Abstract
C-reactive protein (CRP) is an acute phase protein of the pentraxin family that binds ligands in a Ca2+-dependent manner, and activates complement. Knowledge of its oligomeric state in solution and at surfaces is essential for functional studies. Analytical ultracentrifugation showed that CRP in 2 mm Ca2+ exhibits a rapid pentamer-decamer equilibrium. The proportion of decamer decreased with an increase in NaCl concentration. The sedimentation coefficients s20,w0 of pentameric and decameric CRP were 6.4 S and in excess of 7.6 S, respectively. In the absence of Ca2+, CRP partially dissociates into its protomers and the NaCl concentration dependence of the pentamer-decamer equilibrium is much reduced. By x-ray scattering, the radius of gyration RG values ranged from 3.7 nm for the pentamer to above 4.0 nm for the decamer. An averaged KD value of 21 μm in solution (140 mm NaCl, 2 mm Ca2+) was determined by x-ray scattering and modeling based on crystal structures for the pentamer and decamer. Surface plasmon resonance showed that CRP self-associates on a surface with immobilized CRP with a similar KD value of 23 μm (140 mm NaCl, 2 mm Ca2+), whereas CRP aggregates in low salt. It is concluded that CRP is reproducibly observed in a pentamer-decamer equilibrium in physiologically relevant concentrations both in solution and on surfaces. Both 2 mm Ca2+ and 140 mm NaCl are essential for the integrity of CRP in functional studies and understanding the role of CRP in the acute phase response.
Keywords: Blood Coagulation/Complement, Immunology/Innate Immunity, Methods/Computer Modeling, Methods/Surface Plasmon Resonance, Methods/Ultracentrifugation, Methods/X-ray Scattering, Protein/Structure
Introduction
C-reactive protein (CRP)3 is a 115-kDa acute phase protein of the pentraxin family of calcium-dependent ligand-binding proteins in serum (1). CRP binds phosphocholine (2), as well as phosphoethanolamine, microbial surface proteins, chromatin, histones, fibronectin, small nuclear ribonucleoproteins, laminin, and polycations (3). CRP-ligand interactions are important for the recognition of damaged or apoptotic cells and bacterial pathogens. CRP possesses pro-inflammatory effects, where ligand-bound, cross-linked, or aggregated CRP activates the classical pathway of complement by interacting with C1q in a manner similar to antigen-bound or aggregated IgG (3, 4). CRP interacts with immunoglobulin receptors to recruit phagocytic cells, in addition to exerting regulatory effects on neutrophils (5). CRP ligand binding is important for complement activation at cell surfaces (6–8). CRP has been described as also having anti-inflammatory effects. This apparent contradiction in CRP activity resulted from the study of CRP in an abnormally modified form (mCRP), with different biological and antigenic properties (9–13). mCRP consists of dissociated CRP protomers that are produced in vitro under harsh conditions of high acidic pH (2.0), temperature (above 60 °C), or 8 m urea. mCRP may occur naturally in normal or inflamed human and animal tissues (14–17). However, once CRP is denatured, it is rapidly catabolized in normal conditions, and there is no firm evidence for the persistence of monomeric, denatured, or aggregated CRP in vivo (1).
Native CRP is unglycosylated and consists of five identical subunits arranged non-covalently as a cyclic pentamer with a central pore. When co-crystallized with Ca2+ and phosphorylcholine, each subunit has a phosphocholine ligand-binding site proximate to two bound Ca2+ ions held 0.4 nm apart by acidic side chains on the B (B = binding) face of the pentamer (1, 18). The A (A = α) face of the protomer possesses a single α-helix and binds to ligands such as C1q (19). The B face of the pentamer is negatively charged, whereas the A face showed a more varied charge distribution with a ring of negative charge at the central pore (18, 20). This suggests a role for electrostatic effects in both lipid binding at the B face and C1q binding at the A face (19). Native CRP with Ca2+ present exists physiologically as a pentamer according to electron microscopy, gel chromatography, and crystallography (18, 21–23). However, in the presence of Ca2+, CRP shows a marked concentration dependence in its scattering and sedimentation properties, showing that self-association had occurred (24, 25). In the absence of Ca2+, mCRP is formed (26, 27). In contradiction to this, crystallographic studies in 100 mm NaCl in the absence of Ca2+ suggested that CRP formed a decamer through an A face-A face association of two pentamers (20). Although the unfolding of a large Ca2+-binding loop on the B face after Ca2+ depletion may explain the Ca2+-dependent affinity of CRP for lipids, this Ca2+ site is neither located at the protomer interface nor at the A face. Thus the effect of Ca2+ removal on CRP stability is unclear. Even though Ca2+ is present in at least 2 mm in plasma (28), there is much emphasis in structural literature on the effect of Ca2+ depletion on CRP, even though Ca2+ depletion appears unlikely to occur in vivo.
A critical knowledge of the oligomeric state of CRP is essential for understanding its function. Accordingly, a multidisciplinary strategy based on analytical ultracentrifugation (AUC) and x-ray scattering in solution, and surface plasmon resonance (SPR) on surfaces established the self-association state of CRP in a concentration range of 0.1–5.4 mg/ml in 2 mm Ca2+. CRP normally circulates in blood at a median value of 0.0008 mg/ml, which can be as low as 50 ng/ml, but this increases to over 0.5 mg/ml following an acute phase stimulus (a range of 0.4 nm to 4.4 μm) (1). Using previously established methods (29, 30), we show from AUC size distribution analyses c(s) that pentamers and decamers of CRP are readily identified, whereas constrained x-ray scattering modeling quantified the amounts of pentamers and decamers in solution. We show that SPR observes a similar degree of CRP self-association with immobilized CRP at surfaces in 140 mm NaCl (31). This joint application of AUC, scattering, and SPR methods shows that CRP with 2 mm Ca2+ exists in what was previously an unidentified rapid pentamer-decamer equilibrium that is NaCl-concentration dependent. Molecular modeling accounted for the formation of decamers in terms of salt bridges between the pentamers. To clarify previous CRP studies without Ca2+, we show that Ca2+ removal leads to the reduced formation of decamers and the partial dissociation of pentamers into monomers. Our study resolves the outcome of previous studies of CRP and its ligands, and emphasizes the importance of physiological 2 mm Ca2+ and 140 mm NaCl when studying the complex between CRP and complement factor H (see accompanying manuscript (51)).
EXPERIMENTAL PROCEDURES
Purification of CRP
Human CRP was isolated and purified as described previously (32). Samples were extensively dialyzed into 10 mm Tris buffers (with or without 2 mm CaCl2) containing 50 mm NaCl, 140 mm NaCl, or 250 mm NaCl, all at pH 8.0, for AUC and x-ray scattering data acquisition. Size exclusion chromatography was carried out using HEPES buffer (10 mm HEPES, 140 mm NaCl, pH 8.0), with or without 2 mm CaCl2. Samples were routinely analyzed by SDS-PAGE before and after experiments to confirm their integrity. CRP concentrations were determined using an absorption coefficient of 17.5 (1%, 280 nm, 1-cm path length), which is consistent with a value of 19.2 calculated from its composition (33, 34). CRP is unglycosylated and its composition resulted in a calculated molecular mass of 115.0 kDa, an unhydrated volume of 150.3 nm3, a hydrated volume of 197.3 nm3, and a partial specific volume v̄ of 0.7408 ml/g (34). The buffer densities for AUC were measured at 20 °C using an Anton-Paar DMA5000 density meter to be 1.00096 or 1.00092 g/ml (50 mm NaCl), 1.00482 or 1.00455 g/ml (140 mm NaCl), with or without 2 mm CaCl2 in that order, and 1.00934 g/ml (250 mm NaCl).
Analytical Ultracentrifugation Data and Analyses
AUC data for CRP were obtained on two Beckman XL-I instruments equipped with AnTi50 and AnTi60 rotors. Sedimentation velocity data were acquired at 20 °C at a rotor speed of 50,000 × g in two-sector cells with column heights of 12 mm. Sedimentation analysis was performed using direct boundary Lamm fits of up to 300 scans using SEDFIT (version 11.0) (35, 36). SEDFIT resulted in size-distribution analyses c(s) that assume that all macromolecular species have the same frictional ratio, f/fo. The final SEDFIT analyses used a fixed resolution of 200 and optimized the c(s) fit by floating f/fo and the baseline until the overall root mean square deviations and visual appearance of the fits were satisfactory (see Fig. 2). The percentage fraction of pentamers, decamers, or lower oligomers in the total loading concentration was derived using the c(s) integration function (see Fig. 3).
FIGURE 2.
NaCl concentration dependence of CRP decamer formation by AUC. Protein concentrations ranged from 0.2 to 2.5 mg/ml, and are labeled at the right of each c(s) plot. The c(s) plots are displaced vertically and the peak intensities normalized for clarity. The boundary fits corresponding to 0.8 or 0.6 mg/ml are shown in the upper panels, where only every fifth to 10th scan is shown for reason of clarity. Experiments were performed at a rotor speed of 50,000 r.p.m. A–C, buffers containing 2 mm Ca2+ and 50, 140, and 250 mm NaCl. D and E, buffers without Ca2+ and containing 50 mm NaCl and 140 mm NaCl.
FIGURE 3.
NaCl concentration and Ca2+ dependence of the pentamer-decamer equilibrium by AUC. At each CRP concentration, the relative percentage of pentamer and decamer corresponds to data obtained at 50,000 r.p.m. The apparent KD value (KD-app) in 50 mm NaCl is estimated as the point of intersection of the two data sets when fitted to a non-linear regression for an exponential rise (decamer: y = y0 + a(1-bx)) or decay (pentamer: y = y0 + a(10)−bx) as appropriate. A, data correspond to 50 mm NaCl (●), 140 mm NaCl (○), and 250 mm NaCl (Δ) in the presence of 2 mm Ca2+. B, data correspond to 50 and 140 mm NaCl in the absence of Ca2+. The inset shows the percentage contribution of lower oligomers of CRP, which was not seen with Ca2+ present.
X-ray Scattering Data Collection and Analysis
X-ray scattering data were obtained in two beam sessions in 16-bunch mode at the European Synchrotron Radiation Facility (ESRF), Grenoble, France, operating with a ring energy of 6.0 GeV on the Beamline ID02 (37). Storage ring currents ranged from 68 to 77 mA (session 1) and from 70 to 92 mA (session 2). Data were acquired using a recently installed fiber optically coupled high sensitivity and dynamic range CCD detector (FReLoN). Together with a smaller beamstop, this allowed a shorter sample-to-detector distance of 2.0 m to be used and resulted in higher signal-to-noise ratios (38). CRP was studied at concentrations between 0.3 and 5.4 mg/ml (with 2 mm Ca2+) or between 0.1 and 2.5 mg/ml (without Ca2+). Sample volumes of 100 μl were measured in flow cells to reduce radiation damage by moving the sample continuously during beam exposure. Sets of 10 frames, with frame durations of 0.2, 0.3, or 0.5 s each were acquired. Buffers were measured using the same exposure times in alternation with the samples to eliminate background subtraction errors. On-line checks during data acquisition confirmed the absence of radiation damage, after which the 10 frames were averaged. Sample temperatures corresponded to ambient conditions at 20 °C. Other details including the data reduction procedure are described elsewhere (39).
In a given solute-solvent contrast, the radius of gyration (RG) is a measure of structural elongation if the internal inhomogeneity of scattering densities within the protein has no effect. Guinier analyses at low Q (Q = 4π sin θ/λ; 2θ = scattering angle; λ = wavelength) gives the RG and the forward scattering at zero angle I(0) (40).
This expression is valid in a Q·RG range up to 1.5. The RG analyses were performed using an interactive PERL script program SCTPL74 on Silicon Graphics O2 workstations. Indirect transformation of the scattering data I(Q) in reciprocal space into real space to give the distance distribution function P(r) was carried out using the program GNOM (41).
 |
P(r) corresponds to the distribution of distances r between volume elements. For this, the x-ray I(Q) curve utilized up to 562 data points in the Q range between 0.05 and 1.9 nm−1.
Modeling of CRP
All CRP modeling was based on the crystal structure for the pentamer in the presence of Ca2+ (Protein Data Bank code 1b09) (18). The CRP decamer was formed by superimposition of two pentamers face-to-face upon the Ca2+-depleted decamer structure (PDB code 1lj7) (20). The superimposition resulted in a root mean square deviation of 0.460 nm in α-carbon atoms, and overcame the issue of missing residues in the Ca2+-depleted crystal structure. Insight II 98.0 (Accelrys, San Diego, CA) software on Silicon Graphics OCTANE workstations was used for manipulations. A cube side length of 0.532 nm in combination with a cutoff of 4 atoms was used to convert the coordinates into Debye sphere models with 998 and 1997 spheres that corresponded to the unhydrated pentamer and decamer structures, respectively. The hydration shell was created using HYPRO (42), where the optimal total of hydrated spheres in the CRP models were 1310 (pentamer) and 2620 (decamer) (Table 1). The x-ray scattering curves I(Q) for CRP were calculated using the Debye equation adapted to spheres (43). Other details are given elsewhere (44). The pentamer and decamer curves were merged in increments of 1% from 0 to 100% of the decamer for comparison with nine x-ray curves in 140 mm NaCl buffer containing 2 mm Ca2+, and six (50 mm NaCl) or four (140 mm NaCl) curves in buffers without Ca2+. Sedimentation coefficients were calculated from the x-ray hydrated sphere models using the HYDRO program (42, 45), and also from the crystal coordinates using HYDROPRO software (46). The default value of 0.31 nm was used to represent the hydration shell. To generate a putative decamer with the B faces now in contact with each other, one pentamer in the decamer model was translated by 7.4 nm relative to the other along its central symmetry axis.
TABLE 1.
X-ray scattering and sedimentation coefficient data and modeling of CRP
| Spheres | RG | R-factor |
s20,w0 |
KD | ||
|---|---|---|---|---|---|---|
| Pentamer | Decamera | |||||
| nm | % | S | ||||
| Experimental buffer | ||||||
| 50 mm NaCl, 2 mm Ca2+ | 6.5 ± 0.1 | 7.7–8.4 | 19 | |||
| 140 mm NaCl, 2 mm Ca2+ | 3.8–4.5; 3.8–4.3b | 6.4 ± 0.2 | 7.6–8.2 | 22 ± 5 | ||
| 250 mm NaCl, 2 mm Ca2+ | 6.4 ± 0.3 | 7.5–7.7 | NAc | |||
| 50 mm NaCl buffer, no Ca2+ | 3.8–4.1; 3.7–4.1b | 6.4 ± 0.1 | 7.5–8.9 | NA | ||
| 140 mm NaCl buffer, no Ca2+ | 3.6–4.1; 3.6–4.0b | 6.3 ± 0.1 | 7.3–7.7 | NA | ||
| Scattering modeling | ||||||
| CRP pentamer | 1256d | 3.7 | 6.0–6.2 | |||
| CRP decamer | 2492d | 4.2 | 9.8 | |||
| Best fit scattering curves (2 mm Ca2+) | 2.0–3.7 | 23 | ||||
| Best fit scattering curves (no Ca2+) | 1.8–3.5; 1.7–2.8e | |||||
a The experimental value corresponds to the decamer reaction boundary (Fig. 2).
b The first value is from the Guinier RG analyses (Fig. 4, A and B); the second is from the GNOM P(r) analyses (Fig. 5).
c NA, not available.
d The optimum number of hydrated spheres predicted from the sequence are 1310 and 2620 for pentamer and decamer, respectively.
e The first value is for 50 mm NaCl; the second is for 140 mm NaCl.
Surface Plasmon Resonance Data
The self-association of CRP was analyzed by surface plasmon resonance using a Biacore X100 instrument and version 1.1 of its evaluation software (GE Healthcare). Native CRP stored in 140 mm NaCl buffer containing 2 mm Ca2+ was coupled to the flow cell of a carboxylated dextran (CM5) research grade sensor chip via a standard amine coupling procedure according to the manufacturer's protocol. 10 μg/ml of CRP in 10 mm acetate buffer (pH 4.3) in the presence of at least 20 μm Ca2+ was injected over flow cell 2 for 45 min until 150 response units (RU) was attained. Between experiments, the CRP-immobilized chip was stored in a buffer containing 2 mm Ca2+. A control surface was prepared identically on flow cell 1 but without protein immobilization. Binding and equilibrium analyses using these CRP-immobilized chips were performed at 25 °C in duplicate or triplicate runs using the appropriate Biacore X100 wizards at flow rates of 10–30 μl/min. Regeneration after each run was achieved by pulsing 10 mm acetate buffer, 2 m NaCl (pH 4.6) across both flow cells once for 30 s. Running buffer was either Tris-buffered saline (10 mm Tris, 140 mm NaCl, 0.005% P20 surfactant, pH 8.0) or HBS (HEPES-buffered saline, 10 mm HEPES, 50 mm NaCl, 0.005% P20 surfactant, pH 8.0), with or without 2 mm CaCl2.
RESULTS
Size Exclusion Chromatography of CRP
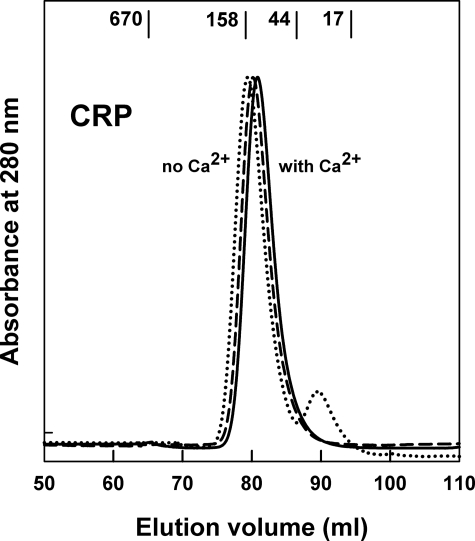
Size exclusion chromatography is a qualitative method that measures the effective hydrodynamic volume of a protein (47). CRP may self-associate into decamers (20) and/or dissociate into protomers when stored in the absence of Ca2+ (26, 27). This was reinvestigated by size exclusion chromatography of CRP in 140 mm NaCl buffer. CRP stored at 1.0 mg/ml at 4 °C in the presence of 2 mm Ca2+ eluted as a single clean peak in both the presence and absence of Ca2+ (Fig. 1). Molecular weight standards indicated that this peak corresponded to an apparent molecular mass of 80–94 kDa, and was therefore assigned to the pentamer. There was no evidence of a decamer. However, when CRP was stored at 4 °C in a buffer lacking Ca2+ for 12 weeks, a second peak was observed at a later elution volume (Fig. 1), in agreement with earlier observations (27). This second peak had an apparent molecular mass of 23 kDa and corresponded to ∼10% of the total eluted protein. It was concluded that partial dissociation into CRP monomers and other lower molecular weight forms occurred slowly in the absence of Ca2+. Small shifts in the elution position of CRP to earlier elution positions were observed with increased storage periods in Ca2+-depleted buffer (Fig. 1). This is attributed to small increases in the amounts of aggregated forms of CRP.
FIGURE 1.
Size exclusion chromatography of 1 mg/ml of CRP in 140 mm NaCl buffers. The elution positions of four globular standards are shown in kDa. CRP stored for over 12 weeks in Ca2+-containing buffer eluted as a single peak (solid line). A fresh sample of CRP eluted with the Ca2+-depleted buffer eluted earlier (dashed line). CRP stored for 12 weeks in Ca2+-depleted buffer (dotted line) eluted earlier still, and showed an additional smaller peak at 90 ml.
Sedimentation Velocity Analyses of CRP
AUC studies macromolecular structures in solution by following their sedimentation behavior in a high centrifugal force (48). The much improved resolution of sedimentation coefficient distribution analyses c(s) from sedimentation velocity experiments using Beckman Proteomelab XL-I instruments compared with older technology was used to monitor CRP self-association. The observed sedimentation boundaries in sedimentation velocity experiments were fitted to yield a c(s) plot according to the Lamm equation using SEDFIT (“Experimental Procedures”). This takes into account all the species present in the sample, although the algorithm assumes that the same frictional ratio applies for all the sedimenting species. The number of species is determined from the peaks observed in the c(s) plot. Macromolecular elongation is monitored through the sedimentation coefficient s20,w0 values. The effect of NaCl concentration on CRP was studied in the concentration range of 0.2 to 2.5 mg/ml in 50, 140, and 250 mm NaCl buffers, all containing 2 mm Ca2+. The c(s) analyses of interference optics data using SEDFIT resulted in good sedimentation boundary fits (top of Fig. 2, A–C). In all three buffers, a single species was observed at the lowest concentration of 0.2 mg/ml, whereas two species were observed above 1 mg/ml. The c(M) mass distribution plot showed a molecular mass of 118 ± 23 kDa for the single species. This was accordingly assigned to pentameric CRP, which has a sequence-calculated molecular mass of 115.0 kDa. The pentamer was consistently observed at s20,w0 values of 6.5 ± 0.1, 6.4 ± 0.2, and 6.4 ± 0.3 S at all concentrations in 50, 140, and 250 mm NaCl buffers, respectively, indicating that the NaCl concentration has no major effect on the CRP conformation (Fig. 2, A–C, Table 1). These values agree with the value of 6.6 S for pentameric CRP in 2 mm Ca2+ obtained using older Beckman Model E technology (25).
A pentamer-decamer equilibrium was identified from the c(s) plots. Above 0.2 mg/ml in 50 mm NaCl, 0.8 mg/ml in 140 mm NaCl, and 1.0 mg/ml in 250 mm NaCl, the c(s) distributions showed steady growth of a second species that indicats a higher oligomer in equilibrium with the pentamer (Fig. 2, A–C). This peak was attributed to the formation of decameric CRP. However, the s20,w0 values for this peak are variable in ranges from 7.7 to 8.4 S in 50 mm NaCl, 7.6 to 8.2 S in 140 mm NaCl, and 7.5 to 7.7 S in 250 mm NaCl. The molecular mass of 138 ± 10 kDa for this peak from c(M) analyses is too low compared with the expected value of 230 kDa. The variation in s20,w0 values and the low molecular mass indicate that this peak is not a second CRP species but is instead a reaction boundary corresponding to a rapid equilibrium between pentameric and decameric CRP on the time scale of sedimentation (49). In fact, a s20,w0 value of 9.5 S has been reported for a stabilized cross-linked form of decameric CRP (25). Integration of the two c(s) peak areas resulted in an estimated dissociation constant KD value of 19 μm for the CRP pentamer-decamer equilibrium in 50 mm NaCl, 2 mm Ca2+ buffer (Fig. 3A). Estimates of the KD values in 140 and 250 mm NaCl buffers were not possible because of poor resolution of the two peaks at lower concentrations. The rapid exchange accounts for the lack of resolution of pentamer and decamer by size exclusion chromatography (Fig. 1).
The removal of Ca2+ at CRP concentrations of 0.1 to 2.4 mg/ml in 50 mm and 140 mm NaCl buffers revealed the presence of smaller oligomers of CRP, together with pentamers and decamers by AUC. Good boundary fits were obtained (Fig. 2, D and E). At all concentrations, pentamers dominated the c(s) analyses with mean s20,w0 values of 6.4 ± 0.1 S in 50 mm NaCl and 6.3 ± 0.1 S in 140 mm NaCl (Table 1). These values were unchanged from those obtained in the presence of Ca2+ (Table 1). The c(M) analyses yielded a mean molecular mass of 119 ± 26 kDa in agreement with the assignment as pentamers. It was concluded that the loss of Ca2+ leads to only small effects in pentameric CRP, in agreement with size exclusion chromatography (Fig. 1). Two further observations were made. First, at all concentrations, one or two additional c(s) peaks were seen at 2.8 ± 0.2 and 4.3 ± 0.3 S in 50 mm NaCl, and at 3.0 ± 0.2 and 4.0 ± 0.3 S in 140 mm NaCl (Fig. 2, D and E). These peaks showed no concentration dependence and were therefore not in equilibrium with the pentamer. Because the c(M) analyses yielded mean molecular masses of 37 ± 5 and 72 ± 7 kDa, these peaks are possible dimer (46 kDa) and trimer (69 kDa) forms of CRP. Thus in the absence of Ca2+, CRP partially dissociates into lower molecular weight oligomers. This agrees with the size exclusion chromatography results after storage in the absence of Ca2+ (Fig. 1). Second, at 1.0 mg/ml and above, the CRP decamer was observable with s20,w0 values ranging from 7.5 to 8.9 and 7.3 to 7.7 S in 50 and 140 mm NaCl buffers, respectively, and a mean apparent molecular mass of 172 ± 59 kDa (Fig. 2, D and E). Peak integrations showed that the apparent KD value for the pentamer-decamer equilibrium is much weakened in higher salt (Fig. 3B). No KD values were estimated because of the presence of 2–33% of the lower molecular weight oligomers. It is concluded that Ca2+ is essential to stabilize the CRP pentamer and formation of decamers.
X-ray Scattering Analyses of CRP
X-ray scattering is a diffraction technique that studies the overall structure of biological macromolecules in random orientations in solution (50). This was used to observe the self-association and conformation of CRP in the presence and absence of Ca2+. Previous x-ray experiments with 2–6 mg/ml of CRP were performed on Station 7.3 at the Synchrotron Radiation Source at Daresbury, Cheshire, which utilized a linear detector with a limited Q range (24). In comparison, instrument ID02 at the European Synchrotron Radiation Facility in Grenoble, France, enabled much improved experiments in a more extended CRP concentration range of 0.3–5.4 mg/ml. ID02 was equipped with a flow cell to eliminate possible radiation effects and a two-dimensional area detector with better sensitivity and larger Q range (see “Experimental Procedures”).
X-ray Guinier analyses confirmed CRP self-association in 140 mm NaCl buffer with 2 mm Ca2+. Guinier analyses of the scattering data I(Q) at low Q values yielded the forward scattered intensity I(0) at zero Q and the radius of gyration RG. The I(0)/c value is proportional to molecular weight (where c is the CRP concentration in mg/ml) and RG monitors the degree of overall macromolecular elongation. Guinier plots gave linear RG fits within appropriate Q·RG limits (Fig. 4A). The I(0)/c values increased from 14.2 to 21.9 units between CRP concentrations from 0.3 to 5.4 mg/ml (Fig. 4C), in agreement with a self-association equilibrium. Assuming that this is a pentamer-decamer equilibrium, extrapolation of I(0)/c to zero concentration gave a value of 13.1 units for the pentamer, thus assigning 26.2 units for the decamer I(0)/c value, and a KD value of 26 μm (3.0 mg/ml) from the mid-point. The concentration dependence of the RG values ranged from 3.8 ± 0.1 to 4.5 ± 0.2 nm (Fig. 4E). Extrapolation to zero concentration yielded an RG value of 3.7 nm for the pentamer. The RG values agreed with the previously reported values of 4.1–4.5 nm at CRP concentrations of 2–6 mg/ml (24).
FIGURE 4.
X-ray scattering Guinier RG analyses for CRP. A, the filled circles correspond to the data points used to obtain RG values using a Q range of 0.06–0.36 nm−1 and the straight lines correspond to the best fit through these points. The Q·RG fit range is indicated by the arrows. In the presence of 2 mm Ca2+, from bottom to top, concentrations of 0.3, 1.3, 2.7, 4.0, and 5.4 mg/ml were used. B, in the absence of Ca2+, the Guinier fits were made in a Q range of 0.18–0.36 nm−1. From bottom to top, the CRP concentrations were 0.1, 0.3, 0.6, and 2.4 mg/ml. C, concentration dependence of the Guinier I(0)/c parameter in 140 mm NaCl buffer containing 2 mm Ca2+. The KD value was determined by a non-linear regression fit (solid line) for an exponential rise to a maximum (y = y0 + a(1-bx)), and the vertical dashed line corresponds to 26 μm. Error bars are shown where visible. D, concentration dependence of the Guinier I(0)/c parameter in 50 mm NaCl (●) and 140 mm NaCl buffer (○) without Ca2+. Note that the I(0)/c values were obtained in a different beam session from that of (c), therefore the I(0)/c values are not comparable. No KD value could be determined. E and F, the corresponding Guinier RG values for CRP in 140 mm NaCl buffer containing 2 mm Ca2+, and in 50 mm NaCl (●) and 140 mm NaCl buffer (○) without Ca2+.
The distance distribution function P(r) in real space was calculated from the I(Q) data, following an assumption of the value of the maximum dimension (Dmax). The P(r) curve represents all the distances between pairs of atoms within CRP, and provides an independent verification of the Guinier RG values. The P(r) curves from Fig. 5A yielded mean RG values of 3.8 ± 0.1 to 4.3 ± 0.1 nm, in good agreement with the Guinier values. Two maxima M1 at r = 3.1 nm and M2 at r = 5.7 nm correspond to the most frequently occurring interatomic distances within CRP. Comparison with the pentamer and decamer crystal structures (PDB codes 1b09 and 1lj7) (18, 20) showed that M1 and M2 could be assigned to the CRP pentamer and decamer, respectively. Although the r values of M1 and M2 remained unchanged with concentration, the intensity of M2 relative to M1 increased with CRP concentration. This showed that the proportion of decameric CRP increased with the concentration, as expected. The maximum dimension L of CRP is the r value when P(r) decreases to 0 at large r. The L values were unchanged with concentrations at 11–13 nm (Fig. 5A). The outermost ring diameter of the CRP pentamer is 11–13 nm by electron microscopy (23). Both are in good agreement with the longest dimensions measured from the unhydrated pentamer and decamer crystal structures of 10.5 and 11.9 nm, respectively, provided that these were increased to 11.1 and 12.5 nm, respectively, by the addition of a uniform shell of water molecules of 0.3 nm thickness on all surfaces (18, 20, 44). The lack of a significant change in L on decamer formation shows that the pentamers are arranged face-to-face within the decamer.
FIGURE 5.
Concentration dependence of the x-ray distance distribution P(r) for CRP. CRP was studied in 140 mm NaCl buffer containing 2 mm Ca2+ (A) and without Ca2+ (B). The two maxima in P(r) are denoted by M1 and M2 at 3.1 and 5.7 nm, respectively, assigned to pentamer and decamer, respectively. The maximum dimension is denoted by L. The CRP concentrations correspond to those in Fig. 4, A and B. The dashed line in A corresponds to 2.7 mg/ml; in B, this is shown again and normalized to M2 using that at 2.4 mg/ml to compare the effect of Ca2+ removal.
X-ray scattering showed that Ca2+ removal from CRP promoted some self-aggregation, and reduced the proportion of CRP decamer, but left the CRP conformation largely unchanged. Ca2+ removal was studied using a CRP concentration range of 0.1–2.5 mg/ml using 50 and 140 mm NaCl buffers without Ca2+. Trace amounts of nonspecific aggregation were readily visible in Guinier plots as non-linear increases in the I(Q) values at very low Q values. Accordingly a reduced Q range was employed for the RG fits (Fig. 4B). The I(0)/c values showed a reduced concentration dependence compared with that in the presence of Ca2+, increasing from 11.7 to 12.9 units and 8.6 to 11.6 units in 50 and 140 mm NaCl, respectively (Fig. 4D). This small increase showed that, even though the pentamer-decamer association is weakened compared with studies in the presence of Ca2+, the pentamer-decamer equilibrium is not eliminated by the removal of Ca2+. The RG values of CRP likewise increased in smaller ranges from 3.8 ± 0.0 to 4.1 ± 0.0 nm in 50 mm NaCl, and from 3.6 ± 0.1 to 4.1 ± 0.0 nm in 140 mm NaCl buffers (Fig. 4F). Extrapolation to zero concentration yielded an RG value of 3.5 nm for the pentamer in 140 mm NaCl (Fig. 4F). This is similar to the RG value of 3.7 nm for the pentamer in 140 mm NaCl with Ca2+ present (Fig. 4E). The P(r) curves yielded RG values of 3.7–4.1 nm in 50 mm NaCl (data not shown) and 3.6–4.0 nm in 140 mm NaCl (Fig. 5B), in agreement with the Guinier analyses. The r values of the peaks M1 (r = 3.1 nm) and M2 (r = 5.7 nm) were unchanged on Ca2+ removal, although the increase in the intensity of the M2 peak relative to M1 was now reduced. The L values of 10–13 nm showed no change from the L values in buffers containing Ca2+. Thus no large conformational change in CRP occurred on Ca2+ removal.
Modeling of Pentameric and Decameric CRP
Constrained modeling starting from crystal structures permits the molecular interpretation of scattering curves (50). Here, modeling was used to determine a KD value for the CRP pentamer-decamer equilibrium and establish the similarity of the solution structure of the CRP decamer to its crystal structure. The scattering curve for the CRP pentamer was calculated from its crystal structure with Ca2+ (PDB code 1b09) (18). Because several residues were missing and Ca2+ was not present in the CRP decamer crystal structure (PDB code 1lj7) (20), the decamer scattering curve with Ca2+ was calculated from a revised decamer crystal structure based on superimposition of two copies of the pentamer crystal structure on it. The modeled pentamer and decamer scattering curves resulted in RG values of 3.7 and 4.2 nm, respectively (Table 1). These agree well with the experimental RG value of 3.7 nm on extrapolation for the pentamer and the observed range of 3.8 ± 0.1 to 4.5 ± 0.2 nm for the pentamer-decamer equilibrium (Fig. 4E). The amounts of pentamer and decamer in each x-ray curve was quantified by comparison with 101 combinations of the modeled pentamer and decamer curves in 1% increments from 0 to 100 and 100 to 0%, respectively. Nine experimental x-ray curves at different concentrations in 140 mm NaCl with Ca2+ were fitted and assessed using a goodness-of-fit R-factor (Fig. 6A). Excellent R-factors of 2.0 to 3.7% were obtained for pentamer-decamer ratios that ranged from 74 to 26% at 0.3 mg/ml to 28 to 72% at 5.4 mg/ml (Fig. 6C). In comparison, the worst possible R-factor was 15.0%. The pentamer-decamer ratios resulted in a KD value of 16 μm (Fig. 6C), which agrees well with the Guinier-determined value of 26 μm (Fig. 4C). The modeled and experimental P(r) curves likewise agree well (Fig. 6E). The curve fits show that the decamer crystal structure with the two A-faces facing each other at the center is a good model for its solution structure.
FIGURE 6.
X-ray scattering curve fits for the CRP pentamer and decamer in the presence and absence of Ca2+. The Q range for the RG analyses is denoted by a horizontal bar. A, in buffer with 2 mm Ca2+, representative best fit curves are shown for CRP concentrations of 1.3 (54% pentamer/46% decamer), 2.7 (41% pentamer/59% decamer), and 5.4 mg/ml (28% pentamer/72% decamer) from bottom to top. B, in Ca2+-depleted buffer, representative best fit curves are shown for CRP concentrations of 0.3 (99% pentamer/1% decamer), 0.6 (96% pentamer/4% decamer), and 2.4 mg/ml (71% pentamer/29% decamer). C, KD determination for the CRP pentamer/decamer equilibrium in the presence of 2 mm Ca2+. Data points correspond to the best fit pentamer/decamer ratio for each CRP concentration between 0.3 and 5.4 mg/ml in 140 mm NaCl buffer. The data were fitted according to Fig. 4C, and the vertical dashed line corresponds to a KD of 16 μm. D, in Ca2+-depleted buffer, no KD value could be determined in either 50 (●) or 140 (○) mm NaCl buffer as this was outside the concentration range studied. E and F, comparison of the modeled (dashed lines) and experimental (full lines) P(r) curves in the presence (E) and absence (F) of Ca2+.
Constrained modeling also confirmed the NaCl concentration dependence of the pentamer-decamer equilibrium in the absence of Ca2+ that was seen by AUC and Guinier analyses. Comparison of the 101 pentamer/decamer curves with five experimental x-ray curves in 50 mm NaCl again resulted in excellent best-fit R-factors of 1.8 to 3.5% (see Table 1). The good quality of the curve fits indicated the absence of a large conformational change in CRP upon Ca2+ removal. The best fit ratios ranged from 100 to 0 to 56 to 44% of the pentamer/decamer (Fig. 6D). Likewise the four experimental curves in the 140 mm NaCl buffer resulted in excellent best fit R-factors of 1.7–2.8% (Fig. 6D and Table 1). The best fit ratios of 100–0 to 71–29% showed that the proportion of decamer had decreased in 140 mm NaCl after Ca2+ removal (Fig. 6D). No KD values were determined because these were too large to be measured. The modeled P(r) analyses showed good agreement with experimental data (Fig. 6F). Although lower molecular weight CRP oligomers will be present according to the c(s) plots by AUC (Fig. 3B), these will have a small effect on curve fits because the scattered intensities are proportional to the square of the molecular weight.
The CRP scattering curves show a minimum Q value of 0.87 nm−1 (Fig. 7A). This minimum is more prominent in the decamer than the pentamer. The mean diameter of the pentameric ring is given by 2π/Q as 7.2 nm, which is in good agreement with the neutron value of 7.4 nm (24) and a value of 7.5 nm measured from the pentamer and decamer crystal structures (Fig. 7B). The agreements support the A face-to-A face crystallographic model for the CRP decamer, in which one pentamer is rotated relative to the other by about 20°, then brought into contact at their A faces (20).
FIGURE 7.
Comparison of the x-ray scattering curves for CRP pentamer and decamer. The arrow corresponds to the minimum position in the scattering curve. A, comparison of the experimental scattering curves for CRP at 2.7 mg/ml in 2 mm Ca2+ buffer (black) and 2.4 mg/ml in Ca2+-depleted buffer (white). The intensity of I(Q) at the minimum (arrow) is lower in the presence of Ca2+, showing that more decamer has formed. B, comparison of the scattering curves for CRP for mixtures of pentamer and decamer calculated from crystal structures. The intensity of the I(Q) minimum decreases as the proportion of decamer increases from 0 to 100%.
Sedimentation coefficients s20,w0 for the pentamer and decamer were calculated from their crystal structure models (Table 1). Those for pentameric CRP were 6.0–6.2 S, in good agreement with the experimental value of 6.4 S. That for the decamer was 9.8 S, for which no experimental value from this study was available for comparison. The value of 9.8 S is in good agreement with the experimental value of 9.5 S for the cross-linked CRP decamer (25).
SPR Analyses of CRP
SPR monitors the interaction between a binding partner in solution (analyte) and a partner (ligand) immobilized at the surface of a sensor chip (31). CRP aggregates at damaged and apoptotic cells by binding to charged groups at their cell surfaces (6–8). Accordingly the self-association of CRP in solution with CRP immobilized on a sensor chip may provide insight on this interaction, provided that Ca2+ is present throughout the SPR experiments including the immobilization stage (“Experimental Procedures”) and that CRP is not denatured.
The NaCl concentration dependence of CRP self-association seen by AUC and scattering was confirmed by SPR, which showed that 140 mm NaCl buffer is essential for this. CRP was flowed across a sensor chip surface containing 150 RU of immobilized CRP, which was prepared in the presence of Ca2+ (“Experimental Procedures”). The binding analysis of 50 μm (5.8 mg/ml) CRP in 50 mm NaCl buffer in the presence of Ca2+ showed that CRP aggregated on the chip surface, as evidenced by the extremely high binding response to greater than 15,000 RU. This was rapidly followed by CRP dissociation to baseline levels (Fig. 8A). On increasing the salt to 140 mm NaCl, the corresponding analysis of 50 μm CRP resulted in a much reduced but more rapid binding response of 34 RU, followed by its rapid dissociation (Fig. 8B). This represents 23% saturation of immobilized CRP. At a lower CRP concentration of 20 μm (2.3 mg/ml), the binding responses were reduced to 245 RU in 50 mm NaCl and 24 RU in 140 mm NaCl (Fig. 8, C and D).
FIGURE 8.
Surface plasmon resonance analysis of CRP self-association. Data correspond to buffers containing 2 mm Ca2+ unless stated otherwise. A and B, 50 μm CRP was flowed over immobilized CRP in 50 (A) and 140 (B) mm NaCl buffers. An abnormally high response (>15,000 RU) was observed in 50 mm NaCl, whereas in 140 mm NaCl the response was normal (∼35 RU). C and D, comparison of binding of CRP in the presence (solid lines) and absence (dotted lines) of Ca2+. 20 μm CRP was flowed over immobilized CRP in buffers containing 50 (C) and 140 (D) mm NaCl. Approximately 10-fold more response was observed in C than D. The dotted lines did not return to baseline levels in C and D. E and F, equilibrium analyses for CRP self-association in 50 (E) and 140 (F) mm NaCl buffer. The binding response increased continuously between 1 and 20 μm in E, whereas saturation was approached in F, from which a KD value of 23 μm was determined from CRP concentrations between 1 and 35 μm. G and H, sensorgrams used for the equilibrium analyses, where panel G corresponds to the data in E, and panel H corresponds to the data in F.
Dissociation constants KD for CRP self-association were determined using equilibrium SPR studies in 50 mm NaCl and 140 mm NaCl buffers with Ca2+ present. At concentrations of 1–20 μm in 50 mm NaCl buffer, CRP showed a continuously increasing binding response that is consistent with aggregate formation (Fig. 8, E and G). A KD value could not be determined because binding saturation was not attained. In 140 mm NaCl buffer, a KD value of 23 μm was determined using CRP concentrations between 1 and 35 μm (Fig. 8, F and H). This agrees well with the KD values of 26 and 16 μm in 140 mm NaCl from the x-ray analyses above, and shows that immobilized CRP self-associated with CRP with the same affinity as CRP in solution when monitored by x-ray scattering.
The corresponding experiments in the absence of Ca2+, but using the same immobilized CRP sensor chip, showed that CRP binding to immobilized CRP was much reduced. Using 20 μm CRP, the binding responses were reduced from 245 to 115 RU in 50 mm NaCl buffer, and from 24 to 19 RU in 140 mm buffer (Fig. 8, C and D). Interestingly, with both buffers, CRP did not completely dissociate from immobilized CRP on the sensor surface back to baseline levels when this was compared with buffers containing Ca2+ (dashed lines in Fig. 8, C and D). This suggested that, in the absence of Ca2+, lower molecular weight oligomers of CRP bind irreversibly to partially dissociated immobilized CRP on the sensor chip surface to form aggregates. This is consistent with our AUC results showing reduced CRP decamer formation and partial dissociation of CRP in Ca2+-depleted buffers.
DISCUSSION
An essential pre-requisite for elucidating CRP function is an understanding of its solution properties. The properties of unbound CRP are important for several biological functional processes involving CRP. Here, we have shown that CRP in 2 mm Ca2+ exists in a rapid pentamer-decamer equilibrium in solution and on surfaces using a combination of three different analytical instrumentation methods with improved resolutions compared with previous studies. To our knowledge, this is the first identification for decameric CRP as a molecular species in physiologically relevant buffer conditions and its dependence on NaCl concentration. The CRP concentration in plasma ranges from as little as 50 ng/ml (0.4 nm) at normal levels to over 0.5 mg/ml (4.4 μm) during acute phase conditions (1). Calcium is present in plasma at 2.5 mm, of which ionic Ca2+ is present at 1.18 mm (28). Only CRP pentamers and decamers are observed in 2 mm Ca2+ in CRP concentrations between 0.1 and 5.4 mg/ml (0.9–47 μm), where these concentrations include the physiological range of the CRP acute phase response. The formation of decameric CRP with a mean KD value of 22 ± 5 μm can be relevant for the interaction of CRP with its ligands in plasma. Most importantly, our AUC data shows that the non-physiological removal of Ca2+ causes CRP to dissociate slowly into lower oligomers and reduces the proportion of decamers. Many previous functional studies of CRP have reported the existence of denatured or monomeric forms of CRP in the absence of Ca2+, and have suggested that these are functionally relevant (9–13). There is currently no rigorous evidence for the existence of monomeric (i.e. denatured) CRP in vivo (1). The standard preparation protocol for CRP in this study involved 2 mm Ca2+ throughout (32). Because our AUC data confirm that pentameric CRP is stable in the presence of Ca2+, functional or structural experiments with CRP in which Ca2+ is omitted or insufficient need to be interpreted with caution.
Functional Relevance of the Pentamer-Decamer Equilibrium
The CRP pentamer-decamer equilibrium is attributed to interactions between two CRP A faces in the pentamer to form decamers (see below) and will be important for the acute phase response when CRP concentrations in plasma reach 4.4 μm. This rapid equilibrium between CRP pentamers and decamers provides a means to reduce nonspecific protein binding to CRP and therefore may maintain the integrity of CRP when CRP is abundant in plasma. This may be functionally important because plasma contains at least 70 mg/ml of other proteins. Further work will be required to establish that the pentamer-decamer exchange occurs in blood, plasma, or serum. Many low-affinity rapid exchange ionic strength-dependent reactions may not occur in the context of IgG antibody, human serum albumin, and other abundant plasma proteins.
Because C1q and complement factor H are presumed to bind to the A face of CRP, whereas lipids bind to its B face, decamer formation will affect only the former ligands. Given that the CRP decamer KD value is 22 μm, CRP decamer formation shows that CRP will only bind to C1q, factor H, and other ligands at its A face if the KD value of these ligand interactions is well below this KD value. Our study of factor H interaction with CRP indeed suggests that CRP decamer formation is inhibited in the presence of factor H, for which the KD value is 4 μm (51). In the decamer, the CRP B faces remain exposed and this will permit lipid binding at damaged host cells or other surfaces irrespective of whether pentamers or decamers are present. At the present time, no direct function is attributable to the decamer form of CRP, although a role in host defense or apoptotic cell clearance cannot be ruled out.
Molecular Explanation for Decamer Formation
In previous studies, self-association of CRP was reported by x-ray and neutron scattering and by AUC, but its occurrence or importance had not been explained (24, 25). Here, the molecular basis of self-association was identified by a combination of AUC c(s) analyses and x-ray scattering modeling. The AUC data revealed a pentamer-decamer equilibrium in the presence of Ca2+. Although pentamers were unambiguously identified as the 6.4 S c(s) peak, the second c(s) peak corresponds to a reaction boundary that could not be assigned to a decamer. This ambiguity was resolved by the excellent x-ray curve fits based on the pentameric and decameric CRP crystal structures. The KD values of 19 μm (50 mm NaCl) by AUC, 26 and 16 μm (140 mm NaCl) by x-ray Guinier fits and modeling, and 23 μm (140 mm NaCl) by SPR show consistency. The mean KD value of 22 ± 5 μm (140 mm NaCl) indicates that 0–15% of CRP will be decameric in its physiological concentration range of 0.4 nm to 4.4 μm if no other factors are involved.
The decamer is formed in 140 mm NaCl from contacts between either the two A faces or the two B faces of the pentamers. The A faces were preferred as the buried interface because this is observed in the Ca2+-free CRP decamer crystal structure, excellent x-ray modeling fits were obtained with the decamer crystal structure (20), and appropriate ionic interactions at the A faces were observed that account for the NaCl concentration dependence of decamer formation. These results imply that the observed 15-mers of CRP in chemically cross-linked CRP are not physiologically relevant (4).
CRP decamer formation is reduced with increased NaCl concentration, and this indicates that the A face-A face interface in the decamer is stabilized by ionic interactions. The partially disordered decamer crystal structure initially suggested that 10 pairs of Thr173 and Pro179 side chains form the closest contact between the A faces (20). These are uncharged residues that cannot form salt bridges. If the Ca2+-bound pentamer crystal structure at higher resolution was superimposed onto the decamer crystal structure (18), it turns out that 10 pairs of charged Asp-169 and Arg-188 residues on the A faces are well positioned to form the requisite ionic bridges (Fig. 9A). No clear candidates for ionic bridges between the B faces have been detected by modeling so far. Lys69–Asp70 and Glu14–Arg47 pairs exist within each B face, however, these cancel each other out. Additionally, there is a surplus of acidic residues on the B face (in addition to those involved with Ca2+ and phosphocholine binding) that may repel each other.
FIGURE 9.
Pentamers and decamers of CRP. A, crystal structures for pentameric CRP with bound Ca2+ (cyan and green ribbons) were superimposed onto the crystal structure of decameric CRP without Ca2+ to reveal 10 pairs of Arg188 (blue) and Asp169 (red) ionic bridges proposed to stabilize CRP decamer formation between the two A faces. Three pairs of bridges are shown in this view. B, the B face of pentameric CRP is viewed in blue with bound Ca2+, and magenta without Ca2+. The Ca2+-binding loop comprising amino acid residues 138–150 in each protomer is colored red (with Ca2+) or orange (without Ca2+). C, pentameric CRP (blue, with bound Ca2+ shown as yellow spheres) self-associates in rapid equilibrium in the presence or absence of Ca2+ to form decamer (green and cyan) through contacts between the A faces in an NaCl concentration-dependent manner. This has a mean dissociation constant KD value of 22 μm determined in 140 mm NaCl containing Ca2+. In the absence of Ca2+, CRP irreversibly dissociates into individual protomers (magenta) and decamer formation is reduced. When Ca2+ is present, pentameric CRP binds to macromolecular ligands to form complexes that interact with C1q to activate the classical pathway of complement.
The NaCl concentration dependence of decamer formation shows that CRP self-association may be affected by the local environment. For example, CRP binding to lipids at the B faces may influence pentamer-decamer association. We observed indefinite CRP aggregation at immobilized CRP in 50 mm NaCl in the SPR analyses. In this context, CRP is known to interact with a variety of polycations, as well as with polyanion-polycation complexes (52–54).
Structural Role of Calcium
The present study has clarified the effect of removal of Ca2+ on CRP. Previous high resolution CRP crystallographic studies identified the role of Ca2+ in binding to the B-face. Ca2+ binds at a double site on the B face (Fig. 9B). Ca2+ site 1 is coordinated by amino acids Asp60, Asn61, Glu138, Asp140, and Leu139 (a total of five ligands), whereas Ca2+ site 2 is coordinated by Gln138, Asp140, Gln150, and possibly Glu147 (18, 22). Both Ca2+-binding sites in CRP possess equal affinity for Ca2+ in solution with a KD of 60 μm (55). A recent study reported a slightly lower KD of 30 μm using SPR (56). The presence of at least 2 mm Ca2+ in plasma is well in excess of the KD value for Ca2+ binding to CRP, and will result in 97–99% occupancy. This is necessary to ensure that CRP is in its native conformation. Although two CRP pentamer crystal structures were reported in the presence of Ca2+ (18, 22), CRP was crystallized as decamers in 100 mm NaCl buffer in the absence of Ca2+ (20). Our AUC data now explains the crystallization of Ca2+-free decamers, simply because decamers were observable by AUC under these conditions. Thus Ca2+ bound to the B face is not structurally involved with decamer formation, and this also argues for decamer formation through A face contacts. In addition, Ca2+ is not structurally related to the contacts between the five protomers in the pentamer, each of which is stabilized by four ionic bridges that span the interface (Glu42–Lys119, Glu101–Lys201, Asp155–Arg118, and Glu197–Lys123). Accordingly our observation of weaker decamer formation and dissociation of smaller oligomers on the removal of Ca2+ is best explained by destabilization of the double β-sheet structure of the CRP protomer. Indeed, comparisons of CRP crystal structures with and without Ca2+ bound (Fig. 9B) show that there is a 0.174-nm root mean square difference in residue positions (18). This conformational sensitivity of the β-sheets to Ca2+ removal provides an explanation for the notable absence of polymorphic forms of CRP (1), because any residue changes will perturb the CRP pentamer structure.
Several studies have proposed that alterations in Ca2+ binding to CRP are important for the mechanism of CRP action. In the absence of Ca2+, CRP is partially dissociated to form lower molecular weight oligomers (Fig. 9C). Our AUC studies show that these smaller oligomers were not in equilibrium with pentameric CRP, and were not dominant. The existence of these lower oligomers of CRP consisting of monomers to trimers (mCRP) has been reported previously in functional studies under extreme conditions of denaturing pH or temperature (9–11) or Ca2+ depletion (26, 27). Given the prevalence of Ca2+ in plasma, an allosteric mechanism involving the removal or addition of Ca2+ during CRP function appears unlikely, as proposed in some studies (20, 25).
Native and Denatured CRP
An understanding of the solution structural behavior of CRP is crucial to elucidate its function in complement and the acute phase response. The interaction of native CRP with complement factor H is described in the accompanying article (51). Here, our main results show that the three key parameters for rigorous experimental studies are 2 mm Ca2+, 140 mm NaCl, and 0.01–4.4 μm CRP. Our data show that native CRP exists as stable pentamers in rapid equilibrium with decamers in 2 mm Ca2+ and 140 mm NaCl (Fig. 9). The spontaneous formation of denatured mCRP under conditions of Ca2+ depletion is slow but observable by AUC. Importantly, our SPR studies of CRP binding to immobilized CRP surfaces show that CRP aggregates in low salt, but not in 140 mm NaCl. The latter results are cautionary in that experimental work with surface-bound CRP can readily lead to artifactual observations. This is attributable to the ease with which Ca2+ can be removed from CRP and the sensitivity of CRP self-association to the NaCl concentration. Cases have been reported when CRP becomes denatured and shows subunit rearrangement on binding to membranes (57). Studies have suggested that mCRP may be produced in normal or inflamed human and animal tissue (14–17), and that mCRP possesses distinct antigenic properties from native CRP (9–12). Further work will be required to establish the existence of mCRP in vivo and a mechanism for its formation. For these studies, a proper appreciation of the oligomeric forms of CRP and the need to work with an appropriate buffer will be essential to understand the role of CRP in the acute phase response and in complement activation.
Acknowledgments
We thank Prof. M. B. Pepys and Dr. Patrizia Mangione for kindly providing purified CRP samples for this study. We thank Dr. Pierre Panine and Dr Anuj Shukla (ESRF, Grenoble) for excellent x-ray instrumental support.
This work was awarded a poster prize at the 18th International Analytical Ultracentrifugation Conference, Uppsala University, Sweden (September 13–18, 2009).
J. T. Eaton and S. J. Perkins, unpublished software.
- CRP
- C-reactive protein
- mCRP
- modified CRP
- AUC
- analytical ultracentrifugation
- RU
- response/resonance unit
- PDB
- Protein Data Bank
- SPR
- surface plasmon resonance.
REFERENCES
- 1.Pepys M. B., Hirschfield G. M. (2003) J. Clin. Invest. 111, 1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volanakis J. E., Kaplan M. H. (1971) Proc. Soc. Exp. Biol. Med. 136, 612–614 [DOI] [PubMed] [Google Scholar]
- 3.Black S., Kushner I., Samols D. (2004) J. Biol. Chem. 279, 48487–48490 [DOI] [PubMed] [Google Scholar]
- 4.Jiang H. X., Siegel J. N., Gewurz H. (1991) J. Immunol. 146, 2324–2330 [PubMed] [Google Scholar]
- 5.Filep J., Földes-Filep E. (1989) Life Sci. 44, 517–524 [DOI] [PubMed] [Google Scholar]
- 6.Volanakis J. E. (1982) Ann. N.Y. Acad. Sci. 389, 235–250 [DOI] [PubMed] [Google Scholar]
- 7.De Beer F. C., Soutar A. K., Baltz M. L., Trayner I. M., Feinstein A., Pepys M. B. (1982) J. Exp. Med. 156, 230–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowe I. F., Soutar A. K., Trayner I. M., Thompson G. R., Pepys M. B. (1984) Clin. Exp. Immunol. 58, 237–244 [PMC free article] [PubMed] [Google Scholar]
- 9.Potempa L. A., Maldonado B. A., Laurent P., Zemel E. S., Gewurz H. (1983) Mol. Immunol. 20, 1165–1175 [DOI] [PubMed] [Google Scholar]
- 10.Potempa L. A., Zeller J. M., Fiedel B. A., Kinoshita C. M., Gewurz H. (1983) Inflammation 12, 391–405 [DOI] [PubMed] [Google Scholar]
- 11.Potempa L. A., Siegel J. N., Fiedel B. A., Potempa R. T., Gewurz H. (1987) Mol. Immunol. 24, 531–541 [DOI] [PubMed] [Google Scholar]
- 12.Khreiss T., József L., Hossain S., Chan J. S., Potempa L. A., Filep J. G. (2002) J. Biol. Chem. 277, 40775–40781 [DOI] [PubMed] [Google Scholar]
- 13.Taylor K. E., van den Berg C. W. (2007) Immunology 120, 404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samols D., MacIntyre S. S., Kushner I. (1985) Biochem. J. 227, 759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees R. F., Gewurz H., Siegel J. N., Coon J., Potempa L. A. (1988) Clin. Immunol. Immunopathol. 48, 95–107 [DOI] [PubMed] [Google Scholar]
- 16.Egenhofer C., Alsdorff K., Fehsel K., Kolb-Bachofen V. (1993) Hepatology 18, 1216–1223 [PubMed] [Google Scholar]
- 17.Diehl E. E., Haines G. K., 3rd, Radosevich J. A., Potempa L. A. (2000) Am. J. Med. Sci. 319, 79–83 [DOI] [PubMed] [Google Scholar]
- 18.Thompson D., Pepys M. B., Wood S. P. (1999) Struct. Fold. Des. 7, 169–177 [DOI] [PubMed] [Google Scholar]
- 19.Gaboriaud C., Thielens N. M., Gregory L. A., Rossi V., Fontecilla-Camps J. C., Arlaud G. J. (2004) Trends Immunol. 25, 368–373 [DOI] [PubMed] [Google Scholar]
- 20.Ramadan M. A., Shrive A. K., Holden D., Myles D. A. A., Volanakis J. E., DeLucas L. J., Greenhough T. J. (2002) Acta Crystallogr. Sect. D 58, 992–1001 [DOI] [PubMed] [Google Scholar]
- 21.Volanakis J. E., Clements W. L., Schrohenloher R. E. (1978) J. Immunol. Methods 23, 285–295 [Google Scholar]
- 22.Shrive A. K., Cheetham G. M., Holden D., Myles D. A., Turnell W. G., Volanakis J. E., Pepys M. B., Bloomer A. C., Greenhough T. J. (1996) Nat. Struct. Biol. 3, 346–354 [DOI] [PubMed] [Google Scholar]
- 23.Osmand A. P., Friedenson B., Gewurz H., Painter R. H., Hofmann T., Shelton E. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 739–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perkins S. J., Pepys M. B. (1986) Protides Biol. Fluids Proc. Colloq. 34, 323–326 [Google Scholar]
- 25.Blizniukov O. P., Kozmin L. D., Falikova V. V., Martynov A. I., Tischenko V. M. (2003) Mol. Biol. 37, 912–918 [PubMed] [Google Scholar]
- 26.Motie M., Brockmeier S., Potempa L. A. (1996) J. Immunol. 156, 4435–4441 [PubMed] [Google Scholar]
- 27.Wu Y., Li S. R., Wang H. W., Sui S. F. (2002) Biochemistry 67, 1377–1382 [DOI] [PubMed] [Google Scholar]
- 28.Hurwitz S. (1996) Crit. Rev. Biochem. Mol. Biol. 31, 41–100 [DOI] [PubMed] [Google Scholar]
- 29.Bonner A., Perrier C., Corthésy B., Perkins S. J. (2007) J. Biol. Chem. 282, 16969–16980 [DOI] [PubMed] [Google Scholar]
- 30.Bonner A., Almogren A., Furtado P. B., Kerr M. A., Perkins S. J. (2009) J. Biol. Chem. 284, 5077–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanious F. A., Nguyen B., Wilson W. D. (2008) Methods Cell Biol. 84, 53–77 [DOI] [PubMed] [Google Scholar]
- 32.De Beer F. C., Pepys M. B. (1982) J. Immunol. Methods 50, 17–31 [DOI] [PubMed] [Google Scholar]
- 33.Nelson S. R., Tennent G. A., Sethi D., Gower P. E., Ballardie F. W., Amatayakul-Chantler S., Pepys M. B. (1991) Clin. Chim. Acta 200, 191–199 [DOI] [PubMed] [Google Scholar]
- 34.Perkins S. J. (1986) Eur. J. Biochem. 157, 169–180 [DOI] [PubMed] [Google Scholar]
- 35.Schuck P. (1998) Biophys. J. 75, 1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayanan T., Diat O., Bösecke P. (2001) Nucl. Instrum. Methods Phys. Res. A 467–468, 1005–1009 [Google Scholar]
- 38.Li K., Okemefuna A. I., Gor J., Hannan J. P., Asokan R., Holers V. M., Perkins S. J. (2008) J. Mol. Biol. 384, 137–150 [DOI] [PubMed] [Google Scholar]
- 39.Gilbert H. E., Eaton J. T., Hannan J. P., Holers V. M., Perkins S. J. (2005) J. Mol. Biol. 346, 859–873 [DOI] [PubMed] [Google Scholar]
- 40.Glatter O., Kratky O. (eds) (1982) Small-angle X-ray Scattering, Academic Press, New York [Google Scholar]
- 41.Semenyuk A. V., Svergun D. I. (1991) J. Appl. Crystallogr. 24, 537–540 [Google Scholar]
- 42.Ashton A. W., Boehm M. K., Gallimore J. R., Pepys M. B., Perkins S. J. (1997) J. Mol. Biol. 272, 408–422 [DOI] [PubMed] [Google Scholar]
- 43.Perkins S. J., Weiss H. (1983) J. Mol. Biol. 168, 847–866 [DOI] [PubMed] [Google Scholar]
- 44.Perkins S. J. (2001) Biophys. Chem. 93, 129–139 [DOI] [PubMed] [Google Scholar]
- 45.Garcia de la Torre J., Navarro S., Lopez Martinez M. C., Diaz F. G., Lopez Cascales J. J. (1994) Biophys. J. 67, 530–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.García de la Torre J., Huertas M. L., Carrasco B. (2000) Biophys. J. 78, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mori S., Barth H. G. (1999) Size Exclusion Chromatography, Springer-Verlag, Berlin [Google Scholar]
- 48.Cole J. L., Lary J. W., Moody T. P., Laue T. M. (2008) Methods Cell Biol. 84, 143–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dam J., Schuck P. (2005) Biophys. J. 89, 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins S. J., Okemefuna A. I., Fernando A. N., Bonner A., Gilbert H. E., Furtado P. B. (2008) Methods Cell Biol. 84, 375–423 [DOI] [PubMed] [Google Scholar]
- 51.Okemefuna A. I., Nan R., Miller A., Gor J., Perkins S. J. (2010) J. Biol. Chem. 285, 1053–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel J., Rent R., Gewurz H. (1974) J. Exp. Med. 140, 631–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegel J., Osmand A. P., Wilson M. F., Gewurz H. (1975) J. Exp. Med. 142, 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Claus D. R., Siegel J., Petras K., Skor D., Osmand A. P., Gewurz H. (1977) J. Immunol. 118, 83–87 [PubMed] [Google Scholar]
- 55.Kinoshita C. M., Ying S. C., Hugli T. E., Siegel J. N., Potempa L. A., Jiang H., Houghten R. A., Gewurz H. (1989) Biochemistry 28, 9840–9848 [DOI] [PubMed] [Google Scholar]
- 56.Christopeit T., Gossas T., Danielson U. H. (2009) Anal. Biochem. 391, 39–44 [DOI] [PubMed] [Google Scholar]
- 57.Wang H. W., Sui S. F. (2001) Biochem. Biophys. Res. Commun. 288, 75–79 [DOI] [PubMed] [Google Scholar]