Abstract
DNA double-strand breaks (DSBs) represent one of the most serious forms of DNA damage that can occur in the genome. Here, we show that the DSB-induced signaling cascade and homologous recombination (HR)-mediated DSB repair pathway can be genetically separated. We demonstrate that the MRE11-RAD50-NBS1 (MRN) complex acts to promote DNA end resection and the generation of single-stranded DNA, which is critically important for HR repair. These functions of the MRN complex can occur independently of the H2AX-mediated DNA damage signaling cascade, which promotes stable accumulation of other signaling and repair proteins such as 53BP1 and BRCA1 to sites of DNA damage. Nevertheless, mild defects in HR repair are observed in H2AX-deficient cells, suggesting that the H2AX-dependent DNA damage-signaling cascade assists DNA repair. We propose that the MRN complex is responsible for the initial recognition of DSBs and works together with both CtIP and the H2AX-dependent DNA damage-signaling cascade to facilitate repair by HR and regulate DNA damage checkpoints.
Keywords: DNA Damage, DNA Recombination, DNA Repair, Protein Translocation, Protein-Protein Interactions, Tumor Suppressor
Introduction
There are two major pathways to repair DSBs,3 the nonhomologous end joining pathway and homologous recombination (HR) pathway (1). The conversion of DNA double-stranded ends to single-stranded DNA (ssDNA) regions is considered to be a key step that controls not only HR repair but also DNA damage checkpoints (2). The MRN complex, comprising MRE11, RAD50, and NBS1, has been implicated in the detection of DSBs, DNA ends resection (3, 4), recombination (5), and S or G2/M checkpoint control (6–8). More recently, CtIP (also known as RBBP8) has been shown to function together with the MRN complex to process DSB ends and generate ssDNA regions (9, 10).
In response to DSBs, the ataxia-telangiectasia mutated (ATM)/ataxia-telangiectasia and Rad3-related (ATR)-dependent phosphorylation of histone variant H2AX creates γH2AX, which is believed to be the initial signal for subsequent accumulation of various signaling and repair proteins to DNA breaks to form so-called ionizing radiation-induced foci (11–14). We and others have shown that MDC1 directly binds γH2AX through its BRCA1 C-terminal (BRCT) domains (15, 16), and the phosphorylation of six SDTDX(D/E) repeats in the MDC1 N terminus functions to recruit NBS1 and regulate the intra-S phase checkpoint in response to DNA damage (17–20). MDC1 also recruits E3 ubiquitin ligase RNF8 in a phosphorylation-dependent manner, and the latter is responsible for tethering 53BP1 and the RAP80-CCDC98-BRCA1 complex at damage sites (21–26). Although the extensive studies noted above have demonstrated that the histone variant H2AX is a central regulator of ionizing radiation-induced focus formation and the stable accumulation of many DNA damage signaling and repair proteins to sites of DNA breaks, surprisingly H2AX seems to be dispensable for the initial recognition of DNA breaks because transient localization of several DNA damage repair proteins were observed in the absence of H2AX (27). The analysis of H2AX-deficient cells also showed that H2AX is not essential for nonhomologous end joining or HR, although it somehow modulates the efficiency of these repair pathways (27–31). Moreover, although disruption of components directly or indirectly involved in HR pathway, such as ATR (32, 33), the MRN complex (34–36), BRCA1 (37), BRCA2 (38), RAD51 (39), and the recently identified CtIP (40), resulted in embryonic lethality, the H2AX−/− mice exhibited relatively mild phenotypes with some degree of genomic instability (12). In fact, mice lacking other key factors involved in the DNA damage signaling cascade such as ATM (41–43), MDC1 (15), and 53BP1 (44, 45) all display increased genomic instability and are prone to tumorigenesis; nevertheless these null mice are viable.
The fact that the effects of H2AX deficiency on DSB repair are subtle suggests that γH2AX might regulate repair of selected DSBs or assist specific repair pathways (46). The role of γH2AX in facilitating DNA repair may be mediated by the contribution of γH2AX to signaling and the associated efficient activation of DNA damage-induced checkpoint response. However, it is unlikely that γH2AX would play a central role in dictating DNA damage repair. These observations prompted us to propose that the HR-mediated DSB repair pathway can be initiated independently of the known H2AX-mediated DNA damage-signaling cascade. Indeed, in this study we showed that the MRN complex can initiate DNA end resection and HR repair in the absence of H2AX. Moreover, the MRN complex is also involved in the recruitment of other signaling and repair factors transiently at DSB sites in H2AX-deficient cells. Together this study highlighted a critical role of the MRN complex at an early stage of DNA damage response.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies against the γH2AX, MDC1, 53BP1, and RAD51 were previously described (47–49). The anti-Myc and anti-BRCA1 antibodies were obtained from Santa Cruz Biotechnology. Anti-NBS1 antibody was obtained from Calbiochem. Anti-MRE11 antibody was purchased from Novus Biologicals. Anti-RPA2 antibody was obtained from Abcam. Anti-γ-tubulin and anti-FLAG (M2) were obtained from Sigma. Andre Nussenzweig (National Institutes of Health, Bethesda, MD) kindly provided us with anti-mouse NBS1 and MRE11 antibodies and Richard Baer (Columbia University, New York, NY) kindly provided us with monoclonal mouse anti-CtIP antibody.
Cell Culture, Transfection, and siRNAs
U2OS and 293T cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, 1 mm sodium pyruvate, and 1% penicillin and streptomycin. Plasmid transfection was performed using Lipofectamine 2000 (Invitrogen) following manufacturer's instruction. The sequence of RAD51 siRNA was CUAAUCAGGUGGUAGCUCAUU; the sequence of H2AX siRNA was CAACAAGAAGACGCGAAUCdTdT; the sequence of NBS1 siRNA was CCAACUAAAUUGCCAAGUAUU; the sequence of MRE11 siRNA was GGAGGUACGUCGUUUCAGAdTdT; and the siRNA targeting mouse NBS1 was CUCCAAAGCUAACAACGUAdTdT. The sequences for MDC1 and CtIP siRNA were previously described (50, 51). The siRNAs transfection was performed using Oligofectamine (Invitrogen) following the manufacturer's instructions. When transfecting mouse NBS1 siRNA into MEFs, the transfection was repeated three times at 24-h intervals to achieve maximal RNA interference effect.
Immunoblotting
The cells were lysed with NETN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) on ice for 30 min. The cleared cell lysates were then collected and boiled in 2× Laemmli buffer and run on SDS-PAGE. The membranes were blocked in 5% milk in TBST buffer and then probed with antibodies as indicated.
Immunostaining
The cells cultured on coverslips were treated with ionizing radiation (IR) followed by recovery for indicated period. The cells were then washed with phosphate-buffered saline, pre-extracted with 0.5% Triton solution for 5 min, and fixed with 3% paraformaldehyde for 12 min. The coverslips were washed with phosphate-buffered saline and then immunostained with primary antibodies in 5% goat serum for 60 min. The coverslips were washed and incubated with secondary antibodies conjugated with rhodemine or fluorescein isothiocyanate for 60 min. The cells were then stained with 4′,6′-diamino-2-phenylindole to visualize nuclear DNA. The coverslips were mounted onto glass slides with anti-fade solution and visualized using a Nikon ECLIPSE E800 fluorescence microscope with a Nikon Plan Fluor 40× oil objective lens (NA 1.30) at room temperature. The cells were photographed and analyzed using a SPOT camera (Diagnostic Instruments, Inc) and Photoshop software (Adobe).
G2/M Checkpoint Assay
G2/M checkpoint assay was performed as described previously (51). Briefly, the cells were treated with 2 Gy IR. One hour later, the cells were fixed with 70% (v/v) ethanol overnight and then stained with anti-phospho-histone H3 (Ser10) antibody and propidium iodide. The samples were analyzed by flow cytometry to determine the percentages of cells in mitosis.
Homologous Recombination Assay
A U2OS cell clone stably expressing HR reporter direct repeat of GFP (DR-GFP) was described previously (52). This reporter consists of two differentially mutated GFP genes oriented as direct repeats. Expression of I-SceI endonuclease will generate a site-specific DSB between the mutated GFP genes, which, when repaired by gene conversion, results in a functional GFP gene. Briefly, 2 days after transfection with siRNA, 1 × 106 U2OS DR-GFP cells were electroporated with 20 μg of pCBASce (an I-SceI expression vector). The cells were harvested 2 days after electroporation and subjected to flow cytometry analysis to determine the percentages of GFP-positive cells, which result from HR repair induced by DNA DSBs. The samples were analyzed in a Becton-Dickinson FACScan on a green (FL1) versus orange (FL2) fluorescence plot.
RESULTS
The MRN Complex Is Responsible for the Recruitment of Repair and Signaling Proteins to DSB Sites in the Absence of H2AX
Although the phosphorylated form of H2AX is required for the retention of DNA damage and repair proteins in the vicinity of DNA lesions, H2AX phosphorylation is dispensable for the initial recognition of DNA breaks because the repair and signaling factors, such as the MRN complex, 53BP1, and BRCA1, can be initially and transiently recruited to DSBs in the absence of H2AX (27). This phenomenon prompted us to explore whether the MRN complex would be involved in the initial recognition of DNA breaks, as suggested by many in vitro biochemical studies (53–57), and responsible for this transient recruitment of signaling and repair proteins in H2AX-deficient cells.
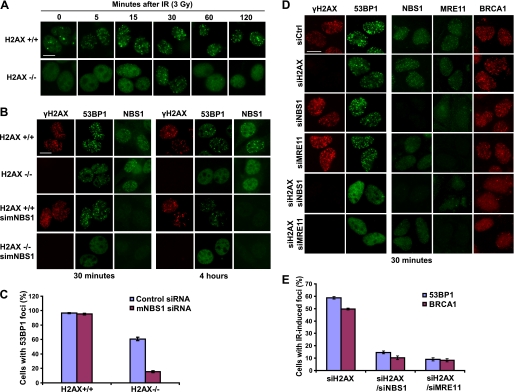
We first used 53BP1 focus formation as a readout. Consistent with a previous report (27), we observed a transient 53BP1 focus formation in H2AX−/− cells (Fig. 1A). Fifteen minutes after IR, we detected a combination of dotted and diffuse staining patterns of 53BP1. At 30 min after IR, clear 53BP1 foci could be observed in ∼60% of H2AX−/− cells. We further treated H2AX−/− cells with different doses of IR; the number of transient 53BP1 foci increased when higher dose of IR was used (supplemental Fig. S1), suggesting that these H2AX-independent 53BP1 foci are also sites of DNA damage. 53BP1 showed uniform nuclear staining 120 min or later after IR (Fig. 1A), which again is in agreement with early study suggesting the transient nature of formation of these foci in the absence of H2AX (27).
FIGURE 1.
The MRN complex is responsible for the initial recruitment of repair and signaling proteins to sites of DNA DSBs. A, 53BP1 transiently localizes to focus structure after IR treatment in H2AX−/− cells. H2AX+/+ and H2AX−/− were irradiated (3 Gy) and fixed at various time points after IR. The cells were immunostained with 53BP1 antibody (green). B, the initial 53BP1 focus formation observed in H2AX−/− cells depends on NBS1. H2AX+/+ and H2AX−/− were transfected with control siRNA or siRNA specifically targeting mouse NBS1 (simNBS1). 72 h later, the cells were irradiated (3 Gy) and fixed at 30 min or 4 h after IR. Immunostaining was carried out with anti-γH2AX, anti-53BP1, or anti-mouse NBS1 antibodies. C, quantification of 53BP1 foci following DNA damage. The data are presented as percentages of cells containing 53BP1 foci. The means and standard deviation (error bars) shown are obtained from three independent experiments in which more than 200 cells were counted. D, the initial focus localization of 53BP1 and BRCA1 requires the MRN complex. Double depletion of H2AX and NBS1 or of H2AX and MRE11 was performed in U2OS cells (see “Experimental Procedures” for details). Following siRNA transfection, the cells were irradiated (5 Gy) and fixed 30 min later. Immunostaining was carried out using the indicated antibodies. E, quantification of 53BP1 and BRCA1 focus formation following DNA damage. The data presented are percentages of cells containing 53BP1 or BRCA1 foci 30 min after DNA damage. The means and standard deviation (error bars) shown are results obtained from three independent experiments in which more than 200 cells were counted. Bars, 10 μm.
To determine whether this initial 53BP1 focus formation would be dependent on the MRN complex, we depleted mouse NBS1 in H2AX−/− cells using mouse specific NBS1 siRNA. As shown in Fig. 1 (B and C), depletion of NBS1 greatly diminished the transient recruitment of 53BP1 into a focus-like structure in H2AX−/− cells at 30 min after IR, but it did not have an obvious effect on 53BP1 focus formation in H2AX+/+ cells. We repeated these experiments using human cells. Similarly, we could observe transient focus localization of NBS1, MRE11, 53BP1, and BRCA1 30 min after IR in U2OS cells following siRNA-mediated depletion of H2AX (Fig. 1D), but formation of the transient foci of 53BP1 and BRCA1 was greatly reduced in cells with co-depletion of H2AX and NBS1 or H2AX and MRE11 (Fig. 1, D and E). Although this transient H2AX-independent recruitment of BRCA1 requires the MRN complex, we noted that the transient recruitment of NBS1 occurred in a manner that is independent of BRCA1 (supplemental Fig. S2). These data, together with in vitro studies suggesting direct binding of the MRN complex to DNA ends, led us to hypothesize that the MRN complex may be the initial proteins that recognize DNA breaks and recruit, at least initially, other DNA damage repair proteins to sites of DNA breaks.
Cells with single or double depletion used above were still viable within the time frame of our experiments, although we observed G2/M checkpoint defects and some changes in cell cycle distribution, which were particularly prominent in cells with NBS1 depletion (supplemental Fig. S3). In addition, damage-induced 53BP1 phosphorylation was greatly impaired in cells depleted of NBS1 or NBS1/H2AX (supplemental Fig. S4), again supporting a critical role for NBS1 in ATM activation and cellular response to DNA damage.
The Transient Recruitment of 53BP1 Depends on NBS1 but Not MDC1 or RNF8
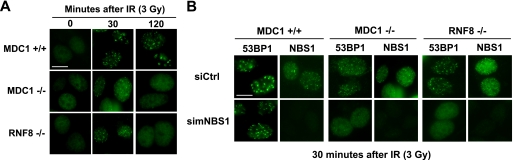
It remains to be determined precisely how the MRN complex may regulate the transient localization of several DNA damage repair proteins in the absence of H2AX. In this regard, we examined 53BP1 focus formation in MDC1−/− and RNF8−/− MEFs. Similar to that observed in H2AX−/− cells, the transient recruitment of 53BP1 was detected in RNF8−/− or MDC1−/− MEFs, although the stable accumulation of 53BP1 was abolished in these cells (Fig. 2A). Again, this transient recruitment of 53BP1 can be greatly diminished by the depletion of NBS1 in these MEFs (Fig. 2B), suggesting that this early recruitment of 53BP1 depends on NBS1 but not MDC1 or RNF8.
FIGURE 2.
The transient recruitment of 53BP1 depends on NBS1 but not MDC1 or RNF8. A, transient recruitment of 53BP1 upon DNA damage still occur in RNF8−/− and MDC1−/− MEFs. B, the transient recruitment of 53BP1 upon DNA damage was abolished by NBS1 depletion in RNF8−/− and MDC1−/− MEFs. The cells with or without siRNA transfection were irradiated and fixed at indicated time points after IR. Immunostaining was carried out with the indicated antibodies. Bars, 10 μm.
MRN Complex and CtIP, but Not H2AX, Are Required for Efficient HR Repair
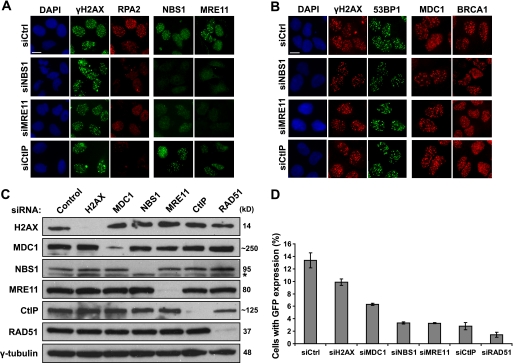
We next examined what would be the likely events regulated by the MRN complex in the absence of H2AX. It is believed that at least a fraction of HR repair is initiated by nucleolytic processing of DSBs, which generate recombination proficient 3′ ssDNA overhangs that are rapidly bound by RPA. Subsequently, Rad51, a key recombinase enzyme, together with its accessory factors displace RPA-ssDNA complexes to form helical nucleoprotein filament, which allows the beginning of homology search and HR repair. Given that the MRN complex but not H2AX is responsible for the initial recruitment of repair and signaling proteins to DSB sites, we speculated that HR repair might be mainly initiated via the action of the MRN complex. To test this hypothesis, we checked the IR-induced RPA focus formation, which can be used as readout of the generation of ssDNA regions following DNA damage. In NBS1-, MRE11-, or CtIP-depleted U2OS cells, the IR-induced RPA2 focus formation was dramatically reduced, supporting the possibility that the MRN complex and CtIP are involved in the generation of ssDNAs (Fig. 3A). On the other hand, the focus formation of other DNA damage cascade components, such as H2AX, 53BP1, MDC1, and BRCA1, was not considerably impaired in cells with NBS1, MRE11, or CtIP depletion (Fig. 3B).
FIGURE 3.
MRN complex and CtIP, but not H2AX, are required for efficient HR repair. A and B, depletion of components of the MRN complex or CtIP impairs RPA focus formation (A) but does not affect focus formation of other DNA damage repair factors (B). U2OS cells were transfected with indicated siRNAs. 48 h later, the cells were irradiated (10 Gy) and allowed to recover for 6 h before fixation and immunostaining with antibodies as indicated. C and D, siRNA-mediated down-regulation of various DNA damage and repair proteins were carried out in U2OS DR-GFP cells. The knockdown efficiency using indicated siRNAs was confirmed by immunoblotting (C). The percentage of GFP positive cells was determined by flow cytometry 48 h after cells were electroporated with pCBASce plasmid (D). The means and standard deviation (error bars) shown are obtained from three independent experiments. Asterisk in C, nonspecific band. Bars, 10 μm. DAPI, 4′,6′-diamino-2-phenylindole.
We further studied HR repair in DR-U2OS cells depleted of H2AX, MDC1, the MRN complex, or CtIP, respectively. The efficient siRNA-mediated knockdown was confirmed by Western blot (Fig. 3C). Significantly, depletion of NBS1, MRE11, or CtIP decreased HR frequencies to levels close to that achieved by the depletion of the key recombinase enzyme RAD51 (Fig. 3D), again supporting an important function for MRN and CtIP in DNA repair. Depletion of H2AX only showed a mild effect on HR repair, whereas depletion of MDC1 showed a 50% reduction in HR efficiency (Fig. 3D). These data suggest that H2AX and MDC1 likely play accessory, but not essential, roles in HR repair.
IR-induced RPA Focus Formation Is Largely Independent of ATM, γH2AX, and MDC1
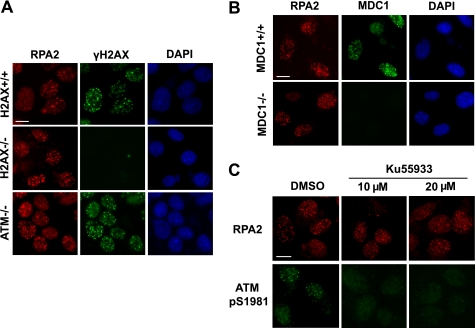
We examined the IR-induced RPA focus formation in H2AX-, MDC1-, or ATM-deficient MEFs. As shown in Fig. 4(A and B), RPA2 focus formation was detected in these deficient cells. We repeated these experiments in human cells using H2AX siRNA. Consistent with that observed in H2AX−/− MEFs, RPA focus formation still occurred in cells following H2AX depletion (supplemental Fig. S5A). We also performed time course experiments and determined the kinetics of RPA focus formation in wild type versus ATM-deficient cells (supplemental Fig. S5B). Again, we failed to observe any obvious difference in RPA focus formation between wild type and ATM-deficient cells at every time point we examined. In addition, DSB end resection (assessed by RPA focus formation) was not severely affected by the inhibition of ATM kinase activity (Fig. 4C). Therefore, we conclude that IR-induced RPA focus formation, which may reflect a long stretch of ssDNA regions, requires the MRN complex and CtIP, but not H2AX, MDC1, or ATM.
FIGURE 4.
IR-induced RPA focus formation is largely independent of ATM, γH2AX, and MDC1. A and B, IR-induced RPA focus formation was observed in ATM-, H2AX-, or MDC1-deficient cells. The cells with the indicated genotypes were irradiated (10 Gy) and allowed to recover for 6 h before fixation and immunostaining. C, inhibiting ATM activity by ATM inhibitor KU55933 did not impair RPA focus formation following DNA damage. U2OS cells were pretreated with dimethyl sulfoxide (DMSO) or KU55933 at the indicated concentrations for 1 h before they were exposed to IR (10 Gy). Immunostaining was performed 6 h after IR using anti-RPA2 and ATM pS1981 antibodies. Bars, 10 μm.
DISCUSSION
Recent studies cumulate and support a model that DNA damage-induced protein accumulation is an intricate part of the DNA damage response (58–60). At first glance, it appears surprising that the H2AX−/− mice exhibited a relatively mild phenotype (12), considering that H2AX is critically important for the accumulation of many if not all of the DNA damage signaling and repair proteins at sites of DNA breaks. Follow-up study suggests that although H2AX is required for the retention of signaling and repair proteins at sites of DNA damage, it is dispensable for the initial recognition of DNA breaks because a few DNA damage repair proteins can at least transiently localize to sites of DNA breaks in the absence of H2AX (27). Similar results were obtained in MDC1−/− cells (61). These observations raise the question of what is the true sensor that initially recognizes DNA breaks.
Our results suggest that at least one of these initial sensor proteins is the MRN complex, because the MRN complex is required for the transient localization of several DNA damage repair proteins in the absence of H2AX. This is also in agreement with the known DNA end binding activity of the MRN complex (53–57). Based on our observations, we believe that the MRN complex carries out at least two distinct functions following DNA damage. One is to promote DNA repair. The other is to activate ATM and ATM-dependent checkpoints (62–66). This idea of two separate functions for MRN in DNA damage response is strongly supported by two recent publications (67, 68), which suggest that although the nuclease activity of MRE11 is essential to initiate DNA repair, it is largely dispensable for ATM activation. Based on our data presented above and previous studies, we would like to propose a modified model of DNA damage signaling cascade (Fig. 5). In this model, the MRN complex is involved in the initial recognition of DNA breaks, which allows the transient localization of many other DNA damage checkpoint and repair proteins via direct or indirect protein-protein interactions. We speculate that this initial recruitment stage is geared up mainly for DNA repair. At the same time, the MRN complex also facilitates ATM activation, H2AX phosphorylation, and initiation of DNA damage checkpoints. If DNA repair occurs rapidly (for example, following a very low dose of ionizing radiation), the cells may resume the cell cycle without any obvious delay and not even utilize these DNA damage signaling or checkpoint pathways. However, if DNA repair cannot be completed quickly, the cells will use the phospho-H2AX-dependent signaling pathway, which permits the build-up of DNA damage checkpoint and repair proteins not only at the vicinity of DNA breaks but also spreading to megabases away from sites of DNA breaks. We image that such stable accumulation of DNA damage and repair proteins at and near the sites of DNA breaks effectively increase the local concentration of these proteins and permit efficient DNA damage signal transduction, cell cycle checkpoint activation, and ultimately DNA repair.
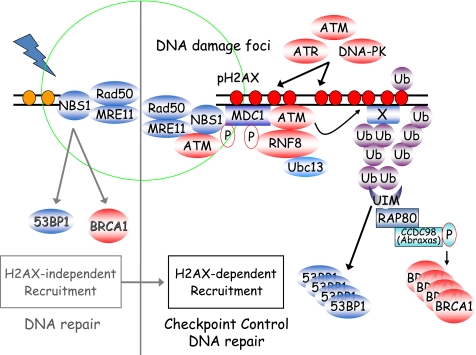
FIGURE 5.
A proposed revised model of mammalian DNA damage response. We hypothesize that there are two separate pathways involved in the recruitment and accumulation of DNA damage checkpoint and repair proteins at DSBs. One of them is the H2AX-independent pathway, which requires the MRN complex and probably limits the localization of these checkpoint and repair proteins at or closer to sites of DNA breaks. We speculate that this pathway is mainly geared toward DNA repair but also initiates ATM activation and cell cycle checkpoint. The second pathway is the well studied H2AX-dependent pathway, which allows the accumulation of DNA repair proteins and the spreading of DNA damage signaling proteins to larger chromatin regions surrounding the damaged sites. This H2AX-dependent pathway carries out at least two functions: one is to facilitate DNA repair, and the other is to promote the amplification of DNA damage signals, which lead to a full checkpoint activation and allow time for the completion of DNA repair.
It is still unknown exactly how the MRN complex initially recruits these DNA damage repair proteins like 53BP1 and BRCA1 to DSB sites. At least for 53BP1, one possible explanation is that NBS1 may bind directly to 53BP1. When we purified NBS1-containing complexes, we identified few 53BP1 peptides by mass spectrometry analysis (data not shown). In addition, we also observed an interaction between NBS1 and 53BP1 when both of these proteins were overexpressed (supplemental Fig. S6A). We further asked whether the FHA or BRCT1 domain of NBS1 is required for its binding to 53BP1. As shown in supplemental Fig. S6B, the NBS1 mutants deleted of FHA or BRCT1 domain showed reduced binding to 53BP1, raising the possibility that both the FHA and BRCT1 domains of NBS1 are involved in its interaction with 53BP1. Of course, future studies will be needed to determine whether this interaction is direct and whether it is required for the transient recruitment of 53BP1 in the absence of H2AX. We speculate that this transient recruitment of 53BP1 by the MRN complex may allow the engagement of a 53BP1-dependent pathway in DNA repair, which should also be assessed by additional experiments.
Although depletion of CtIP, NBS1, or MRE11 can greatly impair RPA focus formation in response to DSBs, the MEFs with H2AX, MDC1, or ATM deficiency exhibit seemingly normal RPA foci when compared with wild type MEFs. Although our data that ATM is dispensable for IR-induced RPA focus formation appear to be contradictory with two previous reports (69, 70), these results are consistent with other publications. For example, DNA damage-induced RPA focus formation was reported to occur independent of γH2AX, because the phosphatidylinositol 3-kinase inhibitor wortmannin could block DNA damage-induced γH2AX but not RPA focus formation (71). Similarly, RAD51 focus formation was also reported to be independent of H2AX (12). Our results also agree with the recent observation that although MRE11 nuclease activity is important for DNA repair, it is dispensable for ATM activation (67, 68). We would like to point out that although we propose that ATM is not essential for DNA repair, this by no means suggests that ATM is not involved in DNA repair. Numerous studies have already indicated that ATM is required for the repair of a subset of DNA breaks. The challenge is to determine whether ATM would play a critical role in certain repair processes, act as an auxiliary factor for multiple DNA repair pathways, modulate repair efficiency and/or fidelity via its kinase activity, influence the outcome of DNA repair by its pro-apoptotic function, or a combination of the above.
As we discussed above, the critical intermediate step during DSB repair is the generation of ssDNA with 3′ overhangs. Such a structure would prevent nonhomologous end joining and promote DSB repair via the HR pathway. Because the RPA complex is known to associate tightly with ssDNA, RPA focus formation has been used as a marker for ssDNA formation (2, 9). Using RPA focus formation as a readout for ssDNA formation, it is obvious that the MRN complex together with CtIP play critical roles in processing DSB ends to form ssDNAs, which are required for efficient HR. Indeed, our HR assay documented that depletion of NBS1, MRE11, or CtIP significantly decreases HR efficiency to levels close to those achieved by the depletion of RAD51. Nevertheless, RAD51 depletion consistently showed an ∼2-fold lower HR efficiency when compared with NBS1, MRE11, or CtIP depletion. This observation would suggest that limited end resection may still occur in the absence of the MRN complex or CtIP. Although these limited ssDNA regions might be difficult to detect, they could still permit at least some HR events. Another interesting observation is that although we did not detect any obvious defect in RPA focus formation in H2AX- or MDC1-deficient cells, we noticed that HR efficiency is reduced in these cells, although not as severely as in cells with MRN or CtIP depletion. This would imply that H2AX and MDC1 may modulate HR repair at a step that is separated from RPA focus formation. For example, MDC1 can bind to and mediate H2AX-dependent HR (72), and MDC1 can interact with Rad51 and facilitate HR (73). More details regarding the complex nature of these regulations will be revealed in the future.
Supplementary Material
Acknowledgments
We thank Dr. Maria Jasin for providing U2OS cells with integrated DR-GFP substrate and DR-GFP and pCBASce constructs. We also thank Dr. Richard Baer for the monoclonal mouse anti-CtIP antibody and Dr. Andre Nussenzweig for anti-mouse NBS1 and MRE11 polyclonal antibodies.
This work was supported, in whole or in part, by a National Institutes of Health Grants CA100109, CA092312, and CA089239 (to J. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- DSB
- double-strand break
- MRN
- MRE11-RAD50-NBS1
- HR
- homologous recombination
- ss
- single-stranded
- E3
- ubiquitin-protein isopeptide ligase
- MEF
- mouse embryonic fibroblast
- siRNA
- small interfering RNA
- GFP
- green fluorescent protein
- IR
- ionizing radiation
- DR
- direct repeat.
REFERENCES
- 1.Pierce A. J., Stark J. M., Araujo F. D., Moynahan M. E., Berwick M., Jasin M. (2001) Trends Cell Biol. 11, S52–59 [DOI] [PubMed] [Google Scholar]
- 2.Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 3.Petrini J. H., Stracker T. H. (2003) Trends Cell Biol. 13, 458–462 [DOI] [PubMed] [Google Scholar]
- 4.Lavin M. F. (2004) DNA Repair 3, 1515–1520 [DOI] [PubMed] [Google Scholar]
- 5.Borde V. (2007) Chromosome Res. 15, 551–563 [DOI] [PubMed] [Google Scholar]
- 6.Falck J., Petrini J. H., Williams B. R., Lukas J., Bartek J. (2002) Nat. Genet. 30, 290–294 [DOI] [PubMed] [Google Scholar]
- 7.Carson C. T., Schwartz R. A., Stracker T. H., Lilley C. E., Lee D. V., Weitzman M. D. (2003) EMBO J. 22, 6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olson E., Nievera C. J., Liu E., Lee A. Y., Chen L., Wu X. (2007) Mol. Cell. Biol. 27, 6053–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S. P. (2007) Nature 450, 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L., Nievera C. J., Lee A. Y., Wu X. (2008) J. Biol. Chem. 283, 7713–7720 [DOI] [PubMed] [Google Scholar]
- 11.Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000) Curr. Biol. 10, 886–895 [DOI] [PubMed] [Google Scholar]
- 12.Celeste A., Petersen S., Romanienko P. J., Fernandez-Capetillo O., Chen H. T., Sedelnikova O. A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M. J., Redon C., Pilch D. R., Olaru A., Eckhaus M., Camerini-Otero R. D., Tessarollo L., Livak F., Manova K., Bonner W. M., Nussenzweig M. C., Nussenzweig A. (2002) Science 296, 922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Celeste A., Difilippantonio S., Difilippantonio M. J., Fernandez-Capetillo O., Pilch D. R., Sedelnikova O. A., Eckhaus M., Ried T., Bonner W. M., Nussenzweig A. (2003) Cell 114, 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassing C. H., Suh H., Ferguson D. O., Chua K. F., Manis J., Eckersdorff M., Gleason M., Bronson R., Lee C., Alt F. W. (2003) Cell 114, 359–370 [DOI] [PubMed] [Google Scholar]
- 15.Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M. A., Celeste A., Manis J. P., van Deursen J., Nussenzweig A., Paull T. T., Alt F. W., Chen J. (2006) Mol. Cell 21, 187–200 [DOI] [PubMed] [Google Scholar]
- 16.Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. (2005) Cell 123, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 17.Wu L., Luo K., Lou Z., Chen J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11200–11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spycher C., Miller E. S., Townsend K., Pavic L., Morrice N. A., Janscak P., Stewart G. S., Stucki M. (2008) J. Cell Biol. 181, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melander F., Bekker-Jensen S., Falck J., Bartek J., Mailand N., Lukas J. (2008) J. Cell Biol. 181, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman J. R., Jackson S. P. (2008) EMBO Rep. 9, 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 24.Wang B., Elledge S. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uanschou C., Siwiec T., Pedrosa-Harand A., Kerzendorfer C., Sanchez-Moran E., Novatchkova M., Akimcheva S., Woglar A., Klein F., Schlögelhofer P. (2007) EMBO J. 26, 5061–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobhian B., Shao G., Lilli D. R., Culhane A. C., Moreau L. A., Xia B., Livingston D. M., Greenberg R. A. (2007) Science 316, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celeste A., Fernandez-Capetillo O., Kruhlak M. J., Pilch D. R., Staudt D. W., Lee A., Bonner R. F., Bonner W. M., Nussenzweig A. (2003) Nat. Cell Biol. 5, 675–679 [DOI] [PubMed] [Google Scholar]
- 28.Petersen S., Casellas R., Reina-San-Martin B., Chen H. T., Difilippantonio M. J., Wilson P. C., Hanitsch L., Celeste A., Muramatsu M., Pilch D. R., Redon C., Ried T., Bonner W. M., Honjo T., Nussenzweig M. C., Nussenzweig A. (2001) Nature 414, 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassing C. H., Chua K. F., Sekiguchi J., Suh H., Whitlow S. R., Fleming J. C., Monroe B. C., Ciccone D. N., Yan C., Vlasakova K., Livingston D. M., Ferguson D. O., Scully R., Alt F. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8173–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R. F., Nussenzweig A., Nussenzweig M. C. (2003) J. Exp. Med. 197, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie A., Puget N., Shim I., Odate S., Jarzyna I., Bassing C. H., Alt F. W., Scully R. (2004) Mol. Cell 16, 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown E. J., Baltimore D. (2000) Genes Dev. 14, 397–402 [PMC free article] [PubMed] [Google Scholar]
- 33.de Klein A., Muijtjens M., van Os R., Verhoeven Y., Smit B., Carr A. M., Lehmann A. R., Hoeijmakers J. H. (2000) Curr. Biol. 10, 479–482 [DOI] [PubMed] [Google Scholar]
- 34.Zhu J., Petersen S., Tessarollo L., Nussenzweig A. (2001) Curr. Biol. 11, 105–109 [DOI] [PubMed] [Google Scholar]
- 35.Xiao Y., Weaver D. T. (1997) Nucleic Acids Res. 25, 2985–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo G., Yao M. S., Bender C. F., Mills M., Bladl A. R., Bradley A., Petrini J. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7376–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludwig T., Chapman D. L., Papaioannou V. E., Efstratiadis A. (1997) Genes Dev. 11, 1226–1241 [DOI] [PubMed] [Google Scholar]
- 38.Sharan S. K., Morimatsu M., Albrecht U., Lim D. S., Regel E., Dinh C., Sands A., Eichele G., Hasty P., Bradley A. (1997) Nature 386, 804–810 [DOI] [PubMed] [Google Scholar]
- 39.Tsuzuki T., Fujii Y., Sakumi K., Tominaga Y., Nakao K., Sekiguchi M., Matsushiro A., Yoshimura Y., Morita T. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6236–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen P. L., Liu F., Cai S., Lin X., Li A., Chen Y., Gu B., Lee E. Y., Lee W. H. (2005) Mol. Cell. Biol. 25, 3535–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J. N., Ried T., Tagle D., Wynshaw-Boris A. (1996) Cell 86, 159–171 [DOI] [PubMed] [Google Scholar]
- 42.Elson A., Wang Y., Daugherty C. J., Morton C. C., Zhou F., Campos-Torres J., Leder P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13084–13089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y., Ashley T., Brainerd E. E., Bronson R. T., Meyn M. S., Baltimore D. (1996) Genes Dev. 10, 2411–2422 [DOI] [PubMed] [Google Scholar]
- 44.Manis J. P., Morales J. C., Xia Z., Kutok J. L., Alt F. W., Carpenter P. B. (2004) Nat. Immunol. 5, 481–487 [DOI] [PubMed] [Google Scholar]
- 45.Ward I. M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J. S., Cascalho M., Chen L., Nussenzweig A., Livak F., Nussenzweig M. C., Chen J. (2004) J. Cell Biol. 165, 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riballo E., Kühne M., Rief N., Doherty A., Smith G. C., Recio M. J., Reis C., Dahm K., Fricke A., Krempler A., Parker A. R., Jackson S. P., Gennery A., Jeggo P. A., Löbrich M. (2004) Mol. Cell 16, 715–724 [DOI] [PubMed] [Google Scholar]
- 47.Lou Z., Minter-Dykhouse K., Wu X., Chen J. (2003) Nature 421, 957–961 [DOI] [PubMed] [Google Scholar]
- 48.Yu X., Fu S., Lai M., Baer R., Chen J. (2006) Genes Dev. 20, 1721–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J., Silver D. P., Walpita D., Cantor S. B., Gazdar A. F., Tomlinson G., Couch F. J., Weber B. L., Ashley T., Livingston D. M., Scully R. (1998) Mol. Cell 2, 317–328 [DOI] [PubMed] [Google Scholar]
- 50.Yu X., Chen J. (2004) Mol. Cell. Biol. 24, 9478–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lou Z., Chini C. C., Minter-Dykhouse K., Chen J. (2003) J. Biol. Chem. 278, 13599–13602 [DOI] [PubMed] [Google Scholar]
- 52.Weinstock D. M., Nakanishi K., Helgadottir H. R., Jasin M. (2006) Methods Enzymol. 409, 524–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Jager M., van Noort J., van Gent D. C., Dekker C., Kanaar R., Wyman C. (2001) Mol. Cell 8, 1129–1135 [DOI] [PubMed] [Google Scholar]
- 54.Moreno-Herrero F., de Jager M., Dekker N. H., Kanaar R., Wyman C., Dekker C. (2005) Nature 437, 440–443 [DOI] [PubMed] [Google Scholar]
- 55.Trujillo K. M., Roh D. H., Chen L., Van Komen S., Tomkinson A., Sung P. (2003) J. Biol. Chem. 278, 48957–48964 [DOI] [PubMed] [Google Scholar]
- 56.Wiltzius J. J., Hohl M., Fleming J. C., Petrini J. H. (2005) Nat. Struct. Mol. Biol. 12, 403–407 [DOI] [PubMed] [Google Scholar]
- 57.de Jager M., Wyman C., van Gent D. C., Kanaar R. (2002) Nucleic Acids Res. 30, 4425–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 59.Huen M. S., Chen J. (2008) Cell Res. 18, 8–16 [DOI] [PubMed] [Google Scholar]
- 60.Wood J. L., Chen J. (2008) Trends Cell Biol. 18, 451–455 [DOI] [PubMed] [Google Scholar]
- 61.Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M. B., Bartek J., Lukas J. (2006) J. Cell Biol. 173, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jazayeri A., Balestrini A., Garner E., Haber J. E., Costanzo V. (2008) EMBO J. 27, 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J. H., Paull T. T. (2005) Science 308, 551–554 [DOI] [PubMed] [Google Scholar]
- 64.Lee J. H., Paull T. T. (2004) Science 304, 93–96 [DOI] [PubMed] [Google Scholar]
- 65.Cerosaletti K., Wright J., Concannon P. (2006) Mol. Cell. Biol. 26, 1691–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cerosaletti K., Concannon P. (2004) J. Biol. Chem. 279, 38813–38819 [DOI] [PubMed] [Google Scholar]
- 67.Williams R. S., Moncalian G., Williams J. S., Yamada Y., Limbo O., Shin D. S., Groocock L. M., Cahill D., Hitomi C., Guenther G., Moiani D., Carney J. P., Russell P., Tainer J. A. (2008) Cell 135, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J. M., Chang S., Ferguson D. O. (2008) Cell 135, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jazayeri A., Falck J., Lukas C., Bartek J., Smith G. C., Lukas J., Jackson S. P. (2006) Nat. Cell Biol. 8, 37–45 [DOI] [PubMed] [Google Scholar]
- 70.Myers J. S., Cortez D. (2006) J. Biol. Chem. 281, 9346–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu J. S., Kuo S. R., Melendy T. (2006) J. Cell. Biochem. 99, 1452–1462 [DOI] [PubMed] [Google Scholar]
- 72.Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F. W., Chen J., Jackson S. P., Scully R. (2007) Mol. Cell 28, 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J., Ma Z., Treszezamsky A., Powell S. N. (2005) Nat. Struct. Mol. Biol. 12, 902–909 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.