV in two steps. Two electrons per each step are supplied from ferredoxin. B, when BV13, an isomer of BV, is used as the substrate, PcyA reduces it to 3Z/3E-iso-PΦB using two electrons. The structural formulae of the bilins are given according to the reference (23).
The structural basis for FDBR reduction regiospecificity has been particularly aided by the availability of high resolution crystal structures of PcyA from Synechocystis sp. PCC6803 and Nostoc sp. PCC7120 (21–23) and by biochemical analysis of site-directed mutants (23, 24). Based on the crystal structure of substrate-bound Synechocystis PcyA (21), we earlier proposed that the conserved Asp-105 side chain could pivot during the catalytic cycle to deliver protons to reduced bilin intermediates. The close apposition of the carboxylic acid side chain of Glu-76 to the exo-vinyl group of BV also provided compelling evidence for its participation in D-ring reduction (21). The latter hypothesis has received strong support from mutagenesis experiments in which mutations in Glu-76 alter the BV vinyl reduction regiospecificity of PcyA (23).
Tu et al. (23) proposed an alternate catalytic cycle for PcyA based on the crystal structure of the substrate-free enzyme from Nostoc sp. PCC7120, a mechanism in which the conserved aspartic residue (Asp-105 in Synechocystis PcyA) does not move during catalysis. Biochemical data from the same group revealed that PcyA can only reduce a single vinyl group of biliverdin XIIIα (BV13), a symmetric BV analog that lacks an exo-vinyl group (Fig. 1B) (17, 18). More recent EPR spectroscopy and density-functional theory computations on the D105N mutant of PcyA revealed potential protonation states of the semireduced BV radical intermediate (25), which, in turn, indirectly implicated the role of proton donors within the active site.
To better understand the catalytic mechanism of cyanobacterial PcyAs, we have determined the crystal structures of 18EtBV- and BV13-bound substrate complexes of Synechocystis PcyA at 1.48 and 1.04 Å resolutions, respectively. In addition, we have determined 1.40 Å resolution structure of the BV substrate-bound complex of the E76Q mutant of Synechocystis PcyA. Our studies not only provide strong support for the catalytic role of Glu-76 for exo-vinyl reduction by PcyA but also shed insight into the protonation state of the bilin substrate within the enzyme that likely specifies the reduction regiospecificity of PcyA. Mechanistic implications for the FDBR family of enzymes in general are also considered in light of these results.
EXPERIMENTAL PROCEDURES
Preparation of 18EtBV and BV13
The synthesis of 18EtBV was performed according to the protocols published for PΦB, PCB, and PCB derivatives. UV-visible (MeOH) λmax = 373 (ϵ = 44,760 m−1 cm−1), 653 (15,380) nm (26–28). BV13 was prepared by oxidizing bilirubin XIIIα with dichlorodicyanoquinone according to McDonagh and Palma (29). Bilirubin XIIIα was prepared by acid scrambling of bilirubin IXα as described (30). BV13 concentrations were determined optically using a molar absorption coefficient of 66,000 m−1 cm−1 (λmax = 370) following dilution of a DMSO stock solution with 5% (v/v) HCl in methanol (31).
Preparation of Ferredoxin and Ferredoxin:NADP+ Oxidoreductase for Enzymatic Assay
The gene for Synechocystis sp. PCC6803 Fd was amplified by PCR with the primers 5′- CATATGGCATCCTATACCGTTAAA-3′ and 5′-GGATCCTTAGTAGAGGTCTTCTTCTTTG-3′ (the underlined sequences indicate the NdeI and BamHI sites, respectively). The PCR product was ligated into pCR2.1-TOPO vector by using a TA cloning kit (Invitrogen), creating the plasmid pCR-fd. Sequence analysis verified that the construct was error-free. The fd gene was excised by digestion with NdeI and BamHI (New England Biolabs) and ligated into the NdeI/BamHI sites of pET-21a vector (Novagen). Escherichia coli strains C41(DE3) (32) were transformed with pET-21a-fd, and the gene was co-expressed with the isc operon to produce holo-Fd in high yield (33, 34). After sonication of the cells in 50 mm Tris-HCl (pH 8.0) and 2 mm EDTA, soluble proteins were precipitated with 80% saturated ammonium sulfate. The resulting supernatant protein fraction was then separated by a HiPrep 16/10 butyl FF column chromatography (GE Healthcare Bio-Sciences). Fd was further purified by a HiLoad 26/10 Q-Sepharose HP column chromatography and a HiPrep 16/60 Sephacryl S-100 HR column chromatography (GE Healthcare Bio-Sciences). Tris-HCl buffer (50 mm, pH 8.0) was used in the purification procedures. The gene for Synechocystis sp. PCC6803 Fd:NADP+ oxidoreductase (FNR) was amplified by PCR with the primers 5′-CATATGTACAGTCCCGGTTACG-3′ and 5′-GGATCCGAGCTCAGTAGGTTTCCACGTGCC-3′ (the underlined sequences indicate the NdeI and BamHI sites, respectively). The fnr gene was inserted into pET-15b vector (Novagen) according to the procedure to prepare pET-21a-fd described earlier. The N-terminal His6-tagged FNR was expressed in E. coli C41(DE3) cells (32) and purified using nickel-nitrilotriacetic acid affinity resin (Sigma) according to the manufacturer's protocol. After the protein was eluted from the resin, His6-FNR was further purified by gel filtration using a HiPrep 16/60 Sephacryl S-200 HR column (GE Healthcare Bio-Sciences).
Preparation of E76Q Mutant Protein of Synechocystis PcyA
Site-directed mutagenesis of Synechocystis PcyA was performed using the QuikChange site-directed mutagenesis kit (Stratagene) with plasmid pET21a-pcyA as the template (21). Two oligonucleotides (5′-CGTTTTACGTAAACGTCAACCGGTTCCACCCG-3′ and 5′-GCCCACCTTGGCCAACTGCAAATGCATTTTGC-3′) were used to introduce the E76Q mutation into pcyA. Sequence analysis verified that the construct was free of errors. The E76Q mutant protein was expressed and purified according to the procedures described for the wild-type PcyA (21).
Enzymatic Assay of the E76Q Mutant Protein
Assays for BV reduction activity were performed as described earlier (18) with the following modifications. The assay solution, which was prepared in an anaerobic chamber, contained 3 μm E76Q mutant protein, 5 μm Fd, 15 nm FNR, and 10 μm BV in 25 mm TES-KOH buffer (pH 8.5). The reaction was initiated in a quartz cuvette by adding 100 μm NADPH (final concentration) to this solution on a laboratory bench, where the air in the cuvette was substituted by 1,1-difluoroethane. The reaction mixture was incubated at 301 K in a capped cuvette, and the absorption spectra were monitored using a SHIMADZU UV-3101PC spectrophotometer.
Crystallization and Data Collection
18EtBV- and BV13-bound forms of PcyA were prepared by adding bilin from a 1 mm stock in DMSO to the purified enzyme on ice. Crystallization conditions for each complex were screened by the hanging-drop vapor-diffusion method using WIZARD I and WIZARD II kits (Jena Bioscience) at 293 K. The solution of each complex (11.5 mg/ml) was mixed with equal volumes of each reservoir solution and equilibrated, where the setups were shielded from light with aluminum foil to avoid photochemical reactions. For both complexes, blue crystals appeared in a few days under several conditions. The optimized conditions included reservoir solutions containing 0.85 m sodium citrate, 0.1 m sodium cacodylate at pH 7.0 for the PcyA-18EtBV complex and the solution containing 2.0 m ammonium sulfate, 0.2 m NaCl, and 0.1 m sodium cacodylate at pH 7.0 for the PcyA-BV13 complex. Preparation and crystallization of the E76Q mutant protein in complex with BV were also performed in a similar way using the reservoir solution containing 1.7 m ammonium sulfate, 2% polyethylene glycol 400, and 0.1 m HEPES (pH 7.0). The crystals were soaked in the crystallization solution supplemented with 15% (v/v) glycerol as a cryo-protectant and flash-cooled under a nitrogen gas stream to 100 K. Diffraction data for both complexes were collected at 100 K using synchrotron radiation and the Quantum 315 detector (Area Detector Systems Corp.) in the BL41XU beamline at SPring-8 (Harima, Japan). The hatch was kept in the dark during the data collection. Diffraction data from E76Q-BV crystal were collected using a Rigaku/MSC Jupiter 210 detector in the BL38B1 beamline at SPring-8. All diffraction data were processed, merged, and scaled with HKL2000 (35). Crystallographic statistics are summarized in Table 1.
TABLE 1.
Diffraction and refinement statistics
| PcyA-18EtBV | PcyA-BV13 | E76Q-BV | |
|---|---|---|---|
| Crystallographic data | |||
| Space group | P21212 | P21212 | P21212 |
| Unit cell dimensions, Å | |||
| a | 70.82 | 70.79 | 70.68 |
| b | 95.09 | 94.80 | 95.70 |
| c | 42.78 | 42.65 | 42.73 |
| Diffraction statisticsa | |||
| Wavelength (Å) | 1.0000 | 0.7085 | 1.0000 |
| Resolution (Å) | 1.48 | 1.04 | 1.40 |
| No. of observations | 160,844 | 801,164 | 384,326 |
| No. of unique reflections | 46,254 | 134,142 | 57,594 |
| Completeness (%) | 95.3 (97.9) | 99.4 (98.9) | 99.4 (98.4) |
| Mean Io/σ (I) | 14.2 | 13.9 | 23.8 |
| Rsymb (%) | 6.6 (30.7) | 4.8 (27.9) | 5.5 (29.5) |
| Refinement statistics | |||
| Rc/Rfreed (%) | 16.4/19.7 | 12.5/14.8 | 17.5/19.2 |
| No. of sulfate ion/water molecules | 0/304 | 1/375 | 0/334 |
| Root mean square deviations from ideal value | |||
| Bond length (Å) | 0.014 | 0.027 | 0.008 |
| Bond angle (°) | 1.47 | 2.75 | 1.22 |
| Ramachandran plot | |||
| Most favored (%) | 95.2 | 93.3 | 93.8 |
| Additionally allowed (%) | 4.3 | 6.2 | 5.7 |
| Generously allowed (%) | 0.5 | 0.5 | 0.5 |
a Values in parentheses correspond to the highest resolution shell (PcyA-18EtBV complex, 1.56–1.48 Å; PcyA-BV13 complex, 1.08–1.04 Å; E76Q-BV, 1.45–1.40 Å).
b Rsym = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl).
c R-factor = Σhkl‖Fo(hkl)| − |Fc(hkl)‖/Σhkl|Fo(hkl)|.
d Rfree is the R-factor calculated for 5% of the data not included in refinements.
Structure Determination and Crystallographic Refinement
Because the crystals of the PcyA-18EtBV, PcyA-BV13, and E76Q-BV complexes appeared to be isomorphous with the PcyA-BV crystal (21), the structures of the protein moiety of this complex (Protein Data Bank (PDB) entry 2D1E) in the respective unit cells were refined with REFMAC5 (36) using diffraction data to 2.0 Å resolution, and then difference Fourier maps were calculated. Each map showed significant electron density at the corresponding site where BV was bound in the PcyA-BV complex, to which the model of the bound bilin substrate was unambiguously fitted. Alternate cycles of the refinement of the coordinates and individual temperature factors with REFMAC5 and water-picking and manual adjustment of the model with Xfit (37) were performed, where the resolution was stepwise-extended. Multiple conformations for some side chains were also taken into account. The Nϵ and Oϵ atoms of the Gln-76 side chain of E76Q-BV were assigned on the basis of the individual temperature factors derived from the refinement.
The structure of the PcyA-BV13 complex was further refined using the conjugate gradient least-squares method with SHELX97 (38) and the diffraction data to 1.04 Å resolution, where anisotropic temperature factors were introduced for the non-hydrogen atoms. Electron density assignable to sulfate ion was found, which was subsequently included in the refinement with occupancy being fixed at 0.5. The Fo − Fc map showed positive densities at expected sites for some hydrogen atoms. The contribution of hydrogen atoms to the structure factors was included in the refinement at riding positions; the hydrogen atoms attached to the terminal carbon, nitrogen, and oxygen atoms in the side chains and those of water molecules were ignored. Restraint was imposed throughout the refinement on the geometries of the protein using the default values (39, 40) but not on those of BV13. Diffraction data and refinement statistics are summarized in Table 1.
RESULTS AND DISCUSSION
Crystallographic Analyses of PcyA-18EtBV, PcyA-BV13, and E76Q-BV Complexes
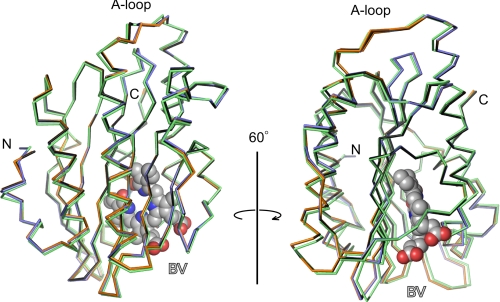
The crystal structures of 18EtBV- and BV13-bound forms of PcyA from Synechocystis sp. PCC6803 and the BV-bound form of the E76Q mutant were refined at 1.48, 1.04, and 1.40 Å resolutions, respectively; the final R (Rfree) values were 0.164 (0.197) for the PcyA-18EtBV complex, 0.125 (0.148) for the PcyA-BV13 complex, and 0.175 (0.192) for the E76Q-BV complex. Residues 1–5 in the PcyA-18EtBV and E76Q-BV complexes and residues 1–4 in the PcyA-BV13 complex could not be modeled due to their poor electron density. The overall structures of all three complexes proved similar to that of the PcyA-BV complex with root mean square distances between the corresponding Cα atoms being 0.27 Å for 242 residues for the PcyA-BV and PcyA-18EtBV pair, 0.20 Å for 243 residues for the PcyA-BV and PcyA-BV13 pair, and 0.21 Å for 243 residues for the PcyA-BV and E76Q-BV pair. Significant deviations of the main-chain atoms in the three complexes were only observed in several residues at the N and C termini and in the A-loop (Val-193–Leu-197) (Fig. 2). The A-loop of the PcyA-BV13 complex adopts two conformations, one of which is nearly identical to the conformation seen in the wild-type PcyA-BV complex, whereas the other is similar to that seen in the PcyA-18EtBV complex. These differences appear to reflect perturbation by the crystal-packing force because these residues are exposed to solvent and have few intramolecular interactions. All of these residues are located far from the substrate-binding site, and hence, these structural variations are not expected to affect the catalytic reactions of PcyA.
FIGURE 2.
Structural comparison of PcyA-BV, PcyA-18EtBV, PcyA-BV13, and E76Q-BV complexes. The Cα traces of PcyA-BV (black), PcyA-18EtBV (blue), PcyA-BV13 (orange), and E76Q-BV (green) are superimposed to minimize the root mean square deviation of the corresponding Cαs with CCP4mg (47, 48). For clarity, only BV in PcyA-BV is shown by the Corey, Pauling, Koltun model. The figure was prepared with PyMOL (49).
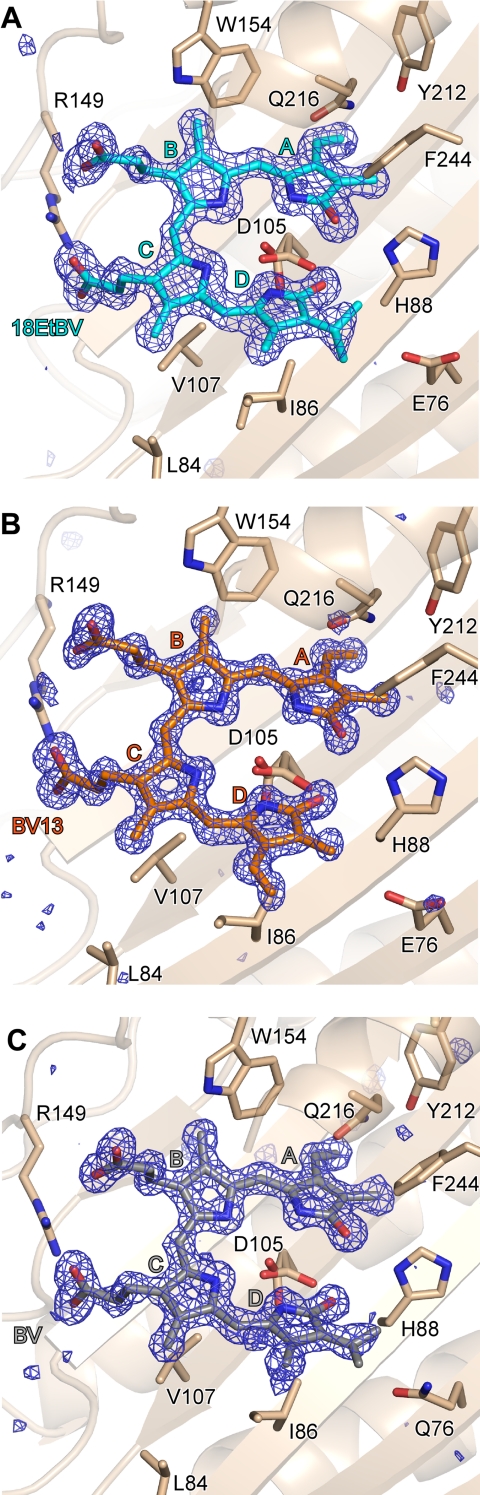
Electron density maps for 18EtBV, BV13, and BV were so clear that their orientations within the protein pocket and their conformations could be unambiguously determined (Fig. 3). All three pigments were strongly bound to PcyA or to its E76Q mutant protein, in contrast with BV bound to apo-myoglobin (41) or to heme oxygenase (42). The tetrapyrrole substrates 18EtBV, BV13, and BV adopted U-shaped conformations similar to that seen in PcyA-BV complex, and all were bound to the protein in a manner nearly indistinguishable from that in the wild-type PcyA-BV complex. Based on the structures of the PcyA-BV13 and E76Q-BV complexes, we can conclude that neither the exchange of the vinyl and methyl groups in the D-ring nor the substitution at Glu-76 by glutamine residue affected the mode of substrate binding to PcyA. The propionate groups of 18EtBV and BV13 also adopt similar conformations to those observed in the wild-type PcyA-BV complex.
FIGURE 3.
Electron density maps for the bound substrates. A, 18EtBV in PcyA at 1.48 Å; B, BV13 in PcyA at 1.04 Å; C, BV in the E76Q mutant protein at 1.40 Å. The Fo − Fc maps contoured at the 4 σ level are superposed on the stick models of the bound substrates. The figure was prepared with PyMOL (49).
Because of the high resolution, crystallographic refinement of PcyA-BV13 permitted glimpses of the lactam/lactim forms and/or the protonation states of the bilin substrate, although the assignment is complicated by the likely presence of multiple tautomeric states of the bound bilin substrate. The latter is implicated by the presence of a long wavelength absorbing shoulder at 724 nm and by the two conformers of Asp-105 seen in the PcyA-BV13 crystal structure. The Fo − Fc map showed positive peaks at the expected sites of the B-, C-, and D-rings of BV13, although the map had considerable noise (supplemental Fig. S1A). The assignment of hydrogen atoms to B-, C-, and D-ring nitrogen atoms is also supported by the C–N–C bond angles derived from a crystallographic refinement that imposed no restraint on the geometry of BV13 (supplemental Fig. S1B); the angles in rings that bear hydrogen atoms (B, 107.2°; C, 108.1°; and D, 107.4°) are smaller than the angles in rings that bear no hydrogen atom (A, 110.2°) (43). Moreover, in the A-ring, the C(1)–N length (1.40 Å) proved longer than the C(4)–N length (1.36 Å). The C–O bonds of the A- and D-rings were co-planar with the respective rings, and the C–O bond lengths were equally 1.27 Å, slightly longer than the typical C=O bond length (1.23 Å) but significantly shorter than the typical C–O bond length (1.43 Å). These data, together with the fact that the carboxyl acid group of Asp-105 for the major conformer adopts a bidentate hydrogen bond with the D-ring (Fig. 3), suggest that the bound BV13 possesses a bislactam structure. The possibility that the presence of multiple bilin tautomers and protonation states could be obscured by averaging leaves the question of substrate tautomeric state unresolved at present.
Interaction between Complexes and Ferredoxin
As evident from the close similarity of the crystal structures among the present three complexes with the PcyA-BV complex, the electrostatic potentials of their molecular surfaces are nearly identical to each other (supplemental Fig. S2). We reported previously that upon BV binding, PcyA changed its main-chain and side-chain conformations by which characteristic perturbation of the electrostatic potential occurred in such a way that positive charge on the surface was more clearly localized around the BV binding site of PcyA (22). The resulting basic patch should facilitate the interaction with acidic Fd for electron transfer. The similar electrostatic potentials of BV-bound Synechocystis PcyA and substrate-free Nostoc PcyA (supplemental Fig. S2) suggest that the localization of positive charge upon BV binding are likely to be shared by all PcyAs. The electrostatic potentials of the present complexes support the conclusion that the interaction between Fd and the substrate complexes are conserved for both reduction steps at least in Synechocystis PcyA.
Mechanistic Implications for the Reduction of the D-ring Exo-vinyl Group by PcyA
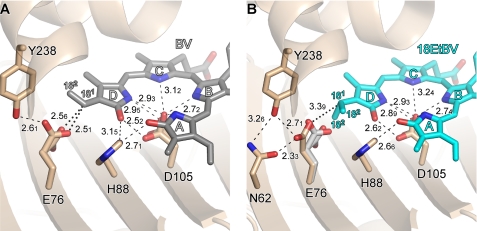
Biochemical studies have revealed that PcyA first catalyzes the reduction of the D-ring exo-vinyl group in BV to yield 18EtBV, the consequence of two one-electron transfers from reduced Fd (17, 18). Upon BV binding to PcyA, subsequent structural studies reveal that significant conformational changes occur around the binding pocket of the BV D-ring (21–23). Notable changes include the side chain of Asp-105, which strongly interacts with His-88 in the substrate-free form but adopts an apparent bidentate interaction with the BV D-ring, and the side chain of Glu-76, which interacts with Tyr-238 in the substrate-free form of PcyA but assumes an alternate conformation in the substrate-bound structure due to strong interaction with the D-ring vinyl group discussed below (Fig. 4A). By contrast, the side chain of Glu-76 in PcyA-18EtBV rotates away from the now reduced ethyl group of the 18EtBV intermediate D-ring to form a hydrogen bond with both Asn-62 and Tyr-238 (the distance between Oϵ(Glu-76) … C(181) is 3.39Å) (Fig. 4B). The side-chain conformation of Asp-105, however, remains unchanged upon vinyl group reduction. The ethyl group in the PcyA-18EtBV did adopt two orientations, both of which are different from that of the vinyl group in wild-type PcyA-BV. This difference may reflect the greater freedom of rotation of the 18EtBV ethyl group when compared with the BV exo-vinyl group whose geometry is biased to co-planarity by conjugation with the bilin π system.
FIGURE 4.
Close-up views around the tetrapyrrole d-rings. A, PcyA-BV complex. B, PcyA-18EtBV complex. Stick models are shown for the substrate and the side chains nearby the D-rings. In B, Glu-76 of PcyA-BV is superimposed in gray on PcyA-18EtBV to clarify the conformational change upon reduction. The side chain of Asn-62 is not shown in A because it does not participate in hydrogen bonding with Glu-76 in the PcyA-BV complex. The Glu-76 side chain in PcyA-BV (A) and the ethyl group of the 18EtBV D-ring in PcyA-18EtBV (B) each adopt two conformations. Hydrogen bonds are drawn with dashed lines, whereas notable interactions are shown with dotted lines. Numerals represent the interatomic distances in Å unit. Due to limited accuracy of the interatomic distances the second decimal numerals are shown in subscript.
These results implicate a direct role for Glu-76 in the reduction of the D-ring vinyl group. In this regard, it is noteworthy that the carboxyl group of Glu-76 is positioned quite close to the vinyl group in wild-type PcyA-BV. Although the side chain of Glu-76 adopts two conformations, the Oϵ(Glu-76) … C(181) distances in both conformers (2.56 and 2.51Å) are shorter than the sum of the van der Waals radii (Fig. 4A). This close interaction suggests that the BV exo-vinyl group and the Glu-76 carboxyl acid are strongly interacting, which is supported from the E76Q-BV structure (see below). One possible interpretation for this close apposition is that an OH-π-type hydrogen bond with the π-electrons of the vinyl group is responsible for this interaction (44). The close interaction between the carboxylic acid group of Glu-76 and the vinyl group was observed only for wild-type PcyA-BV, strongly indicating that the proton transfer to the vinyl group occurs directly from Glu-76 residue. This interpretation is in good agreement with the mechanisms previously proposed (21, 23), i.e. that exo-vinyl group reduction employs Asp-105 to donate a proton to the D-ring lactam oxygen atom, which ultimately rearranges to the C(181) position upon donation of a second proton to C(182) by Glu-76.
A crucial role for Glu-76 in reduction of the exo-vinyl group of BV was previously implicated by Tu et al. (23), who showed that the E73Q mutant of Nostoc PcyA (equivalent to the E76Q mutant of Synechocystis PcyA) yielded the two-electron reduced product 3Z/3E-PΦB. Hence, this mutant appears specifically impaired in exo-vinyl group reduction while retaining partial, i.e. ∼20% catalytic activity for endo-vinyl reduction. To extend these findings, we examined the biochemical properties of the E76Q mutant of Synechocystis PcyA. Fig. 5A depicts the spectrophotometric time course of the reduction of BV catalyzed by the E76Q mutant. These studies show that the UV-visible spectral changes are nearly identical with those reported for the corresponding E73Q mutant of Nostoc PcyA (23). The UV-visible spectrum of the product of BV reduction by the E76Q mutant was identical with that of PΦB reported (45) in the same solvent (data not shown). Thus, our results confirm that Glu-76 is required for exo-vinyl reduction by Synechocystis PcyA but not for endo-vinyl group reduction. Hence, the role of this glutamic acid appears conserved among PcyAs from different cyanobacterial species.
FIGURE 5.
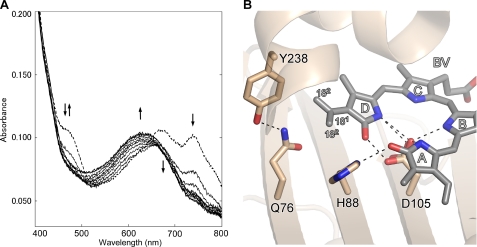
Biochemical reaction of E76Q mutant protein and the close-up view of BV in E76Q-BV complex. A, analysis of BV reduction by E76Q mutant protein from Synechocystis sp. PCC6803 PcyA. Absorption spectra were monitored for 10 min with a 1-min interval. The initial spectrum is shown by a dashed line. The double arrow indicates the increase and decrease of the absorbance at 468 nm. The single arrows indicate the absorbance increase at 630 nm and indicate the absorbance decreases at 670 and 740 nm. B, close-up view around the BV D-ring in the E76Q-PcyA complex. BV, Gln-76, His-88, Asp-105, and Tyr-238 are shown by stick models. The vinyl group of the BV D-ring in E76Q-BV adopted two conformations. Hydrogen bonds are drawn with dashed lines. The interatomic distance between C181 and Gln-76 Oϵ is 3.47Å.
A close-up view of the BV complex of the E76Q mutant shown in Fig. 5B clearly reveals that the side chain of Gln-76 forms a hydrogen bond with Tyr-238 (Nδ … Oη distance is 2.69Å), as seen in one of two conformers of Glu-76 of PcyA-BV. However, the distance between the carbonyl oxygen atom of Gln-76 and the D-ring vinyl group of BV in E76Q-BV complex (Oϵ(Gln-76) … C(18)1) is 3.47Å, which is markedly longer than the distance between Glu-76 and the D-ring vinyl group of BV (2.51Å) seen in the wild-type PcyA-BV complex, clearly showing that the Gln-76 side chain is not in interaction with the vinyl group of the BV D-ring. Interestingly, the vinyl group of the BV D-ring is allowed to adopt two conformations, one of which is identical to that in the wild-type PcyA-BV complex, whereas the other is flipped around the C18–C181 bond. The observation that only one conformation was seen in the D-ring vinyl group of BV for PcyA-BV, in turn, suggests that Glu-76 acts in trapping this vinyl group by an OH-π-type hydrogen bond. The crystal structure of E76Q-BV thus supports the interpretation that the poor activity of the E76Q mutant is due to the inability of Gln-76 to support proton transfer to the exo-vinyl group.
Mechanistic Implications for the Reduction of the A-ring Endo-vinyl Group by PcyA
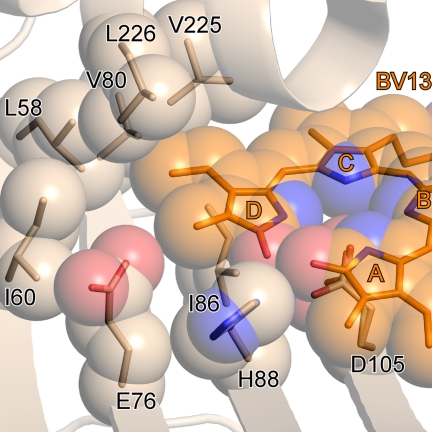
Previous biochemical analyses of the reduction of BV13, a symmetrical isomer of BV in which the D-ring vinyl and methyl groups are exchanged, shows that PcyA can mediate the conversion of BV13 to 3Z/3E-iso-PΦB (Fig. 1B) (17, 18). Structural analysis of the PcyA-BV13 complex shown here indicates that the vinyl group of BV13, which occupies the position corresponding to the D-ring methyl group in the natural BV substrate, is surrounded by only hydrophobic residues, i.e. Leu-58, Ile-60, Val-80, Ile-86, Val-225, and Leu-226 (Fig. 6). Hence, there is no good candidate proton-donating residue near the D-ring vinyl group of BV13 to mediate its reduction. By contrast, the conformation of Asp-105 and its apparent bidentate interaction with the D-ring pyrrole appear indistinguishable from those in PcyA-BV and PcyA-18EtBV. This argues that BV13 conversion to 3Z/3E-iso-PΦB by PcyA occurs by reduction of the endo-vinyl group positioned in the A-ring pocket of the enzyme, a conversion very similar to the reduction of the 18EtBV intermediate.
FIGURE 6.
Close-up view around the tetrapyrrole ring of BV13 in PcyA-BV13 complex. Corey, Pauling, Koltun models for BV13 and the side chains of the residues around the vinyl group are superimposed on the stick models.
As depicted in Fig. 3, Asp-105 in the PcyA-18EtBV complex adopts the same mixture of conformers seen in PcyA-BV, PcyA-BV13, and E76Q-BV complexes, a major conformer forming bidentate hydrogen bonds with the substrate D-ring and a minor conformer sporting a single hydrogen bond with the D-ring nitrogen. The similar dual conformer arrangement of Asp-105 in all three structures supports the conclusion that the Asp-105-bilin interaction is not altered after D-ring reduction (23).
Potential Role of His-88 in the Mechanism of Bilin Reduction by PcyA
Based on structural and biochemical data for Synechocystis PcyA-BV complex (21) and for the substrate-free Nostoc PcyA (24), the highly conserved His-88 residue has been strongly implicated as a key player in the reduction of the A-ring in which the second reduction step (the A-ring reduction) is initiated by proton transfer from His-88 to the oxygen atom of the A-ring of the bound 18EtBV (23). Consistent with this proposal, the His-88 Nδ atom is located close to the A-ring carbonyl oxygen in the 18EtBV complex (Figs. 3 and 4). However, the structure of residues near the A-ring in the 18EtBV complex does not reveal how stereospecific hydrogenation at the C2 atom is achieved.
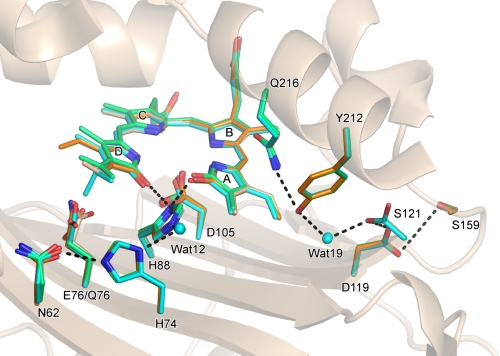
It has been proposed that the second proton for A-ring reduction is derived from the bound water molecule (wat19) (23). This hypothesis is reasonable because a water molecule is present in this position in the crystal structures of all PcyA-substrate complexes (Fig. 7). Moreover, the hydrogen bond network via a water molecule from the molecular surface to the active site was also conserved in the complexes, consistent with the hypothesis that a proton is supplied to the A-ring active site via such a hydrogen bond network.
FIGURE 7.
Hydrogen bond network from the molecular surface to the processing site in the A-ring. The structures of the PcyA-BV (white), PcyA-18EtBV (blue), PcyA-BV13 (orange), and E76Q-BV (green) complexes are superimposed. Bilins, Asn-62, His-74, Glu-76, Gln-76, His-88, Asp-105, Asp-119, Ser-121, Ser-159, Tyr-212, and Gln-216 are shown by stick models. The side chain of Ser-121 in PcyA-18EtBV adopted two conformations. Hydrogen bonds are drawn with heavy dotted lines. For clarity, the hydrogen bonds in which Asp-105 is involved are not shown. The network from Asn-62 to His-88 is conserved in all complexes, but the interactions between Glu-76 Oϵ/Gln-76 Nϵ and Asn-62 Oδ vary; hydrogen bond is present in PcyA-18EtBV (2.33Å) and PcyA-BV13 (2.66 Å) (dotted line is not given), whereas it is absent in PcyA-BV and PcyA-E76Q.
With regard to the issue of reduction regiospecificity, the crystal structure of the BV complex of PebS from cyanophage P-SSM2, another FDBR family enzyme, was recently reported (16). PebS catalyzes the Fd-dependent four-electron conversion of BV to phycoerythrobilin (PEB), an isomer of PCB, via the intermediate 15,16-dihydrobiliverdin. Although the first reduction site in BV by PebS is distinct from that by PcyA, both target the A-ring endo-vinyl group in the second reduction step. Although the structure around the A-ring in all four PebS-BV complexes present in the asymmetric unit is different from that seen in PcyA-BV, there is some similarity in the arrangement of amino acid residues and water molecules near the A-ring, with Asp-105 being conserved in all enzymes. Because His-88 found in all PcyAs is replaced with Asn in PebS, the exact roles and/or functional interchangeability of the residues that participate in the A-ring endo-vinyl reduction remain to be resolved. The ability of PebS to mediate the four-electron conversion of BV to PEB argue that the 15,16-dihydrobiliverdin intermediate remains strongly bound to the enzyme, which contrasts it with the PebA enzyme that must release this intermediate (14).
Molecular Mechanism of the Sequential Two-step Reductions Catalyzed by PcyA and Future Perspectives
PcyA is characteristic not only because PcyA is the only enzyme known so far in FDBR family that is able to reduce the D-ring vinyl group of tetrapyrroles (46) but also because PcyA reduces both vinyl groups of BV. Although individual mechanisms for the catalytic mechanisms of PcyA have been described on the basis of the crystal structures of various complexes, a major question that remains unanswered for PcyA, as for the other FDBR family enzyme of PebS, is how it selects and controls the order of the double bond targeted for reduction. This could reflect a kinetic effect or a thermodynamic effect, or both, and the presently available data do not resolve this issue. We speculate that the unusual interaction between Glu-76 and the D-ring vinyl group is responsible for its preference for reduction; however, the possibility that the proton-containing environment around the A-ring suppresses its reduction remains to be assessed. The switching mechanism by which the latent A-ring reduction activity is manifested following reduction of the D-ring vinyl group, if any, could reside in the change of the proton transfer pathway to His-88. In this regard, His-88 is involved in both the D-ring and the A-ring reductions, indicating that His-88 needs to be reprotonated after the D-ring reduction. Comparative analysis of PcyA complexes with BV and the intermediate 18EtBV suggests that after the reduction of the D-ring vinyl group, a very short contact (2.33Å) between Glu-76 Oϵ and Asn-62 Oδ appears in PcyA-18EtBV (Fig. 4B). This close contact suggests that these two atoms are linked by a low barrier hydrogen bond that may influence the redox chemistry at the A-ring because this hydrogen bond is close to the possible proton transfer pathway from the molecular surface to His-88 (Fig. 7). The mechanistic importance of this observation remains to be investigated. An understanding of the protonation states of the various residues that contribute to the primary and secondary proton transfer steps remains as an important target for future investigation. Neutron crystallography provides one approach to address these questions, an experimental approach that holds much promise for understanding the mechanism and reduction regiospecificity of the FDBR family.
Acknowledgments
We thank Drs. Seiki Baba, Masahide Kawamoto, and Nobutaka Shimizu (Japan Synchrotron Radiation Research Institute) for the aid with data collection using the synchrotron radiation at SPring-8 (Proposal Numbers 2007B1307 and 2008B1079), and we thank Prof. Masato Noguchi and Dr. Hideaki Sato (Kurume University School of Medicine) and Dr. Kei Wada (Osaka University) for advice and encouragement.
This work was supported in part by Grants-in-aid for Scientific Research 20370037 and 18570105 (to K. F.), by Grant-in-aid for Research Fellows of the Japan Society for the Promotion of Science 19·1065 (to Y. H.), and by United States National Science Foundation Grant MCB-0843625 (to Andrew J. Fisher and J. C. L.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 3I8U, 3I94, and 3I95) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PΦB
- phytochromobilin
- BV
- biliverdin IXα
- 18EtBV
- 181,182-dihydrobiliverdin IXα
- BV13
- biliverdin XIIIα
- Fd
- ferredoxin
- FDBR
- Fd-dependent bilin reductase
- FNR
- Fd:NADP+ oxidoreductase
- PcyA
- phycocyanobilin:Fd oxidoreductase
- PCB
- phycocyanobilin
- PEB
- phycoerythrobilin
- PebA
- phycoerythrobilin:ferredoxin oxidoreductase
- PebB
- 15,16-dihydrobiliverdin:ferredoxin oxidoreductase
- PebS
- phycoerythrobilin synthase
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid.
REFERENCES
- 1.Beale S. I. (1993) Chem. Rev. 93, 785–802 [Google Scholar]
- 2.Hughes J., Lamparter T. (1999) Plant Physiol. 121, 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frankenberg N., Lagarias J. C. (2003) in The Porphyrin Handbook (Kadish K. M., Smith K. M., Guilard R. eds) Vol. 13, pp. 211–235, Academic Press, New York [Google Scholar]
- 4.Franklin K. A., Larner V. S., Whitelam G. C. (2005) Int. J. Dev. Biol. 49, 653–664 [DOI] [PubMed] [Google Scholar]
- 5.Karniol B., Viestra R. D. (2005) in Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms (Schäfer E., Nagy F. eds) 3rd Ed., pp. 65–98, Springer, Dordrecht, The Netherlands [Google Scholar]
- 6.Rockwell N. C., Su Y. S., Lagarias J. C. (2006) Annu. Rev. Plant Biol. 57, 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossman A. R., Schaefer M. R., Chiang G. G., Collier J. L. (1993) Microbiol. Rev. 57, 725–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tandeau de Marsac N. (2003) Photosynth. Res. 76, 193–205 [DOI] [PubMed] [Google Scholar]
- 9.Cornejo J., Willows R. D., Beale S. I. (1998) Plant J. 15, 99–107 [DOI] [PubMed] [Google Scholar]
- 10.Migita C. T., Zhang X., Yoshida T. (2003) Eur. J. Biochem. 270, 687–698 [DOI] [PubMed] [Google Scholar]
- 11.Zhang X., Migita C. T., Sato M., Sasahara M., Yoshida T. (2005) FEBS J. 272, 1012–1022 [DOI] [PubMed] [Google Scholar]
- 12.Sugishima M., Migita C. T., Zhang X., Yoshida T., Fukuyama K. (2004) Eur. J. Biochem. 271, 4517–4525 [DOI] [PubMed] [Google Scholar]
- 13.Sugishima M., Hagiwara Y., Zhang X., Yoshida T., Migita C. T., Fukuyama K. (2005) Biochemistry 44, 4257–4266 [DOI] [PubMed] [Google Scholar]
- 14.Frankenberg N., Mukougawa K., Kohchi T., Lagarias J. C. (2001) Plant Cell 13, 965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dammeyer T., Bagby S. C., Sullivan M. B., Chisholm S. W., Frankenberg-Dinkel N. (2008) Curr. Biol. 18, 442–448 [DOI] [PubMed] [Google Scholar]
- 16.Dammeyer T., Hofmann E., Frankenberg-Dinkel N. (2008) J. Biol. Chem. 283, 27547–27554 [DOI] [PubMed] [Google Scholar]
- 17.Tu S. L., Gunn A., Toney M. D., Britt R. D., Lagarias J. C. (2004) J. Am. Chem. Soc. 126, 8682–8693 [DOI] [PubMed] [Google Scholar]
- 18.Frankenberg N., Lagarias J. C. (2003) J. Biol. Chem. 278, 9219–9226 [DOI] [PubMed] [Google Scholar]
- 19.Wüthrich K. L., Bovet L., Hunziker P. E., Donnison I. S., Hörtensteiner S. (2000) Plant J. 21, 189–198 [DOI] [PubMed] [Google Scholar]
- 20.Sugishima M., Kitamori Y., Noguchi M., Kohchi T., Fukuyama K. (2009) J. Mol. Biol. 389, 376–387 [DOI] [PubMed] [Google Scholar]
- 21.Hagiwara Y., Sugishima M., Takahashi Y., Fukuyama K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagiwara Y., Sugishima M., Takahashi Y., Fukuyama K. (2006) FEBS Lett. 580, 3823–3828 [DOI] [PubMed] [Google Scholar]
- 23.Tu S. L., Rockwell N. C., Lagarias J. C., Fisher A. J. (2007) Biochemistry 46, 1484–1494 [DOI] [PubMed] [Google Scholar]
- 24.Tu S. L., Sughrue W., Britt R. D., Lagarias J. C. (2006) J. Biol. Chem. 281, 3127–3136 [DOI] [PubMed] [Google Scholar]
- 25.Stoll S., Gunn A., Brynda M., Sughrue W., Kohler A. C., Ozarowski A., Fisher A. J., Lagarias J. C., Britt R. D. (2009) J. Am. Chem. Soc. 131, 1986–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kakiuchi T., Kinoshita H., Inomata K. (1999) Synlett SI, 901–904 [Google Scholar]
- 27.Kakiuchi T., Kato H., Jaysundera K. P., Higashi T., Watabe K., Sawamoto D., Kinoshita H., Inomata K. (1998) Chem. Lett. 27, 1001–1002 [Google Scholar]
- 28.Inomata K. (2008) Bull. Chem. Soc. Jpn. 81, 25–59 [Google Scholar]
- 29.McDonagh A. F., Palma L. A. (1980) Biochem. J. 189, 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma J. S., Lightner D. A. (1984) J. Heterocyclic Chem. 21, 1005–1008 [Google Scholar]
- 31.McDonagh A. F. (1979) in The Porphyrins (Dolphin D. ed) pp. 293–491, Academic Press, New York [Google Scholar]
- 32.Miroux B., Walker J. E. (1996) J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 33.Nakamura M., Saeki K., Takahashi Y. (1999) J. Biochem. 126, 10–18 [DOI] [PubMed] [Google Scholar]
- 34.Takahashi Y., Nakamura M. (1999) J. Biochem. 126, 917–926 [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 36.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 37.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 38.Sheldrick G. M., Schneider T. R. (1997) Methods Enzymol. 277, 319–343 [PubMed] [Google Scholar]
- 39.Kleywegt G. J., Jones T. A. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 1119–1131 [DOI] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D. 50, 760–76315299374 [Google Scholar]
- 41.Wagner U. G., Müller N., Schmitzberger W., Falk H., Kratky C. (1995) J. Mol. Biol. 247, 326–337 [DOI] [PubMed] [Google Scholar]
- 42.Sugishima M., Sakamoto H., Higashimoto Y., Noguchi M., Fukuyama K. (2003) J. Biol. Chem. 278, 32352–32358 [DOI] [PubMed] [Google Scholar]
- 43.Sheldrick W. S. (1976) J. Chem. Soc. Perkin Trans. 2, 1457–1462 [Google Scholar]
- 44.Afonin A. V., Baikalova L. V., Domnina E. S. (1996) Russ. Chem. Bull. 45, 1137–1141 [Google Scholar]
- 45.Terry M. J., McDowell M. T., Lagarias J. C. (1995) J. Biol. Chem. 270, 11111–11118 [DOI] [PubMed] [Google Scholar]
- 46.Dammeyer T., Frankenberg-Dinkel N. (2008) Photochem. Photobiol. Sci. 7, 1121–1130 [DOI] [PubMed] [Google Scholar]
- 47.Potterton E., McNicholas S., Krissinel E., Cowtan K., Noble M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1955–1957 [DOI] [PubMed] [Google Scholar]
- 48.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 49.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]