Abstract
Norursodeoxycholic acid (norUDCA) exhibits efficient anti-cholestatic properties in an animal model of sclerosing cholangitis. norUDCA is eliminated as a C23-ester glucuronide (norUDCA-23G) in humans. The present study aimed at identifying the human UDP-glucuronosyltransferase (UGT) enzyme(s) involved in hepatic norUDCA glucuronidation and at evaluating the consequences of single nucleotide polymorphisms in the coding region of UGT genes on norUDCA-23G formation. The effects of norUDCA on the formation of the cholestatic lithocholic acid-glucuronide derivative and of rifampicin on hepatic norUDCA glucuronidation were also explored. In vitro glucuronidation assays were performed with microsomes from human tissues (liver and intestine) and HEK293 cells expressing human UGT enzymes and variant allozymes. UGT1A3 was identified as the major hepatic UGT enzyme catalyzing the formation of norUDCA-23G. Correlation studies using samples from a human liver bank (n = 16) indicated that the level of UGT1A3 protein is a strong determinant of in vitro norUDCA glucuronidation. Analyses of the norUDCA-conjugating activity by 11 UGT1A3 variant allozymes identified three phenotypes with high, low, and intermediate capacity. norUDCA is also identified as a competitive inhibitor for the hepatic formation of the pro-cholestatic lithocholic acid-glucuronide derivative, whereas norUDCA glucuronidation is weakly stimulated by rifampicin. This study identifies human UGT1A3 as the major enzyme for the hepatic norUDCA glucuronidation and supports that some coding polymorphisms affecting the conjugating activity of UGT1A3 in vitro may alter the pharmacokinetic properties of norUDCA in cholestasis treatment.
Introduction
Ursodeoxycholic acid (UDCA)7 is the best studied therapy for cholestatic diseases, such as primary biliary cirrhosis and primary sclerosing cholangitis (reviewed in Ref. 1). However, recent analyses have questioned the survival benefits of UDCA treatment in both primary sclerosing cholangitis and primary biliary cirrhosis (2, 3). 24-Norursodeoxycholic acid (norUDCA), a C23 homologue of UDCA (see Fig. 1) (4), has unique therapeutic properties in an in vivo model of cholestasis (5, 6). As such, Fickert et al. (5) reported that norUDCA is superior to UDCA in the treatment of sclerosing cholangitis using the multidrug resistance protein 2 null (Mdr2−/−) mice. This improvement is due to the resistance of norUDCA to N-acyl amidation (i.e. conjugation with taurine and glycine), a major conjugation pathway for UDCA (6, 7). Bile acid (BA) amidation usually involves the carboxyl group at position C24 (8). As a consequence of its resistance to amidation, the corresponding carboxyl group in norUDCA (located at position C23) remains free for reacting with other conjugation reactions, such as glucuronidation (7). Indeed, in human volunteers, the dominant biotransformation product of norUDCA is a C23-ester glucuronide, which is equally recovered in bile and urine (7).
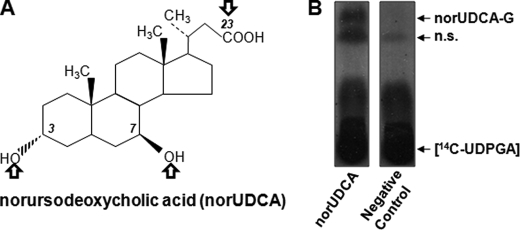
FIGURE 1.
Chemical structure and hepatic glucuronidation of norUDCA. A, norUDCA is a synthetic C23-BA derivative. Arrows indicate the potential sites for glucuronide conjugation. B, norUDCA (200 μm) was incubated for 5 h with human liver microsomes (10 μg) in the presence of radiolabeled [14C]UDPGA. A negative control for the formation of norUDCA glucuronide was performed by incubating human liver microsomes in the absence of norUDCA. The formation of norUDCA-glucuronide was assessed by thin layer chromatography. n.s., not specific.
Glucuronidation is a major drug metabolizing reaction in humans (9). The UDP-glucuronosyltransferase (UGT) enzymes catalyze this enzymatic process, which corresponds to the transfer of glucuronic acid from UDP-glucuronic acid (UDPGA) to hydrophobic molecules (9). The resulting glucuronide conjugates have a high solubility in water and are easily eliminated from the human body into bile or urine (9). Human UGTs are classified into four families: UGT1, UGT2, UGT3, and UGT8. The most important drug conjugating UGTs belong to UGT1 and UGT2 families. Although the UGT1 family is composed of only 1 subfamily, i.e. UGT1A, the members of the UGT2 family are further divided into UGT2A and UGT2B subfamilies. The nine active human UGT1As are produced from a single gene locus located on chromosome 2q37 (10–12). However, an additional common exon 5 (named exon 5b) is alternatively shared to form an additional set of nine UGT1As, designated isoform 2 (i2) (11, 12). These isoforms are enzymatically inactive but act as negative modulators of their active isoform i1 homologue (11). With the exception of UGT1A7, 1A8, and 1A10, all human UGT1A and UGT2B enzymes are expressed in the liver (13).
Although glucuronidation is the major metabolic reaction for norUDCA in humans (7), the UGT enzyme(s) involved in this reaction have never been identified. However, such information may be critical for the future clinical implementation of norUDCA, because some genetic variations of UGT genes exert major impacts on drug efficiency and drug-related toxicities (14, 15). Therefore the primary objectives of the present study were to identify the human UGT(s) involved in norUDCA glucuronidation and to explore how genetic variations in these gene(s) affect norUDCA conjugation. Although glucuronidation is generally viewed as a detoxification mechanism, various studies indicated that the glucuronide conjugates of lithocholic acid (LCA) are at least as pro-cholestatic as the unconjugated lithocholic acid (16, 17). Therefore, the possible interference of norUDCA on the formation of these cholestatic glucuronides was also analyzed in vitro. Finally, because rifampicin is a known UGT inducer used in cholestatic patients to treat pruritus (18), a possible drug-drug interaction with norUDCA has been investigated by analyzing whether rifampicin modulates norUDCA glucuronidation in human hepatocytes.
EXPERIMENTAL PROCEDURES
Materials
Normal and deuterated LCA and/or chenodeoxycholic acid (CDCA) were purchased from Steraloids (Newport, RI) and C/D/N Isotopes (Montréal, Canada), respectively. norUDCA was as described (4, 6). UDPGA and all aglycones were obtained from Sigma and ICN Pharmaceuticals, Inc. (Québec, Canada). [14C]UDPGA (285 mCi/mmol) was obtained from PerkinElmer Life Sciences. Commercially available microsomes from human liver and intestine were from BD Biosciences (Mississauga, Canada). High pressure liquid chromatography grade reagents were from VWR Canada (Mississauga, Canada), Laboratoire MAT (Québec, Canada), Sigma, and ThermoFisher (Ottawa, Canada).
Human Liver Tissues, Cell Culture, and mRNA Level Determination
Human liver samples (n = 16) were from kidney donors and were stored at the University of Toronto Department of Pharmacology as previously described (19, 20).
Human embryonic kidney 293 (HEK293) cells were obtained from the American Type Culture Collection (Manassas, VA) and were grown as reported (20). HEK293 cells expressing human UGTs (UGT-HEK293) and UGT1A3 variant isozymes were engineered as previously described (20, 21).
Cryopreserved human primary hepatocytes (supplemental Table S1) were obtained from Celsis-InVitro Technologies (Baltimore, MD). Hepatocytes were seeded in InVitroGro CP medium at a concentration of 3.75 × 105 cells/well in 12-well plates for 48 h, with medium changes after 24 h as recommended by the supplier. The cells were subsequently incubated in InVitroGrow Hi medium containing rifampicin (20 μm) or Me2SO (0.1%, vehicle) for 48 h. The cells were then homogenized in phosphate-buffered saline containing dithiothreitol (0.5 mm) for glucuronidation assays (see below). In parallel experiments, hepatocytes (2.5 × 105 cells/well in 24-well plates) were also treated with Me2SO or rifampicin (20 μm) for 48 h. Total RNA was isolated according to the Tri-Reagent acid:phenol protocol as specified by the supplier (Molecular Research Center Inc., Cincinnati, OH). The reverse transcription reaction was performed as previously described (22). Cytochrome P450 3A4 mRNA levels were then quantified by real time PCR using 1/500-diluted reverse transcription products and 125 nm of the following primers: sense, 5′-ccaagctatgctcttcaccg, and antisense, 5′-gcaccgtaagtggtgcctga at 64 °C, in a reaction mix previously reported (22). The 28 S RNA was used as the housekeeping gene (23).
Microsome Isolation and Glucuronidation Assays
Microsomes from UGT-HEK293 cells or human liver samples (200 mg) (19, 20) were purified as previously reported (24). Microsomes pellets were resuspended at a concentration of 5 μg/μl.
All of the glucuronidation assays were performed in the previously reported glucuronidation assay buffer (20) complemented with 0.025 μg/ml alamethicin and were ended by adding 100 μl of methanol with 0.02% butylated hydroxytoluene (20).
Assays with human liver microsomes and norUDCA (200 μm) were conducted with 10 μg of microsomes in the assay buffer supplemented with 7.5 μm [14C]UDPGA and 92.5 μm unlabeled UDPGA for 5 h at 37 °C. The formation of norUDCA-glucuronide was assessed by thin layer chromatography.
Quantitative analyses were performed with 100 μg of hepatocytes homogenates or 10 μg of microsomes for the indicated duration and with the indicated concentration of norUDCA in the same conditions as above, except that the assay buffer did not contain [14C]UDPGA and was complemented with 2 mm unlabeled UDPGA.
Inhibition constant (Ki) values were assessed through enzymatic assays using LCA and norUDCA concentrations ranging from 1 to 100 μm and 3 to 200 μm, respectively. The Ki values were estimated using Dixon plots (25).
Determination of Glucuronide Levels by Liquid Chromatography Coupled to Mass Spectrometry (LC-MS/MS)
The chromatographic system consisted of an Agilent 1200 series (Agilent Technologies, Montréal, Canada) equipped with a Synergie Hydro 100 × 4.6 mm × 4 μm column (Phenomenex, Torrance, CA) and coupled to a tandem quadruple mass spectrometer API3200 (Applied Biosystems-Sciex, Concord, Canada). Solvent A corresponded to water with 5 mm ammonium formate, and solvent B corresponded to methanol with 5 mm ammonium formate.
Assays with LCA and norUDCA were analyzed in the presence of [2H4]LCA-24G or [2H4]CDCA-24G, respectively, as internal standards. Analytical and internal standards were enzymatically produced with human liver microsomes as described (26).
Glucuronide conjugate separation was achieved at a flow rate of 0.9 ml/min. The analytes were eluted using the following program: 0–9 min: isocratic 50% A and 50% B; 9–11 min, linear gradient 50 to 60% B; 11–13 min, isocratic 60% B; 13–13.1 min, linear gradient 60–90% B; 13.1–15 min, isocratic 90% B; 15–15.1 min, linear gradient 90–50% B; and 15.1–20 min, isocratic 50% B (column washing). The glucuronide conjugates were monitored in a negative mode, but quantification was achieved by multiple reaction monitoring mode. Negative product multiple reaction monitoring ion pairs were m/z 553.5 → 377.5 for norUDCA-G, m/z 551.5 → 375.5 for LCA-G, 555.5 → 379.5 for [2H4]LCA-24G, and 571.5 → 395.5 for [2H4]CDCA-24G. The limit of quantification of this method was 0.5 ng/ml.
Production of the Anti-UGT1A3 Antibody
An 82-amino acid fragment (64–145) of the UGT1A3 protein fused to the glutathione S-transferase protein has been constructed as previously described (27). Production and purification of the fusion protein and immunization procedures were performed as fully described elsewhere (28). Rabbit sera were tested for the presence of antibodies against UGTs by Western blot using microsomal extracts (10 μg) from human liver and HEK293 cells stably expressing human UGT proteins.
Western Blot Analyses
Microsomes (10 μg) were size-separated by 10% SDS-PAGE, transferred onto nitrocellulose membranes, and then hybridized with the anti-UGT1A3 antibody (1:2,000). An anti-rabbit IgG horse antibody (1:10,000) conjugated with peroxidase (Amersham Biosciences) was used as the second antibody, and the resulting immunocomplexes were visualized and quantified as described (20). The same membrane was then rehybridized with an anti-calnexin (1:5,000) antibody (Stressgen, Ann Arbor, MI) as a loading control assessment.
Determination of UGT1A3 Protein Levels by LC-MS/MS
Microsomes (100 μg) were precipitated with acetone (6 volumes) for 2 h at −20 °C and centrifuged at 13,000 × g for 10 min, and the pellets were resuspended in 50 μl of a 100 mm ammonium carbonate, 8 m urea solution. One-fifth of the solution was reduced in the presence of 5 μl of dithiothreitol (45 mm) for 30 min at 37 °C and alkylated with 5 μl of iodoacetamide (100 mm) for 20 min in the dark. Subsequently, the proteins were digested in the presence of 0.5 μg of trypsin (Promega) in a 50 mm ammonium bicarbonate buffer for 18 h at 37 °C. The reaction was stopped by the addition of formic acid 1%. Peptides were desalted with PepClean C18 Spin columns, evaporated to dryness under speed vacuum, and stored at −20 °C before LC-MS/MS measurements.
A stable isotope internal standard corresponding to the UGT1A3-specific peptide YLSIPTVFFLR containing 13C- and 15N-labeled arginine was ordered from Uvic Genome BC Proteomics Centre (Victoria, Canada).
For LC-MS/MS analyses, the digested proteins were resuspended at a concentration of 0.5 μg/μl in a solution of 0.1% formic acid containing the labeled peptide (5 fmol/μl) as an internal standard. The solution was then injected on a C18 self-packed reverse phase nanoflow column (Picofrit self-packed, 75 μm) packed with Jupiter 5μ 300A C18 particle (Phenomenex) and eluted for 25 min in a gradient of 2–38% acetonitrile with 0.1% formic acid using an Eksigent one-dimensional nanoLC system (Dublin, CA). Each digested sample was injected three times independently, and a blank was eluted between each sample.
The detection of digested peptides and internal standards was performed on an API4000 QTRAP hybrid triple quadrupole linear ion trap mass spectrometer (Applied Biosystems, Foster City, CA) equipped with a nanospray source. The mass spectrometer was operated in positive ion mode with multiple reaction monitoring detection. The identity of the quantified peptide was further ensured through MS/MS analyses using a MIDAS™ workflow software (Applied Biosystems).
The concentration of UGT1A3 proteins in each sample was calculated using the ratio of the area under the curve of the digested peptide versus the area under the curve of the internal standard (labeled peptide) injected at 5 fmol/μl.
Statistical Analyses
Correlation studies as well as parametric Student's t test used to analyze for significant differences between the experimental groups were performed using the JMP V7.0.1 program (SAS Institute, Cary, NC).
RESULTS
Human Liver Microsomes Form Three norUDCA Glucuronide Conjugates
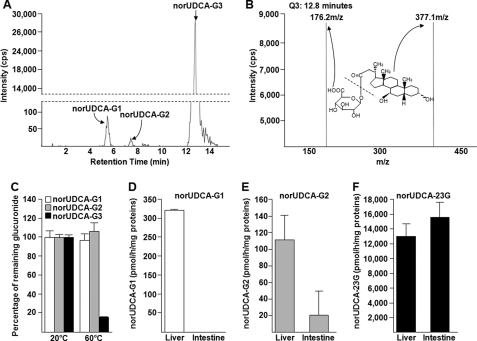
In line with previous findings (7), in vitro glucuronidation assays performed in the presence of [14C]UDPGA revealed the ability of human liver microsomes to form a radiolabeled conjugate of norUDCA (Fig. 1B). As shown on Fig. 1A, norUDCA contains three potential acceptor groups for the glucuronosyl moiety: the carboxyl group at position 23 and two hydroxyl groups at positions 3 and 7. To determine whether one or several groups are used by the hepatic glucuronidation system, norUDCA was reassayed using a sensitive LC-MS/MS detection instead of radiolabeling detection. Three glucuronide conjugates were detected with retention times of 5.8, 7.6, and 12.8 min for norUDCA-G1, -G2, and -G3, respectively (Fig. 2A). The relative intensity of each glucuronide revealed that norUDCA-G3 was far more abundantly formed than the two others. Using a procedure that we established for other BA-glucuronide conjugates (26), specific analytical standards were generated for each of the three glucuronides. MS/MS spectra analyses with low energy collision of the deprotonated ions revealed the formation of only two daughter ions corresponding to norUDCA (m/z 377) and UDPGA (m/z 176), as exemplified with norUDCA-G3 (m/z 553) in Fig. 2B. These observations confirmed the glucuronide conjugate nature of the purified standards. Considering that ester glucuronides are highly susceptible to hydrolysis under elevated temperature conditions (26, 29–31), standards were incubated at 60 °C for 2 h to discriminate between the hydroxyl- and carboxyl-linked glucuronides (Fig. 2C). Under these conditions, only 16% of the initial concentration of norUDCA-G3 was detected, whereas the elevated temperature had only minor effects on the amount of norUDCA-G1 and -G2. Thus, these experiments identify the very abundant norUDCA-G3 as corresponding to the previously identified C23-ester (i.e. acyl) norUDCA glucuronide, norUDCA-23G (7).
FIGURE 2.
norUDCA-23G is the most abundant glucuronide conjugate formed by microsomes from human liver and intestine. A, chromatographic separation of the glucuronide conjugates formed from norUDCA (75 μm) incubated in the presence of microsomal proteins from human liver (10 μg). B, spectra of product ions from low energy collision (Q3) of the deprotonated ions of norUDCA-G3 (eluted at 12.8 min). The chemical structures of norUDCA-3G and the corresponding ions are indicated. C, incubation of the purified glucuronides of norUDCA at 60 °C for 2 h identifies norUDCA-G3 as norUDCA-23G. The values are the means ± S.D. of a duplicate experiment. D–F, human liver or intestine microsomes were incubated in the presence of norUDCA (75 μm) and UDPGA (2 mm) for 1 h at 37 °C. The formation of norUDCA-G1 (D), -G2 (E), and -23G (F) was analyzed by LC-MS/MS. The data represent the means ± S.D. of two different experiments performed in triplicate.
Quantitative experiments were then conducted to ensure the relative production of the three glucuronides using microsomal proteins from human liver and intestine (Fig. 2, D–F). norUDCA-G1 was detected only after incubation with hepatic extracts (Fig. 2D), whereas norUDCA-G2 and -23G were detected in the presence of both hepatic and intestinal microsomes (Fig. 2, E and F). However, norUDCA-G2 was more efficiently synthesized with human liver than with intestine, whereas both microsomal preparations produced similar amounts of norUDCA-23G. More importantly, these quantitative analyses evidenced that the hepatic production of norUDCA-23G is 40 and 120 times higher than those of norUDCA-G1 and -G2, respectively (Fig. 2, D–F). Overall, these results confirm the previous observations by Hofmann et al. (7) that the C23-ester glucuronide is the predominant hepatic glucuronide of norUDCA. The two other conjugates appear as minor catabolites formed exclusively in in vitro assays (7) and have received only minor attention in the following analyses.
Human UGT1A3 Plays a Major Role in the Hepatic Glucuronidation of norUDCA
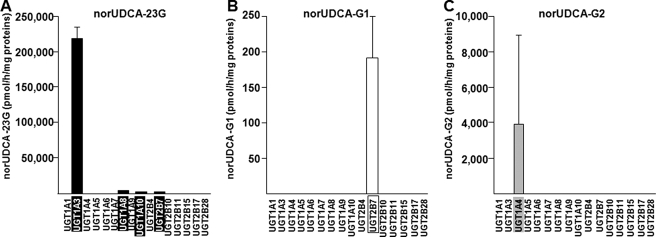
A screening assay realized with microsomes from UGT-HEK293 cells under quantitative conditions (75 μm norUDCA and 1 h of incubation) (20, 26) and using LC-MS/MS detection demonstrated that UGT1A3 exerts a predominant role in producing norUDCA-23G (Fig. 3A). However, the sensitivity of the LC-MS/MS method also allowed the detection of norUDCA-23G production with UGT1A8, 1A10, and 2B7 enzymes but at a much lower rate than UGT1A3. norUDCA-G1 and -G2 were produced only in the presence of microsomes from cells expressing UGT2B7 (Fig. 3B) and UGT1A4 (Fig. 3C), respectively. It is remarkable that the UGT2B7- and UGT1A4-dependent formation of these conjugates appears very low compared with the ability of UGT1A3 to convert norUDCA into norUDCA-23G. Identical results were obtained when replacing the microsomes from UGT-HEK293 cell lines by homogenates from the same cell lines or by commercial bacculosomes (supplemental Fig. S1). On the other hand, similar experiments performed with higher concentrations of norUDCA (200 μm instead of 75 μm) showed similar pattern of glucuronidation activities (data not shown). Finally, norUDCA glucuronidation was also assayed with homogenates from HEK293 cells overexpressing the shorter UGT1A3-i2 isozyme (11), and no glucuronide conjugation was detected (data not shown).
FIGURE 3.
UGT1A3 catalyzes the formation of norUDCA-23G. Microsomes from UGT-HEK293 cells (10 μg) were incubated in the presence of norUDCA (75 μm) for 1 h at 37 °C. The formation of norUDCA-23G (A), -G1 (B), and -G2 (C) was analyzed by LC-MS/MS. The data represent the means ± S.D. of three different experiments performed in triplicate.
Subsequently, norUDCA and LCA, a well characterized UGT2B7 (hydroxyl glucuronidation) and UGT1A3 (carboxyl glucuronidation) substrate (32, 33), were incubated with microsomal preparations from 16 human livers. All of the preparations produced norUDCA-G1, -G2, and -23G as well as LCA-3G and -24G. Furthermore, despite the very limited number of microsome samples, a significant correlation was found between the formation of norUDCA-23G and LCA-24G (Rs = 0.8706, p < 0.0001) and between the formation of LCA-3G and norUDCA-G1 (Rs = 0.7853, p < 0.001) (Table 1). These observations are indicative of the role of UGT2B7 in the hepatic formation of norUDCA-G1 and more importantly support the predominant role played by UGT1A3 in the production of norUDCA-23G by hepatic extracts.
TABLE 1.
Summary of the correlation studies between norUDCA and LCA glucuronidation rates and/or UGT1A3 protein levels in the human liver bank (n = 16)
The correlation analyses were performed using the Spearman rank order correlation (Rs), and the p values for comparisons are indicated. Bold type indicates the significant correlations. WB, Western blotting.
| NorUDCA-G1 |
NorUDCA-G2 |
NorUDCA-23G |
LCA-3G |
LCA-24G |
UGT1A3 (WB) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rs | p | Rs | p | Rs | p | Rs | p | Rs | p | Rs | p | |
| NorUDCA-G2 | 0.0676 | 0.8034 | ||||||||||
| NorUDCA-23G | 0.3029 | 0.2541 | −0.076 | 0.7783 | ||||||||
| LCA-3G | 0.7853 | <0.001 | −0.062 | 0.8202 | 0.4235 | 0.1021 | ||||||
| LCA-24G | 0.4853 | 0.0567 | −0.085 | 0.7535 | 0.8706 | <0.0001 | 0.4882 | 0.0565 | ||||
| UGT1A3 (WB) | 0.2735 | 0.3053 | −0.059 | 0.8287 | 0.9294 | <0.0001 | 0.4529 | 0.0781 | 0.7441 | <0.001 | ||
| UGT1A3 (LC-MS/MS) | 0.0660 | 0.8075 | −0.278 | 0.2969 | 0.6239 | <0.05 | 0.1266 | 0.6404 | 0.6432 | <0.05 | 0.6696 | <0.001 |
To further ascertain the role of UGT1A3 in hepatic norUDCA-23G formation, we next correlated the glucuronidation activity with the content in UGT1A3 protein within the 16 liver microsomal extracts. For this purpose, UGT1A3 protein levels were quantified through a LC-MS/MS method (Fig. 4, A and B). A correlation study revealed that the level of UGT1A3 protein was significantly correlated (Rs = 0.6239, p < 0.05) with the production of norUDCA-23G by liver microsomes (Table 1). As expected the UGT1A3 protein levels determined through LC-MS/MS also correlated with LCA-24G formation (Rs = 0.6432, p < 0.05).
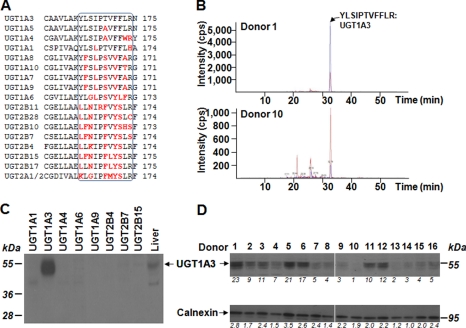
FIGURE 4.
Quantification of the UGT1A3 protein content in 16 human liver samples through LC-MS/MS (A and B) and Western blotting (C and D). A, alignment of the specific UGT1A3 11 amino acids peptide (black box) produced upon trypsin digestion with the corresponding regions in other human UGTs. Letters in red highlight the unconserved amino acids. NCBI Blast analyses confirmed that no other known human protein contains such a peptide (NCBI Blastp). This specific peptide was used to quantify UGT1A3 protein levels by LC-MS/MS. B, extracted ion chromatogram of digested peptide in microsomal proteins from donor 1 (upper panel) and 10 (lower panel) showing that both the digested peptide (blue line) and radiolabeled internal standards (red line) eluate simultaneously. C, a Western blot analysis of the anti-UGT1A3 antibody (1:2,000) with microsomal proteins (10 μg) from the human liver (commercial preparation) and from UGT-HEK293 cells confirmed the specificity of the antibody. D, microsomes from human liver samples were size-separated on SDS-PAGE and hybridized with the anti-UGT1A3 (1:2,000, upper panel) and anti-calnexin (loading control, 1:10,000, lower panel) antibodies. The relative content in proteins was quantified by optical densitometry and expressed as relative to the lower expression sample.
In parallel experiments, the relative UGT1A3 protein level was also determined through Western blotting. For this purpose, a specific rabbit anti-UGT1A3 antibody was generated (Fig. 4C). When assayed with human liver and UGT-HEK293 cells microsomes, this antibody formed a complex only in the presence of microsomes from human liver and UGT1A3-HEK293 cells, thus confirming its specificity for UGT1A3 (Fig. 4C). Subsequently, microsomes from the 16 livers were then immunoblotted with this antibody and with an anti-calnexin antibody (a microsomal protein used as a control for equal loading) (Fig. 4D). UGT1A3 protein levels were subsequently normalized for calnexin levels, and the corrected values were then correlated to norUDCA-23G production. These analyses led to a very strong correlation of Rs = 0.9294 (p < 0.0001) (Table 1). As expected the corrected UGT1A3 protein levels also correlated with the production of LCA-24G (Rs = 0.7441, p < 0.001) (Table 1). Furthermore, a significant correlation (Rs = 0.6696, p < 0.001) was also observed between the two methods used to determine UGT1A3 protein levels (Table 1).
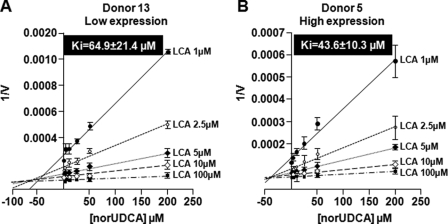
norUDCA Inhibits Hepatic LCA Formation
The present observation that LCA-24G formation by human liver microsomes strongly correlates with norUDCA-23G production confirms that both acids are substrates of the UGT1A3 enzyme in the human liver (32, 33). Therefore, we next investigated whether norUDCA could interfere with the formation of the cholestatic LCA-24G in human liver microsomes and whether such interference may be affected by the level of UGT1A3 expression. For this purpose, hepatic microsomes with high and low UGT1A3 protein contents (from donors 5 and 13, respectively) were incubated in the presence of increasing LCA and norUDCA concentrations (Fig. 5). A marked decrease in the LCA-24G formation was shown to be dose-dependent with the two microsomal preparations tested. The apparent Ki values estimated from Dixon plots were in the same range for donors 5 (43.6 ± 10.3 μm) and 13 (64.9 ± 21.4 μm) (Fig. 5).
FIGURE 5.
norUDCA inhibits LCA-24G production in selected human liver samples. Dixon plots analyses for the inhibition of LCA-24G production by norUDCA in representative human liver samples from patients with low (A) and high (B) UGT1A3 protein levels. The data represent the means ± S.D. of two different experiments performed in triplicate.
Kinetic Parameters for the Formation of norUDCA-23G by UGT1A3 and Altered Glucuronidation by Recombinant Variant Allozymes
The kinetic parameters of the reference UGT1A3 protein (*1) as well as the functional significance of 10 alleles (*2 to *11) previously described for this gene (21) were studied by using stably expressed allozymes in HEK293 cells (21) incubated in vitro in the presence of increasing norUDCA concentrations (1–200 μm) (Table 2). Under these conditions, all of the kinetics fitted the Michaelis-Menten model. However, it should be noted that when higher norUDCA concentrations were analyzed (up to 1 mm), reduced glucuronidation activities were observed. It cannot be excluded that reduced glucuronidation rates observed at such elevated concentrations reflected changes in the chemical properties of the drug or UDPGA depletion in the assay buffer. Therefore these data were not considered for the subsequent calculation of kinetics parameters.
TABLE 2.
Kinetic parameters for the production of norUDCA-23G by UGT1A3 allozymes
CLINT, intrinsic clearance (Km/Vmax); H, high capacity = CLINT > 1000 μl/h/mg of protein; L, low capacity = CLINT < 100 μl/h/mg of protein; I, intermediate clearance = CLINT ≥ 100 and 1000 μl/h/mg of protein.
| UGT1A3 | Haplotypes | Km | Apparent Vmax | Normalized Vmaxa | CLINT | Function |
|---|---|---|---|---|---|---|
| μm | nmol/h/mg of proteins | nmol/h/mg of proteins | μl/h/mg of proteins | |||
| *1 | 14.9 ± 7.1 | 85.7 ± 18.0 | 85.7 ± 18.0 | 5,751.7 | H | |
| *2 | Arg11, Ala47 | 29.2 ± 4.7 | 154.4 ± 51.5 | 57.2 ± 19.1 | 1,958.9 | H |
| *3 | Arg11 | 31.2 ± 6.0 | 187.0 ± 74.7 | 43.5 ± 17.4 | 1,394.2 | H |
| *4 | Trp45 | 114.5 ± 19.1 | 12.8 ± 1.8 | 5.3 ± 0.7 | 46.2 | L |
| *5 | Arg6, Arg11 | 30.6 ± 6.2 | 278.0 ± 47.9 | 17.0 ± 2.9 | 555.5 | I |
| *6 | Arg11, Ala47, Val270 | 320.0 ± 33.9 | 1.0 ± 0.4 | 0.3 ± 0.1 | 0.9 | L |
| *7 | Ile110 | 84.3 ± 37.6 | 129.8 ± 4.7 | 16.6 ± 0.6 | 196.9 | I |
| *8 | Val158 | 295.1 ± 144.5 | 2.1 ± 0.8 | 0.4 ± 0.2 | 1.3 | L |
| *9 | Arg11, Leu208 | 33.8 ± 12.0 | 203.1 ± 44.8 | 27.1 ± 6.0 | 799.4 | I |
| *10 | Ala47 | 49.6 ± 0.6 | 253.8 ± 55.5 | 41.6 ± 9.1 | 838.7 | I |
| *11 | Arg11, Ala47, Ile114 | 42.6 ± 6.5 | 242.9 ± 33.0 | 19.7 ± 2.7 | 462.4 | I |
a For more accurate assessment of the quantitative differences in glucuronidation activity, the absolute Vmax values were normalized for immunoreactive UGT protein levels as previously determined (21).
With the exception of UGT1A3*4, *6, and *8, which presented elevated Km values (114.5, 320.0, and 295.1 μm, respectively), all other values varied within a range of 15–85 μm, with the *1 enzyme showing the lowest values of 14.9 μm (Table 2). After normalization of apparent Vmax by the levels of UGT1A3 expression as established in these cell lines (21), all of the variants demonstrated altered velocity to conjugate norUDCA compared with the reference enzyme *1. Although velocity was reduced by 3–30-fold for allozymes UGT1A3*2, *3, *5, *7, and *9–11, a more dramatic decrease was observed for UGT1A3*4, *6, and *8, reducing by 125-, 6,400-, and 4,400-fold the velocity of the respective protein when compared with UGT1A3*1 (Table 2).
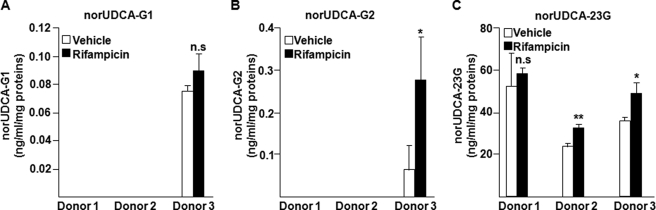
Rifampicin Differentially Modulates norUDCA-23G Production in Human Hepatocytes
Previous studies evidenced that UGT1A3 is a positively regulated target gene of the pregnane X receptor (PXR) in the liver (34). Therefore, we investigated whether rifampicin, which activates PXR, may affect the glucuronide conjugation of norUDCA in three different preparations of human hepatocytes (Fig. 6). The formation of norUDCA-23G was detected in all three preparations and varied from 22 to 58 ng/ml/mg proteins, whereas only cells from donor 3 were able to produce norUDCA-G1 and -G2 at very low levels. Furthermore, rifampicin treatment resulted in a small but significant 1.4-fold increase in norUDCA-23G production only with cells from donors 2 (p < 0.01) and 3 (p < 0.05), whereas the ability to form norUDCA-23G of hepatocytes from donor 1 was not affected by the PXR ligand (Fig. 6). Determination of the mRNA levels of the PXR target gene cytochrome P450 3A4 in parallel mRNA experiments showed that the rifampicin-dependent activation of the receptor strongly modulates its target gene expression in the same hepatocyte preparations (supplemental Fig. S2). These results further support that rifampicin is only a weak inducer of norUDCA glucuronidation.
FIGURE 6.
Effect of rifampicin on norUDCA-23G formation in human hepatocytes. Human hepatocytes from three donors (supplemental Table S1) were treated with vehicle (0.1% Me2SO) or rifampicin (20 μm) for 48 h and subsequently homogenized for in vitro glucuronidation assays with norUDCA (75 μm) for 1 h. The formation of norUDCA-G1 (A), -G2 (B), and -23G (C) was analyzed by LC-MS/MS. Statistically significant differences are indicated by asterisks (Student t test; **, p < 0.01; *, p < 0.05). n.s., not significant.
DISCUSSION
This study identifies UGT1A3 as the major human hepatic UGT enzyme for the formation of norUDCA-23G, the dominant biotransformation product of norUDCA in humans (7). This observation is consistent with previous studies that have established UGT1A3 as an important component of the hepatic BAs glucuronidation machinery (20, 22, 32, 33). Indeed, UGT1A3 catalyzes the ester glucuronide conjugation of BAs, such as CDCA, LCA, and hyodeoxycholic acid (20, 32, 33). The fact that UGT1A3 also conjugates the carboxyl group of norUDCA indicates that even for such a synthetic BA with shortened side chain, UGT1A3 conserves the same stereospecificity as for endogenous BA substrates (20, 32, 33). Such an observation is in line with previous reports indicating that UGT1A3 is able to use the carboxyl position of the short chain homologues of LCA, namely the etianic (C20αβ) and isoetianic (C20ββ) acids (33). Overall, these observations demonstrate that UGT1A3 is highly specialized for the glucuronide conjugation of the carboxyl position found at the extremity of both natural and synthetic BA side chains, whatever the length of this chain. On the other hand, this study also reveals the role that UGT2B7 and UGT1A4 exert in the formation of the two very minor hydroxyl-linked glucuronides norUDCA-G1 and -G2, respectively. Although these metabolites are not detected in vivo (7), it is interesting to note that the same UGT enzymes were also identified as the human enzymes forming the hydroxyl-glucuronides CDCA-3G and LCA-3G (20, 33). Thus, these in vitro observations suggest that the stereospecificity not only of UGT1A3 but also of all BA-glucuronidating enzymes is conserved for the conjugation of the synthetic acid norUDCA.
Accordingly with the predominant role exerted by UGT1A3 in both LCA-24G (33) and norUDCA-23G formation, we observe that norUDCA-23G and LCA-24G formations correlate to each other and, more importantly, that pharmacologically relevant concentrations (7) of norUDCA inhibit the hepatic formation of LCA-24G. Various studies revealed that LCA-glucuronide is a cholestatic agent, exhibiting similar and even higher cholestatic properties as the unconjugated lithocholic acid in rats (16, 17). However, these studies have used LCA-3G, the hydroxyl-linked glucuronide conjugate of LCA. Based on the facts that acyl glucuronides, such as LCA-24G, are extremely hepatotoxic (35) and that human plasma contains similar amount of LCA-3 and -24G (36), it is tempting to speculate that the inhibition of hepatic LCA-24G formation takes place within the series of anti-cholestatic properties exhibited by norUDCA (5, 6). However, such an hypothesis will have to be validated in vivo.
Kinetic parameters analyses revealed that the reference UGT1A3 enzyme (*1) catalyzes the formation of norUDCA-23G with an affinity (Km = 14.9 ± 7.1 μm) almost identical to the Km value (15.2 ± 0.4 μm) reported for the UGT1A3-dependent formation of the C24-ester glucuronide metabolite of the primary BA CDCA (20). This observation suggests that in the presence of the UGT1A3*1 enzyme, norUDCA behaves like natural C24 BAs. On the other hand, when the *1 enzyme was replaced by its naturally occurring variants (*4 to *11), the kinetic parameters of norUDCA glucuronidation were affected for several enzymes. On the basis of these observations, UGT1A3 allozymes could be divided in three groups, namely the enzymes with high (UGT1A3*1, *2, and *3), low (UGT1A3*4, *6, and *8), and intermediate capacity (UGT1A3*5, *7, and *9–11) (Table 2). A similar classification was reported when the consequences of UGT1A3 gene polymorphisms on estrogen metabolism were analyzed (21). A previous pharmacogenomic study established that 5.2% of the Caucasian population carry at least one low activity allele and are expected to present significantly lower glucuronidation profiles of UGT1A3 substrates (21). In the context of a clinical implementation for norUDCA, the results presented here are indicative of the potential need for future pharmacogenetic studies designed to investigate whether UGT1A3 genetic polymorphisms impact the pharmacokinetic profile of norUDCA, particularly in patients carrying low activity alleles.
Rifampicin is the second line therapeutic approach in the treatment of cholestasis-associated pruritus (18); therefore it is likely that a significant part of future norUDCA-treated patients may also be exposed to rifampicin. Only a weak, but significant, induction of norUDCA-23G production was observed in two of the three hepatocyte samples tested. An inducing effect was expected based on the activity of rifampicin as a PXR activator and the previous identification of UGT1A3 as a positively regulated PXR target gene (34). However, the weak induction of norUDCA-23G production observed in human hepatocytes exposed to rifampicin suggests that the antibiotic will only marginally interfere with norUDCA metabolism in co-treated patients. In conclusion, this study identifies UGT1A3 as the major hepatic UGT enzyme for the glucuronide conjugation of norUDCA in humans and provides a strong background for investigating in vivo how glucuronidation interferes with the pharmacological effects and pharmacokinetic properties of norUDCA in cholestasis treatment.
Supplementary Material
Acknowledgments
We thank Prof. Alan F. Hofmann and Dr. Virginie Bocher for critical reading of the manuscript and helpful discussions on this work.
This work was supported by Canadian Institute of Health Research Grant MOP-84338, Canadian Foundation for Innovation Grant 10469, and Austrian Science Foundation Grant P19118-B05.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- UDCA
- ursodeoxycholic acid
- norUDCA
- norursodeoxycholic acid
- UGT
- UDP-glucuronosyltransferase
- LCA
- lithocholic acid
- BA
- bile acid
- UDPGA
- UDP-glucuronic acid
- CDCA
- chenodeoxycholic acid
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- PXR
- pregnane X receptor.
REFERENCES
- 1.Zollner G., Trauner M. (2009) Br. J. Pharmacol. 156, 7–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gong Y., Huang Z. B., Christensen E., Gluud C. (2009) The Cochrane Library http://www.cochrane.org/reviews/en/ab000551.html
- 3.Paumgartner G., Pusl T. (2008) Clin. Liver Dis. 12, 53–80 [DOI] [PubMed] [Google Scholar]
- 4.Schteingart C. D., Hofmann A. F. (1988) J. Lipid Res. 29, 1387–1395 [PubMed] [Google Scholar]
- 5.Fickert P., Wagner M., Marschall H. U., Fuchsbichler A., Zollner G., Tsybrovskyy O., Zatloukal K., Liu J., Waalkes M. P., Cover C., Denk H., Hofmann A. F., Jaeschke H., Trauner M. (2006) Gastroenterology 130, 465–481 [DOI] [PubMed] [Google Scholar]
- 6.Halilbasic E., Fiorotto R., Fickert P., Marschall H. U., Moustafa T., Spirli C., Fuchsbichler A., Gumhold J., Silbert D., Zatloukal K., Langner C., Maitra U., Denk H., Hofmann A. F., Strazzabosco M., Trauner M. (2009) Hepatology 49, 1972–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofmann A. F., Zakko S. F., Lira M., Clerici C., Hagey L. R., Lambert K. K., Steinbach J. H., Schteingart C. D., Olinga P., Groothuis G. M. (2005) Hepatology 42, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 8.Hofmann A. F. (2009) Front. Biosci. 14, 2584–2598 [DOI] [PubMed] [Google Scholar]
- 9.Dutton G. J. (1980) Glucuronidation of Drugs and Other Compounds, CRC Press, Boca Raton, FL [Google Scholar]
- 10.Gong Q. H., Cho J. W., Huang T., Potter C., Gholami N., Basu N. K., Kubota S., Carvalho S., Pennington M. W., Owens I. S., Popescu N. C. (2001) Pharmacogenetics 11, 357–368 [DOI] [PubMed] [Google Scholar]
- 11.Girard H., Lévesque E., Bellemare J., Journault K., Caillier B., Guillemette C. (2007) Pharmacogenet. Genomics 17, 1077–1089 [DOI] [PubMed] [Google Scholar]
- 12.Lévesque E., Girard H., Journault K., Lépine J., Guillemette C. (2007) Hepatology 45, 128–138 [DOI] [PubMed] [Google Scholar]
- 13.Bélanger A., Pelletier G., Labrie F., Barbier O., Chouinard S. (2003) Trends Endocrinol. Metab. 14, 473–479 [DOI] [PubMed] [Google Scholar]
- 14.Guillemette C. (2003) Pharmacogenomics J. 3, 136–158 [DOI] [PubMed] [Google Scholar]
- 15.Nagar S., Remmel R. P. (2006) Oncogene 25, 1659–1672 [DOI] [PubMed] [Google Scholar]
- 16.Oelberg D. G., Chari M. V., Little J. M., Adcock E. W., Lester R. (1984) J. Clin. Invest. 73, 1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takikawa H., Ohki H., Sano N., Kasama T., Yamanaka M. (1991) Biochim. Biophys. Acta 1081, 39–44 [DOI] [PubMed] [Google Scholar]
- 18.Kremer A. E., Beuers U., Oude-Elferink R. P., Pusl T. (2008) Drugs 68, 2163–2182 [DOI] [PubMed] [Google Scholar]
- 19.Campbell M. E., Grant D. M., Inaba T., Kalow W. (1987) Drug Metab. Dispos. 15, 237–249 [PubMed] [Google Scholar]
- 20.Trottier J., Verreault M., Grepper S., Monté D., Bélanger J., Kaeding J., Caron P., Inaba T. T., Barbier O. (2006) Hepatology 44, 1158–1170 [DOI] [PubMed] [Google Scholar]
- 21.Caillier B., Lépine J., Tojcic J., Ménard V., Perusse L., Bélanger A., Barbier O., Guillemette C. (2007) Pharmacogenet. Genomics 17, 481–495 [DOI] [PubMed] [Google Scholar]
- 22.Verreault M., Senekeo-Effenberger K., Trottier J., Bonzo J. A., Bélanger J., Kaeding J., Staels B., Caron P., Tukey R. H., Barbier O. (2006) Hepatology 44, 368–378 [DOI] [PubMed] [Google Scholar]
- 23.Kaeding J., Bélanger J., Caron P., Verreault M., Bélanger A., Barbier O. (2008) Mol. Cancer Ther. 7, 380–390 [DOI] [PubMed] [Google Scholar]
- 24.Barbier O., Albert C., Martineau I., Vallée M., High K., Labrie F., Hum D. W., Labrie C., Bélanger A. (2001) Mol. Pharmacol. 59, 636–645 [DOI] [PubMed] [Google Scholar]
- 25.Bélanger A. S., Caron P., Harvey M., Zimmerman P. A., Mehlotra R. K., Guillemette C. (2009) Drug Metab. Dispos. 37, 1793–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caron P., Trottier J., Verreault M., Bélanger J., Kaeding J., Barbier O. (2006) Mol. Pharm. 3, 293–302 [DOI] [PubMed] [Google Scholar]
- 27.Chouinard S., Pelletier G., Bélanger A., Barbier O. (2004) Endocr. Res. 30, 717–725 [DOI] [PubMed] [Google Scholar]
- 28.Beaulieu M., Lévesque E., Hum D. W., Bélanger A. (1996) J. Biol. Chem. 271, 22855–22862 [DOI] [PubMed] [Google Scholar]
- 29.Griffiths W. J. (2003) Mass Spectrom Rev. 22, 81–152 [DOI] [PubMed] [Google Scholar]
- 30.Hansen S. H., Christiansen I. (2004) Electrophoresis 25, 3277–3281 [DOI] [PubMed] [Google Scholar]
- 31.Shipkova M., Armstrong V. W., Oellerich M., Wieland E. (2003) Ther. Drug Monit. 25, 1–16 [DOI] [PubMed] [Google Scholar]
- 32.Barbier O., Trottier J., Kaeding J., Caron P., Verreault M. (2009) Mol. Cell Biochem. 326, 3–8 [DOI] [PubMed] [Google Scholar]
- 33.Gall W. E., Zawada G., Mojarrabi B., Tephly T. R., Green M. D., Coffman B. L., Mackenzie P. I., Radominska-Pandya A. (1999) J. Steroid Biochem. Mol. Biol. 70, 101–108 [DOI] [PubMed] [Google Scholar]
- 34.Gardner-Stephen D., Heydel J. M., Goyal A., Lu Y., Xie W., Lindblom T., Mackenzie P., Radominska-Pandya A. (2004) Drug Metab. Dispos. 32, 340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritter J. K. (2000) Chem. Biol. Interact. 129, 171–193 [DOI] [PubMed] [Google Scholar]
- 36.Trottier J., Caron P., Straka R. J., Barbier O. (2008) in Hepatology 48, (supple.) 667A [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.