Abstract
Integrin αIIbβ3 is the major membrane protein and adhesion receptor at the surface of blood platelets, which after activation plays a key role in platelet plug formation in hemostasis and thrombosis. Small angle neutron scattering (SANS) and shape reconstruction algorithms allowed formation of a low resolution three-dimensional model of whole αIIbβ3 in Ca2+/detergent solutions. Model projections after 90° rotation along its long axis show an elongated and “arched” form (135°) not observed before and a “handgun” form. This 20-nm-long structure is well defined, despite αIIbβ3 multidomain nature and expected segmental flexibility, with the largest region at the top, followed by two narrower and smaller regions at the bottom. Docking of this SANS envelope into the high resolution structure of αIIbβ3, reconstructed from crystallographic and NMR data, shows that the solution structure is less constrained, allows tentative assignment of the disposition of the αIIb and β3 subunits and their domains within the model, and points out the structural analogies and differences of the SANS model with the crystallographic models of the recombinant ectodomains of αIIbβ3 and αVβ3 and with the cryo-electron microscopy model of whole αIIbβ3. The ectodomain is in the bent configuration at the top of the model, where αIIb and β3 occupy the concave and convex sides, respectively, at the arched projection, with their bent knees at its apex. It follows the narrower transmembrane region and the cytoplasmic domains at the bottom end. αIIbβ3 aggregated in Mn2+/detergent solutions, which impeded to get its SANS model.
Keywords: Cell/Blood, Cell/Adhesion, Membrane/Proteins, Methods/Neutron Scattering, Organisms/Human, Protein/Adhesion, Protein/Structure, Receptors/Membrane
Introduction
Blood platelets have distinct mechanisms to respond to the different agonists of platelet activation, all of which end up with the acquisition by the integrin αIIbβ3 or glycoprotein IIb/IIIa of the capacity to recognize and bind fibrinogen and other adhesive proteins. Binding of fibrinogen, the major adhesive protein in plasma, to activated αIIbβ3 and the binding of fibronectin, the major adhesive protein in the extracellular matrix of subendothelium, to resting or activated αIIbβ3 lead to the formation of interplatelet cross-linking and to platelet adhesion to the subendothelium, respectively, and, eventually, to the formation of the platelet plug. Thus, knowledge of the fibrinogen receptor and the mechanisms of induction and modulation of its activity is determinant to understand hemostasis and the pathophysiological paths leading to thrombosis, as well as to develop new ways to prevent, detect, and treat thrombosis effectively, securely, and selectively. After 20 years of molecular genetics and 10 years of high resolution structural biology of integrins, the disposition of an integrin in the membrane is unknown, the modifications in the cytoplasmic domains and the transmembrane segments related with the receptor activation are still not definitively defined, and the molecular mechanism of activation and molecular properties of the activated receptor are still under discussion (1–4). Structural and cell biological information reported recently suggests different models (the switchblade, the deadbolt, and the disulfide reduction models) (5–8) for the mechanism of signal transduction between the cytoplasmic domains and the extracellular ligand binding domains, as well as different activation states (9), which require both, to clear the disagreement between some experimental observations and to carry out more comprehensive experimental tests at different levels of structural and functional organization.
Previous x-ray crystallographic experiments showed that the recombinant ectodomains of integrin αVβ3 (r-αVβ3 ectodomain) and αIIbβ3 (r-αIIbβ3 ectodomain) and the complex of r-αVβ3 ectodomain with an RGD peptide are in the bent configuration (10–12). In addition, the soluble Mn2+-bound r-αVβ3 ectodomain, the complex of this ectodomain with a fibronectin fragment (with type III domains 7–10 and the extradomain B), and Mn2+-activated αIIbβ3 were equally in the bent configuration, as determined by electron microscopy (EM)2 (13, 14). Finally, αIIbβ3 in resting and physiologically activated platelets were also in the bent configuration, as found by epitope mapping and competition among monoclonal antibodies directed to the C-terminal region of the αIIb subunit and to the N-terminal domain of the β3 subunit (9) and by fluorescence intramolecular energy transfer (15). However, all of these results are inconsistent to some extent with the extended conformation found: in whole αIIbβ3 observed by cryo-EM (16); in different recombinant ectodomain products of αVβ3 in the presence of Mn2+ alone or Mn2+ with an RGD peptide or other ligand-mimetic antagonists, observed by EM (5); and in experiments of fluorescence intermolecular energy transfer between a fluorescent peptide bound to integrin α4β1, expressed at the surface of Mn2+ or chemokine-activated monoblastoid cells, and a fluorescent lipophilic probe incorporated into their membrane (17).
To observe the low resolution structure of native membrane proteins in detergent solution and their functional modifications, small angle neutron scattering (SANS) is an alternative technique to EM. SANS in combination with shape reconstruction algorithms allows us to obtain a low resolution three-dimensional model of the protein in detergent solution, if the scattering of the detergent micelles is contrast-matched (18). The main advantages over EM are that it does not require perturbation of the sample, such as drying, staining, or freezing, and the radiation damage is very low. In addition, the large difference between the scattering amplitudes of hydrogen and deuterium allows the use of D2O to obtain favorable contrast and background conditions; in our case, we have contrast-matched the detergent micelles by equilibration of the protein solutions with the detergent buffer at the required D2O concentration. The disadvantages are the high incoherent background from hydrogen in aqueous solutions, the relatively low incident neutron flux, the high concentration of homogeneous material required, and the limited beam time available. In the case of whole membrane proteins, the low resolution SANS models in solution are an alternative to the x-ray diffraction models, given that SANS does not require crystallization, a limiting step in membrane proteins. In addition, if SANS is combined with protein reconstitution in deuterate lipid vesicles, it could provide low resolution information on the structure and disposition of the protein sitting in the bilayer and on its structural modifications in different functional states.
In the present paper, we used human platelet αIIbβ3 in Triton X-100 solutions and SANS. The obtained low resolution model of the integrin has an average maximum length of 20 nm, and its projections after counterclockwise 90° rotations along its long axis show successively an elongated and “arched” form (135°) and a “handgun” form with a 45° bend and three well differentiated parts. At the top of the handgun projections, there is a large region with a maximum length of 14 nm and variable width and thickness that we assign to the ectodomain of the integrin, with particular features tentatively identified structurally. At the bottom, there is a small region of 3 × 2.5 × 2 nm on average, which we assign to the intracellular domains of the αIIb and β3 subunits, and connecting these two regions there is a cylinder of about 3 × 2 × 2 nm, which we assign to the transmembrane domains of the subunits. This model of the integrin is unequivocally consistent with the bent conformation observed before for the r-αVβ3 and r-αIIbβ3 ectodomains by x-ray crystallography (10, 12) and for the r-αVβ3 and r-αIIbβ3 ectodomains (5, 12, 13) by EM. In this way, we also demonstrate that SANS at the match point can be used to distinguish clearly between the bent and the contentious extended configuration of integrins in solution, to which the integrin resting and the activated states, respectively, have been assigned.
EXPERIMENTAL PROCEDURES
Preparation of Integrin αIIbβ3 from the Human Platelet Plasma Membrane for SANS Measurements
The integrin αIIbβ3 was prepared from the plasma membrane fraction of outdated human platelets as described elsewhere (19). The purified protein, free from salts and detergent by dialysis, was freeze-dried and stored in liquid nitrogen until its use. αIIbβ3 (228 kDa) binds 0.38 mg of Triton X-100/mg of protein, and it is stable in its monomer form at 0.2% (w/v) Triton X-100 in 50 mm Tris-HCl, 0.1 mm CaCl2, pH 7.4, aqueous buffer (20). In the first SANS experiments, pure αIIbβ3 was solubilized with a slight excess of Triton X-100, at a protein concentration of 30 mg/ml in 50 mm Tris-HCl, 0.1 mm CaCl2, 0.2% (w/v), pH 7.4, Triton X-100 aqueous buffer (column buffer) and centrifuged at 12,000 rpm in a Beckman microfuge, to get rid of any dust. To avoid any protein aggregate and excess of detergent, this solution was fractionated on a Superdex 200 column (1 × 150 cm) equilibrated with the same buffer at 4 °C, and the column elution was monitored at 280 nm. The monomer fraction at the maximum was concentrated by ultrafiltration through an Amicon YM-100 membrane (molecular mass cut-off of 100 kDa) to concentrate the protein but not the detergent. The protein concentration was measured by the method of Markwell et al. (21) and corrected for the Tris content of the samples adjusted to 4 mg/ml with column buffer, and the concentrated fraction was dialyzed extensively at 4 °C against the same buffer, but now with 16% D2O (v/v) (SANS buffer), i.e. the concentration of D2O at which we found the contrast match point for 0.2% Triton X-100 (see below). The equilibrated sample, after five changes of the external deuterated buffer, was checked by protein, Triton X-100 (22), and SDS-PAGE analyses, before and after the SANS measurements. Protein solutions of lower concentration were prepared by dilution from the 4 mg/ml solution, using the last external deuterated buffer used for sample equilibration. When we measured the Triton X-100 bound to the protein eluted from the column and to the ultrafiltrated protein, we found that although the protein solution coming directly from the column had the expected excess of Triton X-100 caused by the protein-bound Triton X-100 (20), that is, it was equilibrated with the column buffer, the ultrafiltrated protein solution had a larger excess because of a moderate concentration of the detergent micelles (molecular mass, 95 kDa) during ultrafiltration, which was not totally corrected by the extensive dialysis. This caused difficulties for buffer subtraction during the data reduction of the SANS experiments. By increasing the protein concentration of the sample loaded into the column up to 40–50 mg/ml, we could obtain over 4 mg/ml of integrin solution at the elution maximum of the monomer fractions, avoiding the step of concentration by ultrafiltration. Finally, these fractions were subjected to equilibrium dialysis against SANS buffer at 4 °C, as described above. On three occasions, aliquots of the integrin were also equilibrated at 4 °C against the SANS buffer with 1 mm Mn2+ instead of 0.1 mm Ca2+.
SANS Measurements
The neutron scattering measurements were carried out at Beam Line D22 of the Institute Laue-Langevin (Grenoble, France). Intensity is scaled to absolute units by measuring the direct beam intensity. The contrast match point of the detergent (the D2O concentration at which the intensity of the forward scattering at zero angle, I(0), approaches zero) was determined from the scattering profiles of 0.2% (w/v) Triton X-100 dissolved in the sample buffer referred to above, at 0, 10, 20, 30, 40, 50, 75, and 100% (v/v) D2O in H2O. The I(0) values were determined by extrapolation of the scattering profiles to zero angle, and the normalized intensities were plotted as a function of the D2O concentration. The intercept on the abscissa of the linear dependence indicated that the scattering intensity was matched at 16% (v/v) D2O.
The scattering of solutions of the integrin αIIbβ3 at 4 mg/ml in SANS buffer at the match point was measured in round quartz cells of 1-mm path length and 320-μl capacity, at sample to detector distances of 2 and 8 m, after positioning them in the sample rack of the instrument kept at 25 °C. The SANS measurements were done in five or six cycles, and the integrity and size of the protein in the samples were stable during the 24 h of measuring time, as assessed by the measurement of the actual radius of gyration (Rg) and molecular mass, which did not change along the five or six cycles of measurement in each experiment and, afterward, by SDS-PAGE (supplemental Fig. S1) and analytical gel filtration analyses of the samples. A neutron wavelength (λ) of 6Å and a circular aperture 16 mm in diameter were used. To correct for the incoherent scattering of the background and to obtain the coherent scattering curves where the structural information is found, the scattering curve of the SANS buffer at the match point was subtracted from the scattering curves of the sample solutions. The fulfillment of the Porod approximation was applied to the subtracted curves (23).
Data reduction was performed using the standard ILL GRASansP software. For every experiment, the data acquired at both sample-detector distances were merged to obtain the corresponding experimental scattering curve. The individual scattering curves from different experiments for a given type of sample were normalized, considering their individual protein concentration, to obtain the average scattering curve for that sample, which was used for further calculations. The resulting q range was 0.007–0.35 Å−1 (q = 4π sin θ/λ, where 2θ is the scattering angle).
Analysis of the Reduced Scattering Data
A first estimation of the Rg and molecular mass of αIIbβ3 was done by the Guinier approximation at the low q region (q·Rg < 1.3) of the scattering curves (23). In addition, the Rg, the forward scattering, I(0), and the pair-distance distribution function P(r) were calculated from the scattering intensities I(q), using the software GNOM (25). P(r) corresponds to the distribution of the distance r between any two volume elements within one particle, weighted by the product of their scattering length density relative to that of the solvent. The P(r) can be used to calculate Rg and I(0), taking into account all of the collected data and not only the small region at very low angles, as used for the Guinier plot approximation. P(r) gives also the maximum dimension of the integrin (Dmax), as the distance where the P(r) function approaches zero.
In our conditions, the molecular mass (Mm) of the integrin in solution is proportional to I(0) (26), according the following equation,
 |
where Σb is the sum of the coherent scattering lengths (cm) over all nuclei in each protein molecule; N (n = cNA St/Mm) is the number of protein molecules; V (V = Mm ῡp/NA) is the volume of the protein; ρs is the scattering length/unit volume of the solvent; A is a factor that includes the incident beam intensity, the size of the sample, its transmission, and all the geometrical parameters of the experiment; c is the protein concentration (g/dm3); NA is Avogadro's number; S × t is the cross-sectional area × sample path length (cm3); and ῡp is the partial specific volume of the protein (cm3 g−1).
Low resolution models of the integrin were obtained from the experimental data by ab initio modeling using DAMMIN, a program of simulated annealing algorithms (27). The scattering curve up to qmax = 0.35 Å was used for fitting, corresponding to a resolution of 18 Å (2π/qmax). The program calculates the scattering intensities from a multiphase model of a particle constructed of a finite number of dummy beads. The bead radius used for the ab initio modeling of the solubilized integrin was set to 10.5 Å, chosen taking into account the 230 Å Dmax (see above) and the 18 Å resolution and some constraints within the program. The process to obtain 100 models and to select the 20 more representative ones was as followings: 100 models were obtained and automatically averaged independently in 20 groups of five models each by the program DAMAVER (28). The average normalized spatial discrepancy (NSD) value for each model was calculated with respect to the rest of the set. The result of those runs in each set were analyzed using DAMSUPP, where the five models of each group were superimposed in all possible pairs by using the program SUPCOMB (29). The model with the lowest average NSD is considered to be the most probable within each group, and the model with the highest NSD is considered as an outlier. From the nondiscarded models, an average envelope is constructed. A new DAMMIN run is performed, using the lowest NSD average envelope as starting state, to obtain a more accurate model. This averaging process is performed with the rest of the five model sets. After these 20 runs, the outputs are compared based on their NSD, and they are ordered in terms of this parameter. Finally, the entire assembly of beads was remapped onto a densely packed grid of beads, where each grid point was characterized by its occupancy factor. The portion of points with higher non-zero occupancy was selected to yield the volume equal to the average excluded volume of the models. The same averaging process was repeated again with the 20 models resulting from the previous averaging. This last model was considered as the most representative one. The computation of the theoretical solution scattering curves from the reconstructed atomic models of αIIbβ3 was done using CRYSON (30).
RESULTS
SANS Data
The average SANS intensity curves as a function of the scattering vector q using GRASansP software, obtained from six experiments of integrin αIIbβ3 equilibrated in SANS buffer with 0.1 mm Ca 2+ and from three experiments of the same integrin equilibrated in the same buffer but with 1 mm Mn2+ instead of Ca2+, are shown in Fig. 1a. The linearity of the Guinier plot at the low q region (23) (Fig. 1a, inset) and the values of Rg and scattering intensity at zero angle I(0) derived from it, 5.16 ± 0.04 nm and 4.67 × 10−2 cm−1, respectively, indicate that αIIbβ3 in Ca2+/detergent solutions was homogeneous and in monomer form. On the contrary, αIIbβ3 in Mn2+/detergent solutions at low q shows a scattering curve that turns upwards at low angle and points to a nonlinear Guinier plot at very low q, which we interpret as being due to protein aggregation and particle heterogeneity. This interpretation is supported by the values for both Rg (7.30 ± 0.2 nm) and I(0) (6.48 × 10−2 cm−1) obtained from the Guinier approximation in the qRg range of <1.3, which are too large compared with those obtained for the monomeric integrin in Ca2+ buffer and by the constantly increasing curvature up at low angle.
FIGURE 1.
a, experimental SANS curve from αIIbβ3 in detergent solution equilibrated in SANS buffer containing 0.1 mm Ca2+ (○) and from αIIbβ3 in detergent solution equilibrated in SANS buffer containing 1 mm Mn2+ (▵). For the sake of clarity, the data from the integrin in Mn2+ buffer has been multiplied by 10. The continuous line shows the fit of the scattering profile computed from the average ab initio model (30) to the experimental data. Inset, Guinier plot (23) of the SANS data. The straight line corresponds to the best fit through the linear behavior plot. b, distance distribution functions calculated for the SANS data using GNOM (25). The maximum distance of the P(r) function is denoted by Dmax.
From the P(r) function (25) (Fig. 1b), the maximum dimension of αIIbβ3 in Ca2+/detergent solutions (Dmax = 23 ± 2 nm) can be estimated, and a more accurate estimation of Rg (5.8 ± 0.1 nm) and I(0) (4.17.10−2 cm−1) is obtained, compared with the values for these parameters obtained from the Guinier plot. The forward scattering intensity is directly proportional to the number of particles in the sample (26). From I(0), after scaling by the concentration, we obtained a molecular mass of 238 ± 5 kDa, which is in good agreement with the molecular mass of αIIbβ3 (228 kDa) calculated from the amino acid and sugar composition of the αIIb and β3 subunits and from previous hydrodynamic and equilibrium measurements (20) and confirms the monomeric state of the protein in solution. However, for the integrin in Mn2+/detergent solutions, the I(0) value obtained from the Guinier approximation gives a molecular mass of 370 ± 5kDa, which, as above, we interpret as being due to integrin aggregation and particle heterogeneity in Mn2+ solutions.
Reconstruction of the High Resolution Structural Model of Whole αIIbβ3 in Its Bent Configuration and in Its Extended Form
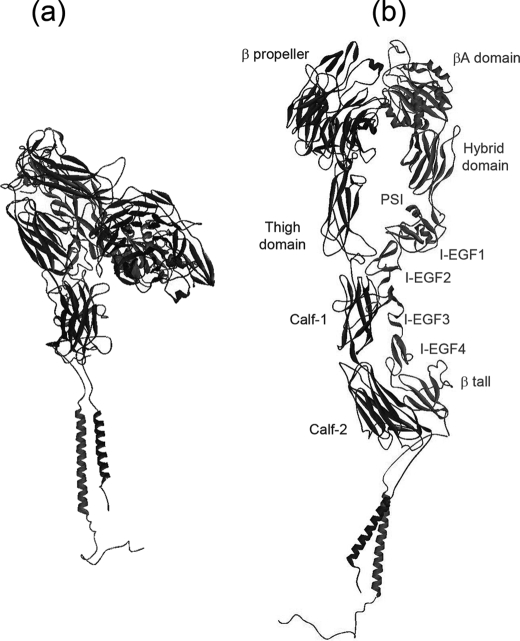
We used the crystallographic model of r-αIIbβ3 (12) and the NMR structural models of the transmembrane segments (31) and cytoplasmic domains (32) of the αIIb and β3 subunits to reconstruct the best possible high resolution structural model of whole αIIbβ3 in its bent configuration (Fig. 2a) and in what would be its extended form (12) (Fig. 2b). The reconstructed model of the bent configuration was used to calculate its theoretical SANS curve and to dock it into the SANS low resolution model, which allowed us to assess the degree of fitting of both the theoretical scattering curve with the experimental SANS curve and the reconstructed high resolution models with the SANS model. Equally, the theoretical scattering curves of the fully extended form and the ectodomain of αIIbβ3 were calculated from the reconstructed high resolution model and from the crystallographic model, respectively, to compare them with the experimental SANS curve of αIIbβ3.
FIGURE 2.
Reconstruction of the best possible high resolution structural model of whole αIIbβ3 made from the crystallographic model of the r-αIIbβ3 ectodomain (Protein Data Bank 3FCS) (12) and the NMR structural models of the transmembrane segments (Protein Data Bank 2K9J) (31) and cytoplasmic domains of the αIIb and β3 subunits (Protein Data Bank 1M8O) (32), considering the bent (a) and what would be the fully extended (b) conformations.
Low Resolution Structure of αIIbβ3
SANS in combination with shape reconstruction algorithms allowed us to obtain an average low resolution three-dimensional sphere model (NSD = 0.712 ± 0.024) of human integrin αIIbβ3 in detergent solutions with Ca2+. The fit of the scattering profile computed from this model to the experimental data (χ2 = 1.85 ± 0.05) is shown in Fig. 1a (continuous line). Fig. 3 shows different side projections of three models selected among the three-dimensional models obtained by DAMMIN (27). They indicate that the αIIbβ3 structure in solution is well defined, despite its multidomain nature and the expected segmental flexibility between their different regions and between their different domains within each region. The main features of the average model are also observed in the SANS envelope enclosing all the initial models obtained (supplemental Figs. S2 and S3).
FIGURE 3.
Different projections after successive counterclockwise 90° rotation along the long axis of three examples of reconstructed three-dimensional ab initio sphere models of αIIbβ3 integrin in detergent solution with Ca2+, obtained by DAMMIN (27).
In Fig. 4(a–d), we show four side projections, after successive counterclockwise 90° rotations along the long axis, of the optimum docking of the reconstructed high resolution structure of whole αIIbβ3 into the average ab initio low resolution envelope of αIIbβ3, together with its top and bottom views (Fig. 4, e and f). This allowed us to assign tentatively the most probable disposition of the known integrin domains within the low resolution model and to point out the structural differences between the crystallographic model of the r-αIIbβ3 and r-αVβ3 ectodomains and the SANS model of the whole integrin in solution, as well as the differences between the whole integrin models obtained from cryo-EM and SANS.
FIGURE 4.
Optimum docking of the reconstructed high resolution structure of αIIbβ3 (Fig. 2a) into the average ab initio low resolution envelope of αIIbβ3 (gray surface), calculated from the SANS data (27, 28–30), where it is shown. a–d, the handgun projection (a and c) and the arched projection (b and d) obtained after successive counterclockwise 90° rotations along the long axis. e and f, the top and bottom projections.
In Fig. 4, projections b and d show an elongated and slightly arched form, not observed before, which inscribes an angle of 135°, and projections a and c show a handgun-shaped form, with three well differentiated parts. The model has an average maximum length of 20 nm, and its average width and thickness decreases from 11 × 7 nm at the top, down to 2 × 2 nm at the bottom, respectively. At the top of the handgun projections is the largest region of 14 nm of length, which contains most of the integrin mass and has two differentiated parts: the “butt end” and the “cylinder.” The butt end, with its long axis at 45° with respect to the long axis of the model and an average length, width, and thickness of 10 × 7.5 × 6 nm, respectively, contains the integrin headpiece and includes, from its lower end to its top end, most the “β-propeller” domain of αIIb, the βA domain of β3, the upper αIIb (“thigh domain”), and β3 (“hybrid,” “PSI,” and “I-EGF-1” domains) legs, the αIIb and β3 bent knees (at the apex of the model) and most of the upper part of the lower legs of αIIb (“Calf-1” domain) and β3 (“I-EGF-2” and “I-EGF-3”). The cylinder of 5.5 × 5 × 4 nm on average contains most of the lower αIIb (“Calf-2” domain) and β3 (“I-EGF-4” and “β-tail” domains) legs. This largest region, which we assign to the ectodomain of the integrin, shows particular structural features that are not identified unequivocally yet that reflect their multidomain organization. The αIIb and the β3 subunits occupy mainly the concave and convex side, respectively, of the arched projections. Some segments of the β-propeller, Calf-1, and Calf-2 domains of αIIb and of βA, PSI, I-EGF1, and C-terminal cytoplasmic domain of β3 are outside the low resolution envelope.
At the bottom end of the integrin envelope is the “gun barrel,” with a cylindrical region of about 3 × 2 × 2 nm on average, which we assign to the integrin transmembrane domain with the helical transmembrane segments of the αIIb and β3 subunits, followed by a small region of 3 × 2.5 × 2.2 nm on average, which should contain the intracellular domains of the αIIb and β3 subunits.
In agreement with what is observed on docking the reconstructed high resolution model of αIIbβ3 into the low resolution envelope, the calculated scattering curve for the reconstructed model of the bent configuration fits fairly well to the experimental SANS data (Fig. 5). However, the theoretical scattering curve, calculated for what would be the αIIbβ3 fully extended form, fits very poorly to the experimental SANS data, and it deviates very significantly from the calculated scattering curve for the bent configuration. These results strengthen the interpretation that αIIbβ3 in solution is in the bent configuration.
FIGURE 5.
Superposition on the experimental SANS data (○) of the theoretical scattering curve (continuous line) (30), obtained from the reconstructed high resolution structural model of whole αIIbβ3 in its bent configuration (Fig. 2a). The discontinuous line represents the theoretical scattering curve computed from the reconstructed model of whole αIIbβ3 in what would be its fully extended conformation (Fig. 2b) (12).
In supplemental Fig. S4, we superimposed the experimental SANS curve on the calculated scattering curves for the reconstructed model of the bent configuration of αIIbβ3 and for the crystallographic model of αIIbβ3 ectodomain. It is evident that at low q, the scattering curve for the ectodomain deviates from the other two curves, which points to a significantly lower Rg, as expected, and therefore to a more globular structure of the ectodomain, given the small difference in mass between whole αIIbβ3 and its ectodomain. So SANS was able to reveal the transmembrane and cytoplasmic domains of αIIbβ3.
To find out whether the integrin structure acquired the extended conformation in the presence of Mn2+, as described before (5), three SANS experiments of αIIbβ3 in SANS buffer with 1 mm Mn2+ instead of Ca2+ were done in parallel with control measurements of the same integrin preparation in SANS buffer with Ca2+. Unfortunately, as shown above, protein aggregation and particle size heterogeneity were observed, which made it impossible to obtain the configuration of αIIbβ3 in solutions with Mn2+ as planned. This observation is in agreement with the earlier chromatographic elution of αIIbβ3 in Mn2+ buffer with respect to αIIbβ3 in Ca2+ buffer and with previous observations that Mn2+, but not Ca2+, can induce formation of dimers and multimers of whole αIIbβ3 and r-αVβ3 (13, 33).
DISCUSSION
The Low Resolution SANS Model of αIIbβ3 in Solution
The most relevant features of the SANS low resolution model at 2-nm resolution of the whole human integrin αIIbβ3 in solution are: an average length of 20 nm and a decreasing average width and thickness from top (11 × 7 nm) to bottom (2 × 2 nm); the model projections after counterclockwise 90° rotations along its long axis show successively a handgun form with a 45° bend and an elongated arched form (135°), as well as three differentiated parts, from top to bottom; the butt end at the top contains most of the molecular mass of the heterodimer, its long axis forms a 45° angle with the long axis of the molecule and together with the cylinder in the middle has 14 nm of average length; the gun barrel at the bottom has two differentiated parts linearly disposed of about 3 nm in length each. Optimization of the docking of the reconstituted high resolution model on the SANS model allowed tentative assignment of the different domains of the former to the major features of the latter. So the head of the protein together with the upper legs, the knees, and the upper part of the lower legs of the αIIb and β3 subunits are mainly accommodated within the butt end. The lower part of the lower legs of αIIb and β3 are located in the cylinder and the transmembrane helices and the cytoplasmic domains of both subunits occupy the proximal and distal parts of the gun barrel, respectively. Equally, the β3 subunit occupies the convex side of the arched projection, the αIIb subunit is located in the concave side, and at the apex of the model are the bent knees of both subunits.
According to this model, the αIIbβ3 structure in solution is well defined, despite its multidomain nature and the expected segmental flexibility between their different regions and between their different domains within each region. The role of the disulfide bonds to maintain the intradomain and interdomain structure in αIIb is obvious, after looking at their distribution: three intradomain disulfides in the β-propeller (integrins 1–451) and two intradomain disulfides in each of the other extracellular thigh (residues 452–601), Calf-1 (integrins 602–743), and Calf-2 (residues 744–960) domains (34). The disulfide Cys826–Cys877 cross-links the α-chain to the β-chain of αIIb in the whole integrin. These chains are products of the proteolytic processing of this subunit in the megakaryocytes, which do not take place in the soluble r-αIIbβ3 ectodomains. The role of the 28 disulfide bonds of the β-subunit is even larger in maintaining its intradomain and interdomain structure of this subunit. Although there are still discrepancies in the precise cysteine cross-linking pattern in the human platelet β3 subunit found by protein chemistry (35) and the pattern found in r-αVβ3 and r-αIIbβ3 ectodomains by x-ray crystallography (10, 12, 36), there is agreement on the presence of a long range disulfide bond cross-linking the N-terminal of the PSI domain (1–56) to Cys435, between the C terminus of the hybrid domain (residues 157–108 and 353–433) and the N terminus of the I-EGF-1 domain (residues 437–472) and on the intradomain and/or interdomain cross-linking in these and the rest of the extracellular domains of the β3 subunit.
As could be expected, the docking of the crystallographic model into the SANS model (Fig. 4) and the fit of the calculated scattering curve for the reconstructed model of the bent configuration to the experimental SANS data (Fig. 5, continuous line) are not perfect, with the cystallographic structure more constrained than the structure in solution. On the one hand, some segments of the β-propeller, Calf-1, and Calf-2 domains of αIIb and of the βA, PSI, I-EGF-1, and the C-terminal cytoplasmic domains of β3 in the reconstructed crystallographic structure are outside the integrin envelope. However, the SANS model of αIIbβ3 is unequivocally consistent with the bent conformations of the r-αVβ3 and r-αIIbβ3 ectodomains observed before by x-ray crystallography (10–12). On the other hand, the distal part of the gun cylinder is empty of components of the reconstructed structure. To fill this volume, given that the transmembrane segments cannot be moved from the cylindrical region where they have been assigned, the distal parts of the lower legs of the αIIb and β3 subunits should move 1–2 nm downwards to connect with their corresponding transmembrane helices. We interpret these appreciable differences between the reconstructed high resolution model and the SANS model to be due to the differences between the crystal and the solution structure, as well as to the fact that the solution structure was derived from the whole integrin, whereas the crystal structure was derived from the r-αIIbβ3 ectodomain. This recombinant ectodomain lacks the sequence αIIb 840–873, where the proteolytic processing of human platelet αIIbβ3 takes place, besides the transmembrane and cytoplasmic domains of both subunits.
Unexpectedly, the SANS model for αIIbβ3 in solution is not consistent with the cryo-EM model for αIIbβ3 in a frozen sample (16). The cryo-EM model is shorter (18 nm), and in a projection along the long axis, the head and the stalk are in an extended form, and the lower legs form a 90° bend with the transmembrane and cytoplasmic domains, whereas in the projection after a 90° rotation along the long axis, the different domains are linearly aligned (16). So besides the differences in dimensions between the two models, the jackknifed conformation of the ectodomain and the arched projection observed in the SANS model do not fit into the three-dimensional cryo-EM model.
However, the SANS model of whole αIIbβ3 in solution seems consistent with the compact triangular shape of the negative staining EM models available for the r-αVβ3 ectodomain in the presence of Ca2+ or Mn2+ in its liganded and unliganded forms (13) and the r-αVβ3 and r-αIIbβ3 ectodomains in the presence of Ca2+ (5, 12). Equally, the SANS model of αIIbβ3 is also unequivocally consistent with the bent conformation of αIIbβ3 in resting and physiologically activated platelets, as observed earlier by epitope mapping and competition among monoclonal antibodies directed to the C-terminal region of the αIIb subunit and to the N-terminal domain of the β3 subunit (9) and by fluorescence intramolecular energy transfer between Fab fragments bound to these regions (15). Finally, the SANS model is consistent with the bent conformation of αVβ3 in resting and Mn2+-activated K562 cells, observed by intermolecular energy transfer (37).
The tentative assignment of the different domains of the reconstituted high resolution structure could be checked by SANS measurements of the complexes of αIIbβ3 with high affinity ligands (specific monoclonal antibodies, adhesive proteins, fragments derived from them, competitive inhibitors, etc), which bind to well defined sites. We are analyzing the SANS data from experiments using the Fab from an anti-αIIbβ3 monoclonal antibody that activates and aggregates platelets.3
Functional Aspects and Integrin Activation Models: the Role of Mn2+ and the Transmembrane Segments Inserted in the Bilayer
In our opinion, attempts to understand integrin activation have been both supported and hindered by abundant and sometimes conflicting data. So we should mention that the activation of the r-αIIbβ3 and r-αIIbβ3 ectodomains in solution with Mn2+ is a controversial issue. On the one hand, Mn2+ is neither a physiological agonist of platelet activation and aggregation, nor an agonist of endothelial cell activation and binding of serum adhesive proteins. On the other hand, given the invoked central role of the transmembrane segments of the α and β subunits (3, 5–7, 31, 38) in bidirectional signaling across the plasma membrane, the use of recombinant ectodomains to study integrin activation is questionable. So one would expect that the simplest system to check the structural modifications following integrin activation would be the whole integrin reconstituted in single bilayer lipid vesicles. Furthermore, although αIIbβ3-fibrinogen complexes in detergent solution in the presence of Ca2+ or Mn2+ have been seen by EM (33, 39), these observations were not quantitative. So it was observed by sedimentation equilibrium that fibrinogen in the presence of Ca2+ and Mn2+ did not bind to αIIbβ3 (KD > 1 mm) (40), and the attempts to isolate αIIbβ3-fibrinogen complexes in detergent solution at physiological concentrations of fibrinogen by size exclusion chromatography, in the presence of Mn2+ or the Fab of an anti-αIIbβ3 antibody, which induces platelet activation, fibrinogen binding, and platelet aggregation, always failed.4 Equally, the height of αIIbβ3 reconstituted in liposomes does not change in the presence of Mn2+, as observed by cryoelectron tomography (14), and preliminary results of SANS experiments suggest that the binding of the Fab fragments of one of these monoclonal antibodies to αIIbβ3 in detergent solution is not followed by the extension of the integrin structure.3 So to clear the conflicting experimental observations on the Mn2+ activation of αIIbβ3 and on whether its activated state is in the bent or the extended configuration, the reconstitution of the integrin in single bilayer vesicles made using deuterated lipids and its activation using Mn2+ or an Fab of an anti-αIIbβ3 activating antibody, in the absence or in the presence of equimolecular fibrinogen concentration, is one of the first experiments to do to try to make progress in this direction. As was theoretically shown above, the SANS measurements are able to distinguish between the bent and the contentious extended conformation of αIIbβ3 (Fig. 5) and between the whole integrin and its ectodomain (supplemental Fig. S4).
Finally, at present there are high resolution crystallographic models for very few whole membrane proteins, because of the severe limitations on how to crystallize them. This limiting situation could be eased by SANS low resolution models of membrane proteins. In the future, the low resolution structure of membrane proteins reconstituted in single lipid bilayer vesicles, their disposition with respect to the membrane and their structural modifications in different functional conditions could be obtained by SANS, if the lipid vesicles were made invisible by deuteration (24).
Supplementary Material
Acknowledgments
We thank Roland P. May, our former local contact at ILL during the first sets of experiments. We are also grateful to the Institute Laue-Langevin for the allocation of the beam time used.
This work was supported by Grants SAF 2003-042/66 (2003–2007) and Intramural Frontera 200680F-0083 (2007–2008) (to J. G.-R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
A. Nogales, C. García, T. A. Ezquerra, and J. González-Rodríguez, manuscript in preparation.
J. González-Rodríguez, unpublished observations.
- EM
- electron microscopy; r-αIIbβ3 and r-αVβ3 ectodomains, recombinant products of the extracellular domains of human integrins αIIbβ3 and αVβ3, respectively
- SANS
- small angle neutron scattering
- NSD
- normalized spatial discrepancy
- PSI
- plexin-semaphorin- integrin.
REFERENCES
- 1.Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2.Humphries M. J., McEwan P. A., Barton S. J., Buckley P. A., Bella J., Mould A. P. (2003) Trend. Biochem. Sci. 28, 313–320 [DOI] [PubMed] [Google Scholar]
- 3.Arnaout M. A., Mahalingam B., Xiong J. P. (2005) Annu. Rev. Cell Dev. Biol. 21, 381–410 [DOI] [PubMed] [Google Scholar]
- 4.Luo B. H., Springer T. A. (2006) Curr. Opin. Cell Biol. 18, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takagi J., Petre B. M., Walz T., Springer T. A. (2002) Cell 110, 599–611 [DOI] [PubMed] [Google Scholar]
- 6.Xiong J. P., Stehle T., Goodman S. L., Arnaout M. A. (2003) Blood 102, 1155–1159 [DOI] [PubMed] [Google Scholar]
- 7.Xiao T., Takagi J., Coller B. S., Wang J. H., Springer T. A. (2004) Nature 432, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan B., Smith J. W. (2001) Biochemistry 40, 8861–8867 [DOI] [PubMed] [Google Scholar]
- 9.Calzada M. J., Alvarez M. V., Gonzalez-Rodriguez J. (2002) J. Biol. Chem. 277, 39899–39908 [DOI] [PubMed] [Google Scholar]
- 10.Xiong J. P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D. L., Joachimiak A., Goodman S. L., Arnaout M. A. (2001) Science 294, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. (2002) Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 12.Zhu J., Luo B. H., Xiao T., Zhang C., Nishida N., Springer T. A. (2008) Molecular Cell 32, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adair B. D., Xiong J. P., Maddock C., Goodman S. L., Arnaout M. A., Yeager M. (2005) J. Cell Biol. 168, 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye F., Liu J., Winkler H., Taylor K. A. (2008) J. Mol. Biol. 378, 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutinho A., García C., González-Rodríguez J., Lillo M. P. (2007) Biophys. Chem. 130, 76–87 [DOI] [PubMed] [Google Scholar]
- 16.Adair B. D., Yeager M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14059–14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chigaev A., Buranda T., Dwyer D. C., Prossnitz E. R., Sklar L. A. (2003) Biophys. J. 85, 3951–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johs A., Hammel M., Waldner I., May R. P., Laggner P., Prassl R. (2006) J. Biol. Chem. 281, 19732–19739 [DOI] [PubMed] [Google Scholar]
- 19.Rivas G. A., Calvete J. J., González-Rodríguez J. (1991) Protein Expr. Purif. 2, 248–255 [DOI] [PubMed] [Google Scholar]
- 20.Rivas G. A., Aznárez J. A., Usobiaga P., Saiz J. L., González-Rodríguez J. (1991) Eur. Biophys. J. 19, 335–345 [DOI] [PubMed] [Google Scholar]
- 21.Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. (1978) Anal. Biochem. 87, 206–210 [DOI] [PubMed] [Google Scholar]
- 22.Garewal H. S. (1973) Anal. Biochem. 54, 319–324 [DOI] [PubMed] [Google Scholar]
- 23.Guinier A., Fourner G. (1955) Small Angle X Ray Scattering, John Wiley & Sons, New York [Google Scholar]
- 24.Hunt J. F., McCrea P. D., Zaccai G., Engelman M. (1997) J. Mol. Biol. 273, 1004–1019 [DOI] [PubMed] [Google Scholar]
- 25.Svergun D. I. (1992) J. Appl. Crystallog 25, 495–503 [Google Scholar]
- 26.Jacrot B., Zaccai G. (1981) Biopolymers 20, 2413–2426 [Google Scholar]
- 27.Svergun D. I. (1999) Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallog 36, 860–864 [Google Scholar]
- 29.Kozin M. B., Svergun D. I. (2001) J. Appl. Crystallog 34, 33–41 [Google Scholar]
- 30.Svergun D. I., Richard S., Koch M. H., Sayers Z., Kuprin S., Zaccai G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2267–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau T. L., Kim C., Ginsberg M. H., Ulmer T. S. (2009) EMBO J. 28, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinogradova O., Velyvis A., Velyviene A., Hu B., Haas T., Plow E., Qin J. (2002) Cell 110, 587–597 [DOI] [PubMed] [Google Scholar]
- 33.Litvinov R. I., Nagaswami C., Vilaire G., Shuman H., Bennett J. S., Weisel J. W. (2004) Blood 104, 3979–3985 [DOI] [PubMed] [Google Scholar]
- 34.Calvete J. J., Henschen A., González-Rodríguez J. (1989) Biochem. J. 261, 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calvete J. J., Henschen A., González-Rodríguez J. (1991) Biochem. J. 274, 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiong J. P., Stehle T., Goodman S. L., Arnaout M. A. (2004) J. Biol. Chem. 279, 40252–40254 [DOI] [PubMed] [Google Scholar]
- 37.Xiong J. P., Mahalingham B., Alonso J. L., Borrelli L. A., Rui X., Anand S., Hyman B. T., Rysiok T., Müller-Pompalla D., Goodman S. L., Arnaout M. A. (2009) J. Cell Biol. 186, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennett J. S. (2005) J. Clin. Invest. 115, 3363–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisel J. W., Nagaswami C., Vilaire G., Bennett J. S. (1992) J. Biol. Chem. 267, 16637–16643 [PubMed] [Google Scholar]
- 40.Rivas G., Tangemann K., Minton A. P., Engel J. (1996) J. Mol. Recognit. 9, 31–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.