Abstract
Here, we report the overexpression, purification, and characterization of the transcriptional activator fumarate and nitrate reductase regulator from the pathogenic bacterium Neisseria meningitidis (NmFNR). Like its homologue from Escherichia coli (EcFNR), NmFNR binds a 4Fe-4S cluster, which breaks down in the presence of oxygen to a 2Fe-2S cluster and subsequently to apo-FNR. The kinetics of NmFNR cluster disassembly in the presence of oxygen are 2–3× slower than those previously reported for wild-type EcFNR, but similar to constitutively active EcFNR* mutants, consistent with earlier work in which we reported that the activity of FNR-dependent promoters in N. meningitidis is only weakly inhibited by the presence of oxygen (Rock, J. D., Thomson, M. J., Read, R. C., and Moir, J. W. (2007) J. Bacteriol. 189, 1138–1144). NmFNR binds to DNA containing a consensus FNR box sequence, and this binding stabilizes the iron-sulfur cluster in the presence of oxygen. Partial degradation of the 4Fe-4S cluster to a 3Fe-4S occurs, and this form remains bound to the DNA. The 3Fe-4S cluster is converted spontaneously back to a 4Fe-4S cluster under subsequent anaerobic reducing conditions in the presence of ferrous iron. The finding that binding to DNA stabilizes FNR in the presence of oxygen such that it has a half-life of ∼30 min on the DNA has implications for our appreciation of how oxygen switches off FNR activatable genes in vivo.
Keywords: Cytochromes, Extracellular Matrix Proteins, Fibrinogen, Ligand Binding Protein, MyD88, Myeloid Cell, Polyadenylation, Sorting
Introduction
Neisseria meningitidis is a Gram-negative bacterium, forming part of the normal flora of a significant minority of the human population at any given time. The bacterium is able to cause disease following entry into the bloodstream (1), where it is capable of multiplying, thus causing septicemia, and/or crossing the blood brain barrier and multiplying in the cerebrospinal fluid, thus causing meningitis (2). Both of these diseases continue to have considerable mortality rates despite the availability of effective antibiotic therapies. In its natural environment in the nasopharynx, N. meningitidis shares an environment with both aerobic and anaerobic bacteria (3). Consequently, N. meningitidis is able to adapt its gene expression to up-regulate the gene aniA, encoding nitrite reductase, that enables it to denitrify nitrite to nitrous oxide as an alternative to oxygen respiration (4). This oxygen-dependent regulation requires fumarate and nitrate reduction regulator (FNR)3 (encoded by gene NMB0380 of the N. meningitidis serogroup B (MC58) genome) (4), a protein that is well established as the chief regulator in response to anaerobiosis in Escherichia coli (5).
FNR of E. coli is a transcriptional activator that controls transcription by binding to promoters under anaerobic conditions. The presence of an oxygen-sensitive 4Fe-4S cluster is correlated with protein dimerization (6), which enables it to bind promoter DNA (7, 8). The iron-sulfur cluster is believed to be ligated by four cysteine residues, three toward the N terminus and one centrally located within the FNR polypeptide, which are required for FNR function (9, 10). In the presence of oxygen, the cluster breaks down into a 3Fe-4S cluster and then a 2Fe-2S cluster (11). FNR containing a 2Fe-2S cluster is unable to bind DNA (8), but it is unknown whether the 3Fe-4S cluster-containing intermediate is able to bind DNA. FNR binds to a consensus palindromic sequence TTGA(T/C)NNNN(A/G)TCAA, which must be located appropriately with relation to the RNA polymerase binding site to allow effective promoter activation (12). FNR homologues have been identified in many other bacteria, where their roles also appear to be in regulating gene expression in response to oxygen (13).
fnr mutants of N. meningitidis fail to express the nitrite reductase aniA under oxygen limitation and are thus not able to exploit nitrite as an alternative respiratory electron acceptor when oxygen is absent (4). As well as a strict requirement for FNR, the aniA promoter is controlled in part by the nitric oxide-sensing repressor (14). In wild-type N. meningitidis MC58, expression of nitrite reductase is tightly regulated by oxygen. In mutants lacking the nitric oxide-sensing repressor, expression of aniA is retained even in the presence of 60–80% air saturation (14). Retention of some activity in the presence of oxygen may be related to the physiology of N. meningitidis, which appears to be adapted for a microaerobic lifestyle and yet is unable to grow well under strictly anaerobic conditions. The promoter for aniA in N. meningitidis contains an FNR consensus binding site (TTGACTTAAATCAA, consensus sequence underlined) centered 42.5 bases upstream from the experimentally determined transcriptional start site, consistent with FNR binding here and forming class II promoter-type interactions with the RNA polymerase (15).
Given the apparently unusual response to oxygen of FNR in N. meningitidis, we set out to establish the biochemical properties of its FNR (NmFNR). Our results indicate that NmFNR is less sensitive to oxygen than E. coli FNR (EcFNR). Furthermore, we report that binding of NmFNR to DNA stabilizes its iron-sulfur cluster in the presence of oxygen, which has implications for our understanding of oxygen-dependent regulation via FNR.
EXPERIMENTAL PROCEDURES
Bacterial Strains
E. coli DH5α was used as a general cloning vehicle. E. coli BL21 (DE3) (Novagen) was used to overexpress the FNR protein. Cultures were routinely grown in LB with kanamycin at a final concentration of 100 μg ml−1.
Cloning of FNR
The FNR gene (NMB0380) from N. meningitidis MC58 (16) was amplified using the forward primer 5′-CCAGGGACCAGCAATGGCTTCGCATAATACTACACATC-3′ and reverse primer 5′-GAGGAGAAGGCGCGTCAAATGGCGTGCGAGCAGCCG-3′, which incorporated linker regions (underlined) suitable for ligation independent cloning into the plasmid pET-YSBLIC3C using a method described previously (17). The resulting NmFNR expression construct was transformed into E. coli BL21 (DE3) cells and frozen in aliquots at −80 °C.
Recombinant Expression of NmFNR
Recombinant protein expression was performed using autoinduction media based on a previously described method (18). Starter cultures of E. coli BL21 (DE3) containing the NmFNR expression construct were grown overnight in LB medium at 37 °C, shaken at 200 rpm and used to inoculate 590 ml of autoinduction medium. Cultures were grown for 24 h at 25 °C with shaking at 150 rpm until A600 reached ∼8. Cells were then harvested in a Sorvall Evolution centrifuge at 4,000 × g before being transferred to an anaerobic chamber (Coy Laboratory Products) containing a 95% nitrogen, 5% hydrogen atmosphere with O2 concentrations maintained below 1 ppm in the presence of a palladium catalyst. Cells were resuspended in anaerobic buffer A (20 mm K2HPO4, 300 mm NaCl, 20 mm imidazole, pH 7.4) and 50 mg lysozyme and stirred in a 250 ml Duran bottle for 30 min. Cells were then transferred to a 50 ml Falcon tube before being broken using a Misonix 3000 sonicator for 2 min with a 10 s on/off cycle with an amplitude of 7. The crude lysate was clarified by centrifugation in an Allegra X-22R centrifuge at 9,500 × g for 30 min before being passed through a 0.45-μm filter prior to nickel affinity purification.
Purification of NmFNR
Purification was performed anaerobically using a 1-ml HiTrap chelating column (GE Healthcare) charged with 0.1 m nickel sulfate. All buffers were filtered before an overnight equilibration in the anaerobic chamber to become anoxic. The nickel column was equilibrated with 5 column volumes of buffer A before 5 column volumes of soluble protein was applied, and a dark brown band was seen to bind to the top of the column. The column was washed with 5 column volumes of buffer B (as buffer A but with 80 mm imidazole) before eluting with buffer C (as buffer A but with 500 mm imidazole). Dark brown fractions were collected and immediately desalted into 1× phosphate-buffered saline (PBS), pH 7.0, buffer using a PD-10 column (GE Healthcare) and eluted into sealed glass-stopped bottles. An overnight incubation with polyhistidine-tagged protease 3C at 4 °C was performed to cleave the polyhistidine tag from the recombinant FNR. The following day, the FNR-3C protein mix was applied back to a nickel column using the same method as before, and the flow containing the polyhistidine tag cleaved FNR was collected, desalted again into 1× PBS to remove imidazole and stored in 1× PBS and 2 mm dithiothreitol (DTT) at 4 °C until ready for use.
Protein, Iron, and Labile Sulfide Assays
Protein assay was performed using a Bradford assay (Bio-Rad) according to the manufacturer's instructions. Typical protein concentrations before dilution were ∼100 μm. Iron was assayed colorimetrically using Ferene S as described previously (19) using FeSO4 as a standard. Sulfide assay was adapted from Ref. 20. Briefly, 300 μl of sample was mixed with 1 ml 55 mm zinc acetate, to which 50 μl 3 m NaOH was then added. Subsequently, 250 μl of 7.4 mm N,N-dimethyl-1,4-phenylenediamine in 5 m HCl and 50 μl 0.23 m FeCl3 in 1.2 m HCl were added. The mixture was incubated at room temperature for 30 min with regular vortexing, the precipitate was removed by centrifugation, and the absorbance of the supernatant was read at 670 nm. Na2S was used as a standard.
UV-visible Spectrophotometry
Spectra were obtained using a Shimadzu 1601 spectrophotometer. Purified NmFNR samples were maintained under anaerobic conditions in cuvettes fitted with silicone seals secured in place by screw caps (Hellma). Spectra were obtained from 250 to 700 nm or at fixed wavelengths over a time course. The impact of oxygen on the spectral characteristics of NmFNR was evaluated by adding 800 μl of air saturated buffer to 200 μl of NmFNR in a sealed cuvette.
Fluorescence Anisotropy
Fluorescence anisotropy was used to measure FNR binding to the predicted binding site from the promoter for aniA. The FNR consensus site was located centrally within a pair of complementary 40-base primers, of which the forward primer was 5′-labeled with fluorescein isothiocyanate. All primers were ordered from MWG Eurofins and resuspended in filtered distilled H2O. Primers represented the wild-type N. meningitidis aniA promoter region containing the putative FNR site (5′-TTTTATGAATTATTTGACTTAAATCAAAATGCCCCCAATG-3′ and the reverse complement of this oligonucleotide; the consensus FNR box is underlined) and a control region in which the FNR consensus sequence is disrupted (5′-TTTTATGAATTATTCTGCTTAAACTGAAATGCCCCCAATG-3′ and the reverse complement of this oligonucleotide; deviations from the FNR box are in bold). Sealed florescence cuvettes (Hellma) were prepared anaerobically containing PBS and 5 mm DTT, 2 nm annealed primers, 360 μg acetylated bovine serum albumin and 0.0026 μg polydeoxyinosinic-deoxycytidylic acid. Fluorescence anisotropy was measured at 25.4 °C in a FluoroMax-3 fluorimeter fitted with autopolarizers (Jobin Yvon Horiba) using an excitation of 485 nm and detecting emission at 520 nm. FNR was titrated in to concentrations up to 300 nm using an airtight syringe (Hamilton). Fluorescence anisotropy was calculated in triplicate for each [FNR], and the experiment was repeated using both the FNR consensus and the control 40-mers.
Anaerobic Electrophoretic Mobility Gel Shift Assay (EMSA)
Gel shift assays were performed anaerobically to visualize the FNR-DNA dissociation in the presence of O2. 10 ml gels were poured containing 8% bis-acrylamide (Bio-Rad), 2.5% (v/v) glycerol, and 2 ml running buffer (50 mm Tris, 380 mm glycine, pH 8.0). The apparatus was assembled with running buffer and transferred to the anaerobic chamber to equilibrate overnight before prerunning the gel for 2–3 h at 40 V. Once loaded, the gel was run at 40 V for 5 h at 20 °C, and the DNA subsequently visualized by staining with ethidium bromide. Samples were prepared by mixing 10 μl of 92.5 μm FNR (calculated as a dimer) with 10 μl of duplex DNA at a concentration of 100 μm. The DNA sequence used was the same as for anisotropy except lacking the fluorescein isothiocyanate label. Where a FNR-DNA complex was formed prior to oxygen exposure, identical 20 μl samples were incubated anaerobically for 30 min to form a complex before removal from the anaerobic chamber and exposure to atmospheric oxygen by rapid mixing. An aliquot (20 μl) of each time point was returned to the anaerobic chamber before mixing with 10 μl of anaerobic filtered 1× PBS, pH 7.0, and 10 μl of anaerobic EMSA buffer (0.5 m Tris pH 6.5, 30% (v/v) glycerol, 0.5% (w/v) bromphenol blue, 5 mm β-mercaptoethanol). Where free FNR was first exposed to oxygen, the protocol was the same except that 10 μl of 92.5 μm FNR was first mixed with 10 μl anaerobic 1× PBS, pH 7.0, before oxygen exposure. Upon returning to the anaerobic environment, 10 μl of duplex DNA was added to FNR and allowed to incubate for 30 min, and after this time, 10 μl of EMSA load buffer was added to the sample.
Data Analysis
Rates of spectral change were fitted using the freely available R statistical package. The raw empirical results were compared with an exactly solvable, two step mass action kinetics chemical reaction by a nonlinear least squares fit for the five free parameters (two rate constants and the values of the absorbance for the three reactants involved) (supplemental Fig. 1). Typical r values for the fits were in the region of 0.97 indicated a high degree of consistency of this model to the data.
Oxygen Utilization
Oxygen concentration was followed in a Clark-type oxygen electrode (Rank Brothers, Bottisham, UK). A chamber containing 2 ml of solution was fitted with a cap through which samples could be added via a Hamilton syringe. A stable base line for the oxygen content of PBS and 2 mm DTT was measured, and, subsequently, solutions containing NmFNR (±DNA) were added, and the change in oxygen concentration was followed over time.
RESULTS
Purification and Characterization of NmFNR
FNR from N. meningitidis (NmFNR) was purified following heterologous expression in E. coli as described in the “Experimental Procedures.” Cultures were grown aerobically, but given that the final optical density of cultures was A600 ≈ 8, it is likely that the cultures were oxygen-limited. Enhanced yields of holo-FNR were obtained by incubating cell extracts under anaerobic conditions for 30 min prior to purification by nickel affinity chromatography, after which preparations of protein exhibited A400/A280 purity ratios of between 0.36 and 0.45 and appeared >90% pure by SDS-PAGE analysis (supplemental Fig. 2).
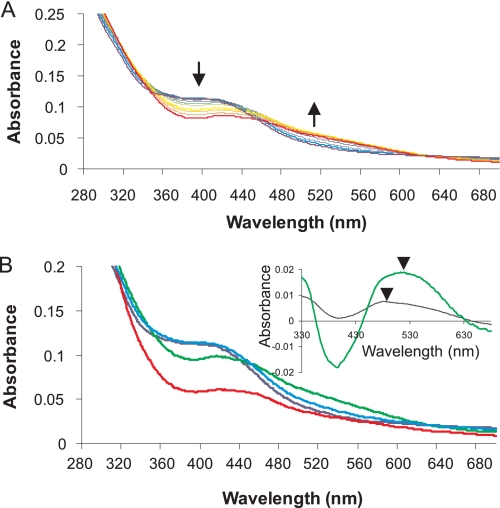
The purified NmFNR has a distinctive brown color that disappears under aerobic conditions. A spectral scan of purified NmFNR (Fig. 1 and supplemental Fig. 2) displays features typical of an iron-sulfur cluster, with a prominent feature ∼405 nm and a shoulder ∼310 nm, very similar to the spectra of the 4Fe-4S form of EcFNR (11). The stoichiometry of protein:iron:sulfur was assayed for different preparations of NmFNR, yielding ratios of 4.5 ± 0.15 iron and 3.4 ± 0.31 S2− per protein, consistent with NmFNR containing a 4Fe-4S cluster. Apo-NmFNR was purified by removing the anaerobic incubation step that allows in vitro assembly of the iron-sulfur cluster. Under these conditions, a purified NmFNR was obtained that was colorless and showed no spectral features due to an iron-sulfur cluster (supplemental Fig. 2).
FIGURE 1.
Effect of O2 on spectral features of NmFNR. A, spectrum of 20 μm anoxic NmFNR is shown in bold in violet. Spectral scans taken periodically during 1,040 s following the introduction of 200 μm oxygen into 20 μm NmFNR are displayed in purple, blue, green, yellow, orange, and red, respectively, with increasing time. The absorbance at ∼400 nm decreases, and the absorbance at ∼520 nm increases over time as marked by arrows. B, different spectral forms of NmFNR over time. Spectra at time 0 (dark blue), 120 (light blue), 600 (green), and 3,600 s (red). Inset, difference spectrum 120 s minus time 0 (black line) and 600 s minus time 0 (green line). Arrows mark the different absorbance peaks at 490 nm (120–0 s) and 520 nm (600–0 s).
Sensitivity of NmFNR Cluster to Oxygen
The effect of oxygen on the iron-sulfur cluster of NmFNR was followed by UV-visible spectroscopy. Spectral changes were observed, which indicated a rapid conversion from the native 4Fe-4S form, via a short-lived intermediate, to a more stable form with a lower absorbance and with features typical of the 2Fe-2S form of EcFNR (Fig. 1). During the first ∼2 min following introduction of oxygen the absorbance increased in the visible region and was red-shifted slightly. Subsequently, the absorbance around 400 nm decreased, and the absorbance between 450 nm and 600 nm increased to give a broad featureless signal in this region. The change within the first 2 min is quite distinct from the overall change within the first 10 min (see Fig. 1B, inset), indicating that a short-lived intermediate species appears then disappears over this time. This intermediate is most likely a 3Fe-4S cluster, as seen following the oxygen-dependent step in EcFNR cluster breakdown. In EcFNR, the [3Fe-4S]+ cluster is detectable by EPR. Using the same EPR parameters as used by Crack et al. (11), no EPR detectable signal was observable during breakdown of the NmFNR, which may be due to the inclusion of DTT reducing the NmFNR iron-sulfur cluster to the EPR-silent [3Fe-4S]0 state. Between 10 min and 1 h after adding oxygen, the absorbance at all visible wavelengths decreased, but the spectral line shape stayed the same, consistent with the breakdown of the 2Fe-2S form of NmFNR into apo-NmFNR.
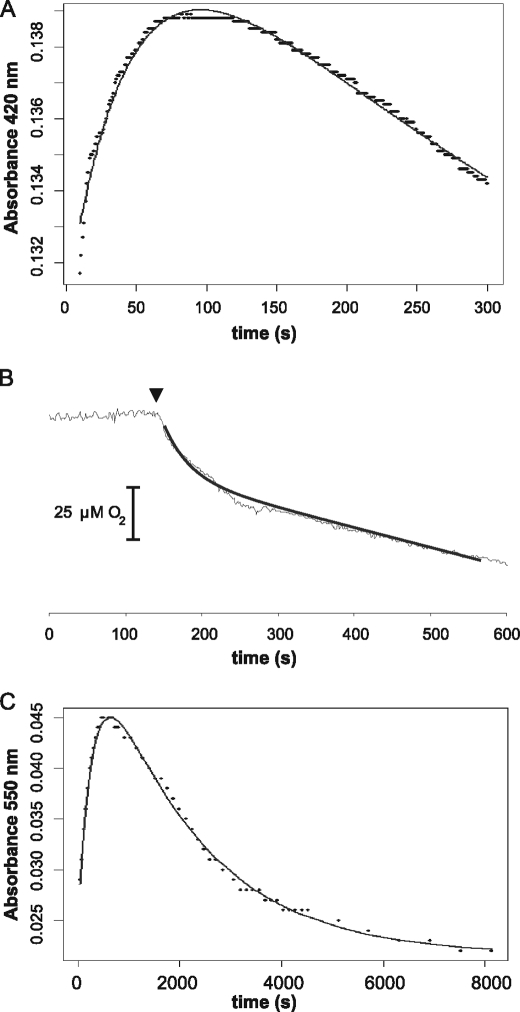
To determine the kinetics of the fast process occurring in response to oxygen, the change in absorbance was monitored at 420 nm for 5 min (Fig. 2A). At this wavelength, the absorbance at 420 nm increases as 4Fe-4S breaks down to the intermediate (3Fe-4S) and then decreases as this intermediate breaks down further to 2Fe-2S. The fast step was determined to have an apparent first order rate constant of 0.021 s−1. To determine whether this step is the oxygen-dependent step, we followed oxygen concentration in a Clark-type oxygen electrode and monitored the change following introduction of anaerobic NmFNR (Fig. 2B). Oxygen uptake occurred immediately following the introduction of NmFNR, and ∼20 μm oxygen was consumed over the subsequent 100 s with the same rate constant (0.02 s−1) as the rate of the fast spectroscopic change. The continued linear oxygen uptake after this period is a chemical process; the same rate of oxygen uptake was observed in PBS and 2 mm DTT and 20 μm ferrous iron. The fast spectral change and oxygen uptake rates are consistent with the fast step being an oxygen-dependent breakdown of 4Fe-4S to 3Fe-4S, as seen with EcFNR, but the rate here is slower with a second order rate constant (k1) of 105 m−1 s−1 compared with ∼278 m−1 s−1 for EcFNR (21). Dependence of this fast step on [O2] was confirmed by treating NmFNR with a range of O2 concentrations (data not shown). The slower decrease in absorbance at 420 nm is due to two further linked exponential decay processes. To fit the rates of these slower steps in NmFNR, spectral data were fitted to two linked exponentials using the data recorded for spectral time points at 550 nm (Fig. 2C). At this wavelength, there is an increase in absorbance as the initial 4Fe-4S NmFNR is converted via the 3Fe-4S intermediate to a 2Fe-2S cluster. Breakdown of the 2Fe-2S cluster leads to a decrease in absorbance at this wavelength (Fig. 1). Rate constants were calculated for the breakdown of the 3Fe-4S intermediate to the 2Fe-2S (k2 = 0.0033 s−1) and from 2Fe-2S to apo-FNR (k3 = 0.00051 s−1). To confirm that calculated k2 was due to the conversion from 3Fe-4S to 2Fe-2S and that the conversion of 4Fe-4S to 3Fe-4S was not a contributing factor, we removed data from the first 2 min of the incubation (during which time >90% of the fast 4Fe-4S to 3Fe-4S reaction had occurred) and found that calculated values of k2 (and k3) were unaffected. In EcFNR, the rate constant k2 is ∼0.0087 s−1 (21), i.e. 2–3× faster than the rate of breakdown measured here for NmFNR.
FIGURE 2.
Kinetics of 20 μmNmFNR breakdown in response to 200 μm oxygen in the absence of DNA. A, the fast spectral changes occurring in the first 2 min following introduction of oxygen were followed at 420 nm. The initial increase in absorbance had a rate constant of 0.021 s−1. B, disappearance of oxygen caused by FNR was monitored using a Clark electrode. 20 μm NmFNR was introduced as marked by an arrowhead. Oxygen uptake occurred over the same time scale (100 s) as the fast spectral change in A. C, spectral changes in NmFNR treated with oxygen over 8,000 s were monitored at 550 nm. Two rate constants were calculated from this fit, 0.0033 s−1 and 0.00051 s−1.
NmFNR Binds Specifically to DNA Containing an FNR Consensus Sequence
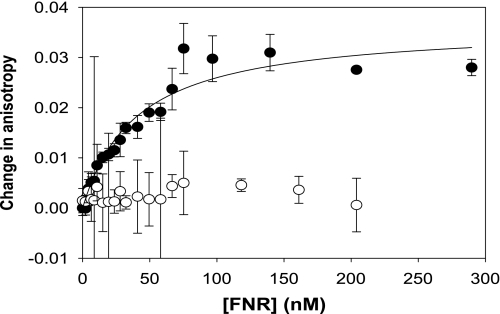
Fluorescence anisotropy was used to investigate the binding of NmFNR to a target DNA sequence containing the FNR consensus site and a control in which the sequence of the consensus site had been scrambled. NmFNR binds to the consensus site but not the random DNA (Fig. 3), and the NmFNR protein was calculated to have a dissociation constant (Kd) of 40 nm for the consensus DNA sequence.
FIGURE 3.
Binding of NmFNR to DNA measured using fluorescence anisotropy. The change in fluorescence anisotropy of 2 nm duplex 40-mer DNA containing a consensus FNR box (filled circles) or lacking an FNR consensus sequence (open circles) was measured on titrating increasing concentrations of anaerobic NmFNR. An increase in anisotropy was observed with the DNA containing the FNR consensus sequence and the data fitted to a hyperbolic curve. Error bars represent S.D. from the mean.
Impact of DNA Binding on Sensitivity of NmFNR to Oxygen
NmFNR pretreated with oxygen for 30 min was unable to bind DNA as judged by fluorescence anisotropy and by EMSA (Fig. 4A). However, if oxygen was introduced into a fluorescence cuvette after the anaerobic titration of NmFNR with DNA, then no loss of fluorescence anisotropy was measurable (supplemental Fig. S3). Similarly, NmFNR remained bound to DNA, as judged by EMSA, if oxygen was added after the protein-DNA complex had formed, for up to an hour (Fig. 4B). Given this unexpected apparent resistance to oxygen of the FNR-DNA complex, we investigated this phenomenon further.
FIGURE 4.
Electrophoretic mobility shift assays show that the NmFNR-DNA complex is relatively oxygen resistant. A, NmFNR was treated with oxygen for various periods and then allowed to complex with DNA containing a consensus FNR site, prior to separating DNA-containing fragments by 8% acrylamide electrophoresis and staining gels for DNA with ethidium bromide. Lane 1, DNA only; lane 2, anaerobic NmFNR and DNA; lanes 3–6, NmFNR made aerobic for 1, 5, 30, and 60 min, respectively, then allowed to complex with DNA. Free DNA is indicated with an asterisk, and an arrowhead indicates where a protein-DNA complex forms for anaerobic NmFNR and NmFNR made aerobic for 1–5 min but not longer periods. The same procedure was followed for B as in A except that the NmFNR-DNA complex was generated prior to treatment with oxygen. An FNR-DNA complex persists after 30 or 60 min under aerobic conditions. Reactions consisted of 10 μl of FNR (92.5 μm, assuming binding as a dimer) and 10 μl of DNA (100 μm duplex) either mixed for 30 min anaerobically prior to O2 exposure (B) or the DNA was added after each time point subsequent to the aerobic exposure of FNR (A).
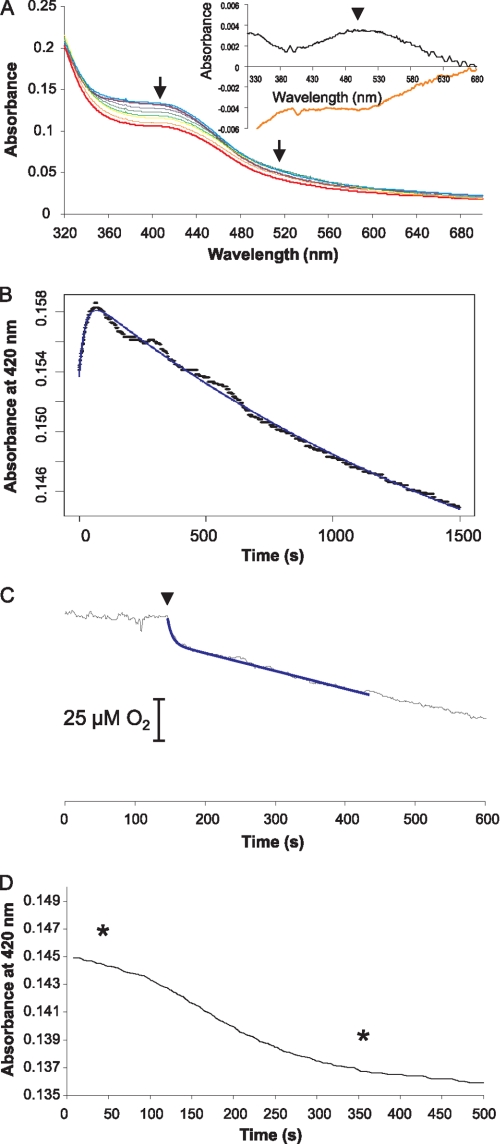
Spectra were measured under anaerobic conditions for NmFNR in the presence of a 1.2-fold molar excess of duplex DNA containing the FNR consensus binding site. The spectral features of the complex were the same as those of the pure protein. In the presence of oxygen, the spectral features of the iron-sulfur cluster change over time. Within a minute the spectral feature at 400 nm increased and was red-shifted slightly (Fig. 5A) as seen in the absence of DNA (inset shows the difference spectrum between time 0 and 1 min, as seen in the absence of DNA), consistent again with the conversion from 4Fe-4S to 3Fe-4S. Subsequent events, however, were dissimilar to what was seen in the absence of DNA. The spectrum slowly decreased in intensity, but there was no red shift of the absorbance around 400 nm, and no appearance of the featureless sloping spectral signal between 500 and 600 nm, typical of a 2Fe-2S cluster. The shape of the spectrum did not change, but there was a steady decline in absorbance across the wavelength range, i.e. there was a gradual decay of the 3Fe-4S cluster to a colorless product.
FIGURE 5.
Spectral changes and oxygen consumption with an NmFNR-DNA complex. A, spectrum of the anoxic NmFNR-DNA complex is shown in bold in violet. Spectral scans taken periodically over 2,040 s following the introduction of 200 μm oxygen are displayed in purple, blue, green, yellow, orange, and red, respectively, with increasing time. Arrows indicate the trend of absorbance change over the period (i.e. absorbance decreases at 400 nm and between 500 and 600 nm). The inset shows the difference spectrum calculated from the spectrum at time = 1 min minus time 0 (black line) and the difference spectrum following a return to anaerobiosis of an oxygen-treated NmFNR-DNA complex (orange line, spectra from time points indicated in D, below). B, absorbance change at 420 nm monitored over time. The spectral changes fit to two linked exponentials with rate constants of 0.043 s−1 and 0.00051 s−1. C, disappearance of oxygen caused by FNR was monitored using a Clark electrode. 20 μm NmFNR-DNA was introduced as marked by an arrowhead. D, NmFNR-DNA complex was exposed to oxygen for 30 min, after which time oxygen depletion was completed. A drop in absorbance at 420 nm was observed following this return to anoxia. Spectra were measured using an equivalent experiment at the times marked by asterisks. The difference between these spectra is shown in the inset to A. Arrowhead indicates the maximum absorbance differences.
The change in absorbance over time was measured at 420 nm. After an increase during the first minute, a very gradual decrease in absorbance was observed for greater than 30 min (Fig. 5B). This data fitted to two linked exponential events with apparent first order rate constants of 0.043 and 0.00051 s−1. The fast rate corresponds to the conversion between 4Fe-4S and 3Fe-4S. While the apparent rate of this step was twice that measured in the absence of DNA, the extent of spectral change was approximately half of that observed in the absence of DNA. Furthermore, oxygen uptake occurred over this time scale (Fig. 5C), and half an oxygen molecule was consumed per FNR monomer, indicating that only half of the 4Fe-4S clusters of FNR are quickly converted to 3Fe-4S. The slow rate of decrease of absorbance at 420 nm is similar to k3 (conversion of 2Fe-2S cluster to apoprotein) measured in the absence of DNA. No spectrum due to a 2Fe-2S cluster was observed, indicating that the conversion of 3Fe-4S cluster to 2Fe-2S cluster is inhibited by binding to DNA, such that its rate is no faster than the rate of conversion of 2Fe-2S cluster to apoprotein, i.e. the presence of DNA stabilizes NmFNR in the presence of oxygen. As a control, the spectral features of NmFNR were monitored in the presence of oxygen and duplex DNA not containing an FNR consensus sequence. In this case, the NmFNR spectral features changed with the same kinetics as NmFNR in the absence of DNA. The protection of NmFNR from oxygen-dependent breakdown (as observed in the presence of the 40-mer DNA sequence from the N. meningitidis aniA promoter) was also seen when NmFNR was incubated in the presence of a 34-bp DNA duplex containing an FNR consensus used previously for studies with EcFNR (27).
Following treatment of the NmFNR-DNA complex with oxygen for 40 min, oxygen was allowed to become depleted. Subsequently to oxygen depletion, the absorbance at 420 nm due to the iron-sulfur cluster decreases (Fig. 5D), and difference spectra before and after oxygen depletion show that a feature ∼490 nm disappears, mirroring the change that occurs on initial introduction of oxygen to NmFNR-DNA complex (Fig. 5A, inset) or NmFNR in the absence of DNA (Fig. 1B, inset), indicating that the partial breakdown of the NmFNR iron-sulfur cluster in the presence of DNA and oxygen is reversible.
DISCUSSION
The overexpression, purification, and characterization of FNR from N. meningitidis reported here indicates that this regulator has similar overall features to its homologue from E. coli. In particular, the proteins recognize the same DNA consensus sequence, and they both contain a labile 4Fe-4S cluster that breaks down in a two-step process to a 2Fe-2S cluster. The first step of this breakdown is the oxygen-dependent step, shown here directly by measuring FNR breakdown in an oxygen electrode. A form of NmFNR, with spectral properties distinct from both the 4Fe-4S cluster and the 2Fe-2S cluster, forms over the same time scale as oxygen utilization occurs. For EcFNR, this intermediate can be identified by EPR as a [3Fe-4S]+ cluster (11), but no EPR-detectable intermediate was identifiable for NmFNR here. The lack of an EPR detectable signal intermediately suggests that for NmFNR, an EPR-silent [3Fe-4S]0 intermediate is formed due to DTT (the inclusion of which is required to maintain the stability of NmFNR, which contains eight Cys residues compared with only five in EcFNR) resulting in the reduction from [3Fe-4S]+ to the EPR-silent [3Fe-4S]0 cluster, as seen in this redox potential range for other 3Fe-4S proteins (23–25).
N. meningitidis FNR Is Relatively Oxygen-insensitive Compared with E. coli FNR
Previously, we showed that an FNR activatable promoter in N. meningitidis is only weakly inhibited by oxygen in vivo (14). This is consistent with the microaerobic lifestyle of N. meningitidis, in which alternatives to oxygen respiration need to be activated even in the presence of finite oxygen content. Here, it is shown that this lack of oxygen sensitivity can be explained by the relative oxygen insensitivity of NmFNR protein itself compared with its homologue from E. coli. The rates of conversion of 4Fe-4S to the presumed 3Fe-4S intermediate, and from that to the 2Fe-2S form are both ∼2.5–3 times slower in NmFNR than EcFNR. This difference in sensitivity to oxygen at first appears to be relatively modest, but is similar to the difference in oxygen sensitivity between wild-type EcFNR and the constitutively active EcFNR* mutant S24F (26). The FNR* mutant of E. coli is able to support FNR-dependent transcription activation aerobically in intact E. coli (26), as observed for wild-type N. meningitidis. The sequence of NmFNR does not reveal any obvious reason for the decreased sensitivity to oxygen in the protein from N. meningitidis (serine is conserved at the position equivalent to Ser-24 in NmFNR), but in the absence of a crystal structure, it is not possible to predict the precise impact of differences in the polypeptide chain on the geometry and environment of the iron-sulfur cluster.
DNA Binding Affects Oxygen Sensitivity of NmFNR
In this paper, we have explored the impact of DNA binding on the sensitivity of NmFNR to oxygen. Fluorescence anisotropy and EMSAs show that an NmFNR-DNA complex is resistant to oxygen treatment. Furthermore, spectroscopic analysis of the NmFNR-DNA complex shows that the breakdown of the iron-sulfur cluster is inhibited compared with the protein in the absence of DNA. The finding that FNR is able to remain bound to its cognate DNA for periods of >30 min (a period similar to the duplication time of a rapidly growing N. meningitidis culture) in air-saturated solutions has obvious implications when considering the mechanism of oxygen sensing by FNR. Sutton et al. (27) showed for EcFNR that DNA has no effect on the rate of iron release from anaerobic FNR following exposure to oxygen in the presence of an iron chelating agent. That experiment effectively measured the conversion of 4Fe-4S cluster to 3Fe-4S cluster (with release of ferrous iron) and is in keeping with our findings here with NmFNR as the rate of 4Fe-4S cluster conversion to 3Fe-4S is unaffected by DNA. It would be interesting to know whether the conversion from 3Fe-4S cluster to 2Fe-2S cluster is affected by DNA for EcFNR as it is for NmFNR.
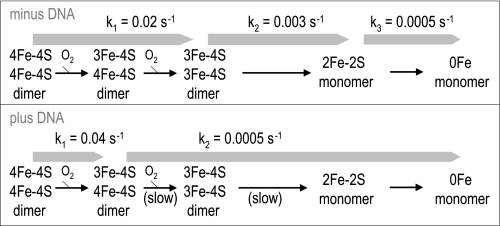
Here, it is found that the rate of the initial oxygen-dependent reorganization of the NmFNR iron-sulfur cluster is unaffected by the presence of DNA, but the extent of oxygen utilization and iron-sulfur cluster spectroscopic change is halved. This leads immediately to the suggestion that half of the 4Fe-4S clusters of NmFNR are converted rapidly to 3Fe-4S on exposure to oxygen, but that half of the clusters are unaffected. Given that FNR binds DNA as a dimer, it is likely that one subunit in each 4Fe-4S cluster-containing dimer is converted to 3Fe-4S, but that the presence of a bound DNA molecule prevents the second 4Fe-4S cluster from breaking down (or increases the back reaction in which iron is reinserted into a 3Fe-4S cluster) due to some long range steric effect of DNA binding (noting that the iron-sulfur cluster is predicted to be located at a site distant from the DNA-binding face of FNR). Implicit in this proposition is that the oxygen dependent breakdown of 4Fe-4S cluster to 3Fe-4S cluster requires a conformational change (at least in the presence of DNA), and that a (partial) 3Fe-4S cluster-containing FNR dimer can bind DNA. In the presence of DNA, no intermediate 2Fe-2S form of the protein is identifiable spectroscopically, indicating that the rate at which the 4Fe-4S/3Fe-4S intermediate breaks down to 2Fe-2S is no faster than the subsequent breakdown of the 2Fe-2S cluster into the apoprotein. The rate of the slow step of breakdown of NmFNR in the presence of DNA is the same as the rate of breakdown from 2Fe-2S cluster to apoprotein in the absence of DNA. Following the resumption of anaerobiosis, the spectral features of a DNA-bound NmFNR mirror the changes occurring immediately after adding oxygen, suggesting that the 3Fe-4S cluster is reassembled to a 4Fe-4S cluster, indicating that this step in the assembly of the iron-sulfur cluster is spontaneous (in the presence of available ferrous iron). Spontaneous conversion from 3Fe-4S to 4Fe-4S in the presence of endogenous iron has been observed for other iron-sulfur proteins previously (28, 29). A model for FNR cluster disassembly is shown in Fig. 6.
FIGURE 6.
Model of FNR disassembly in the presence of oxygen ± DNA. Steps that become significantly slower in the presence of DNA are marked (slow). Rate constants for each step calculated from spectroscopic data in this paper are shown.
In N. meningitidis, the FNR-dependent promoter that has been subject to most detailed study is that for the nitrite reductase (aniA). In this case, a nitric oxide sensing regulator, nitric oxide-sensing repressor, represses FNR dependent activation, probably by binding to an adjacent site that is likely to restrict FNR binding (14, 30). Promoters for other confirmed FNR-dependent genes, such as maltose phosphorylase and other sugar metabolism genes (31), do not contain complete FNR consensus sites. In N. meningitidis, repressors and deviation from an FNR consensus recognition sequence appear to be mechanisms to prevent excessively high expression of FNR-dependent genes. Whether mechanisms are required in vivo in other bacterial species such as E. coli to facilitate removal of FNR from its target DNA is as yet unclear.
To conclude, we have shown here that the oxygen sensitivity of the FNR protein from N. meningitidis is altered significantly by binding to cognate DNA. Many other DNA binding proteins contain sensory iron-sulfur clusters. It has recently been shown that in at least one other case, the superoxide sensitive SoxR, DNA binding has a major impact on the iron-sulfur cluster properties (22). If we wish to appreciate fully the mechanism of action of sensory DNA-binding proteins, it is crucial to investigate their sensory properties in the presence of cognate DNA.
Supplementary Material
Acknowledgment
We are grateful to Clive Butler (University of Exeter) for carrying out EPR experiments with the purified NmFNR.
This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/F000952/1 (to J. W. B. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- FNR
- fumarate and nitrate reduction regulator
- NmFNR
- Neisseria meningitidis FNR
- EcFNR
- Escherichia coli FNR
- PBS
- phosphate-buffered saline
- DTT
- dithiothreitol
- EMSA
- electrophoretic mobility gel shift assay.
REFERENCES
- 1.Yazdankhah S. P., Caugant D. A. (2004) J. Med. Microbiol. 53, 821–832 [DOI] [PubMed] [Google Scholar]
- 2.Can Deuren M., Brandtzaeg P., van der Meer J. W. (2000) Clin. Microbiol. Rev. 13, 144–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brook I. (2003) Int. J. Pediatr. Otorhinolaryngol. 67, 1047–1053 [DOI] [PubMed] [Google Scholar]
- 4.Rock J. D., Mahnane M. R., Anjum M. F., Shaw J. G., Read R. C., Moir J. W. (2005) Mol. Microbiol. 58, 800–809 [DOI] [PubMed] [Google Scholar]
- 5.Kiley P. J., Beinert H. (2003) Curr. Opin. Microbiol. 6, 181–185 [DOI] [PubMed] [Google Scholar]
- 6.Moore L. J., Kiley P. J. (2001) J. Biol. Chem. 276, 45744–45750 [DOI] [PubMed] [Google Scholar]
- 7.Green J., Bennett B., Jordan P., Ralph E. T., Thomson A. J., Guest J. R. (1996) Biochem. J. 316, 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khoroshilova N., Popescu C., Münck E., Beinert H., Kiley P. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6087–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharrocks A. D., Green J., Guest J. R. (1990) FEBS Lett. 270, 119–122 [DOI] [PubMed] [Google Scholar]
- 10.Melville S. B., Gunsalus R. P. (1990) J. Biol. Chem. 265, 18733–18736 [PubMed] [Google Scholar]
- 11.Crack J., Green J., Thomson A. J. (2004) J. Biol. Chem. 279, 9278–9286 [DOI] [PubMed] [Google Scholar]
- 12.Spiro S., Guest J. R. (1990) FEMS Microbiol. Rev. 6, 399–428 [DOI] [PubMed] [Google Scholar]
- 13.Sawers G. (1999) Curr. Opin. Microbiol. 2, 181–187 [DOI] [PubMed] [Google Scholar]
- 14.Rock J. D., Thomson M. J., Read R. C., Moir J. W. (2007) J. Bacteriol. 189, 1138–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delany I., Rappuoli R., Scarlato V. (2004) Mol. Microbiol. 52, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 16.McGuinness B. T., Clarke I. N., Lambden P. R., Barlow A. K., Poolman J. T., Jones D. M., Heckels J. E. (1991) Lancet 337, 514–517 [DOI] [PubMed] [Google Scholar]
- 17.Bonsor D., Butz S. F., Solomons J., Grant S., Fairlamb I. J., Fogg M. J., Grogan G. (2006) Org. Biomol. Chem. 4, 1252–1260 [DOI] [PubMed] [Google Scholar]
- 18.Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 19.Moir J. W., Wehrfritz J. M., Spiro S., Richardson D. J. (1996) Biochem. J. 319, 823–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brumby P. E., Miller R. W., Massey V. (1965) J. Biol. Chem. 240, 2222–2228 [PubMed] [Google Scholar]
- 21.Crack J. C., Green J., Le Brun N. E., Thomson A. J. (2006) J. Biol. Chem. 281, 18909–18913 [DOI] [PubMed] [Google Scholar]
- 22.Gorodetsky A. A., Dietrich L. E., Lee P. E., Demple B., Newman D. K., Barton J. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean K. J., Warman A. J., Seward H. E., Marshall K. R., Girvan H. M., Cheesman M. R., Waterman M. R., Munro A. W. (2006) Biochemistry 45, 8427–8443 [DOI] [PubMed] [Google Scholar]
- 24.Condon C., Cammack R., Patil D. S., Owen P. (1985) J. Biol. Chem. 260, 9427–9434 [PubMed] [Google Scholar]
- 25.Petoukhov M. V., Svergun D. I., Konarev P. V., Ravasio S., van den Heuvel R. H., Curti B., Vanoni M. A. (2003) J. Biol. Chem. 278, 29933–29939 [DOI] [PubMed] [Google Scholar]
- 26.Jervis A. J., Crack J. C., White G., Artymiuk P. J., Cheesman M. R., Thomson A. J., Le Brun N. E., Green J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4659–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutton V. R., Mettert E. L., Beinert H., Kiley P. J. (2004) J. Bacteriol. 186, 8018–8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu A., Gräslund A. (2000) J. Biol. Chem. 275, 12367–12373 [DOI] [PubMed] [Google Scholar]
- 29.Gao-Sheridan H. S., Kemper M. A., Khayat R., Tilley G. J., Armstrong F. A., Sridhar V., Prasad G. S., Stout C. D., Burgess B. K. (1998) J. Biol. Chem. 273, 33692–33701 [DOI] [PubMed] [Google Scholar]
- 30.Heurlier K., Thomson M. J., Aziz N., Moir J. W. (2008) J. Bacteriol. 190, 2488–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartolini E., Frigimelica E., Giovinazzi S., Galli G., Shaik Y., Genco C., Welsch J. A., Granoff D. M., Grandi G., Grifantini R. (2006) Mol. Microbiol. 60, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.