Abstract
The human monoclonal antibody 2G12 is a member of a small group of broadly neutralizing antibodies against human immunodeficiency virus type 1. 2G12 adopts a unique variable heavy domain-exchanged dimeric configuration that results in an extensive multivalent binding surface and the ability to bind with high affinity to densely clustered high mannose oligosaccharides on the “silent” face of the gp120 envelope glycoprotein. Here, we further define the amino acids responsible for this extraordinary domain-swapping event in 2G12.
Keywords: Methods/Circular Dichroism CD, Methods/Immunochemistry, 2G12, HIV-1, Antibody Structure, Domain Swapping, gp120
Introduction
Several mechanisms have been proposed to be involved in three-dimensional domain swapping in proteins. These include 1) destabilization of the monomer, 2) accumulation of mutations in the dimer interface, and 3) modifications (e.g. deletions or mutations) in the hinge loop (1–3). All such modifications have been suggested to play a role in the domain-swapping capability of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 (4–7). The crystal structure of Fab 2G12 revealed a tightly packed Fab dimer created by three-dimensional swapping of the variable heavy (VH)3 domains. This uncommon assembly results in an additional antigen-binding site at the novel VH/VH′ interface in addition to two “conventional” VH/VL-binding sites (6, 8, 9). Several somatic mutations have been identified, particularly in the heavy chain, which could be responsible for promoting domain exchange in this antibody (6). These amino acid substitutions are mostly localized to the VH/VH′ interface and “elbow” region of 2G12. We have generated three IgG1 2G12 heavy chain mutants based on the Fab 2G12 crystal structure and tested their potential for domain swapping. We discovered that the domain exchange-enabling properties of ProH113 at the elbow region of 2G12 can be emulated by other residues. Our results furthermore support a combination of up to four somatic mutations at the VH/VH′ interface and elbow region as being responsible for promoting domain exchange in 2G12.
EXPERIMENTAL PROCEDURES
Expression and Purification of IgG1 2G12 Mutants
The modified genes of the IgG1 2G12 mutants were codon-optimized and synthesized by GeneArt (Regensburg, Germany) and stably transfected into protein-free cultivated Chinese hamster ovary dehydrofolate reductase-deficient suspension cells (ATCC CRL-9096) as described earlier (10). 24 h after transfection with polyethyleneimine/DNA, selection was started with hypoxanthine/thymidine-deficient medium. Stably transfected clones were propagated and purified over a protein A column. Quality control of purified IgG fractions was assessed by SDS-PAGE analysis. Final IgG concentrations were determined by A280 measurement.
Circular Dichroism (CD) Spectrometry
CD analysis was performed on a PiStar-180 spectrometer equipped with a thermostatic cell holder from Applied Photophysics (Leatherhead, U.K.). For recording near-UV spectra (240–340 nm) the quartz cuvette had a path length of 10 mm. Spectral bandwidth was 2 nm; step size, 1 nm; scan time, 21 min; protein concentration, 0.1 mg/ml. All CD measurements were performed in 5 mm PBS, pH 7.3, at 25 °C (11). Each spectrum was automatically corrected with the base line to remove birefringence of the cell. The instrument was flushed with nitrogen with a flow rate of 5 liters min−1.
Electron Microscopy
For the preparation of the 2G12 IgG samples, freeze drying and heavy metal shadowing were performed according to Kellenberger and Kistler (12). Electron micrographs of the antibody preparations were taken on a CM 12 electron microscope (Philips, Eindhoven, The Netherlands) at an acceleration voltage of 80 kV.
Papain Digestion
Buffer-exchanged (1 mm EDTA, 50 mm sodium phosphate buffer, pH 6.3) and concentrated (1 mg/ml) 2G12 IgG samples were mixed with activated papain solution (1 mm EDTA, 10 mm cysteine, 50 mm sodium phosphate buffer, pH 7.0) to give a final papain/antibody ratio of 4% (w/w). The resulting mixtures were incubated for 2 h at 37 °C and stopped by adding 30 mm iodoacetamide. Subsequently, the cleaved antibody solution mix was buffer-exchanged (PBS) and incubated twice with PBS-preequilibrated protein A-Sepharose beads for 1 h at room temperature under gentle agitation on a shaker. Finally the Fab-containing supernatants were collected and further characterized by SDS-PAGE.
Gel Electrophoresis
Each purified antibody sample (IgG and Fab) was mixed with Laemmli sample buffer (Bio-Rad), heated for 5 min at 100 °C, and examined by denaturing gel electrophoresis in a Bio-Rad Mini-Protean 3 cell system. Subsequently, the 2G12 samples were visualized with SimplyBlue SafeStain (Invitrogen) by following the manufacturer's instructions.
ELISA
96-well microplates (eBioscience) were sensitized with recombinant gp160 MN (100 ng/well) overnight at 4 °C. All of the following steps were carried out at room temperature. After washing the plates with TPBS (PBS containing 0.01% Tween 20), the wells were blocked for 1 h with 4% nonfat dry milk in TPBS. The 2G12 Fab samples (2 μg/ml) were serially diluted in 1% nonfat dry milk/TPBS and added to the antigen-coated wells. After 1 h, bound 2G12 Fab was detected with a peroxidase-conjugated goat anti-human IgG Fab conjugate (Sigma) diluted 1:1,000 in 1% nonfat dry milk/TPBS. Finally, bound conjugate was visualized with tetramethylbenzidine substrate (Pierce) and measured at 450 nm.
Gel Filtration
The papain-digested 2G12 Fab variants (100 μg/ml) were loaded in PBS (50 μl) onto a PBS-equilibrated Superdex 75 10/30 column (GE Healthcare). The protein was eluted in PBS at a flow rate of 0.5 ml/min and detected by UV absorbance.
Virus Neutralization Assay
JR-FL pseudovirus was generated as described previously (13) by co-transfection of a gp160 JR-FL envelope with the pSG3ΔEnv backbone. Pseudotyped virus was added at a 1:1 ratio to serially diluted (1:3) 2G12 variants (starting at 200 μg/ml) and incubated at 37 °C, for 1 h. TZM-bl cells were then seeded (1:1 by volume) at 1 × 104 cells/well in a final concentration of 10 μg/ml DEAE-dextran. After a 48-h incubation at 37 °C, the cells were washed, lysed, and finally developed with luciferase assay reagent according to the manufacturer's instructions (Promega). Luminescence in relative light units was measured using an Orion microplate luminometer (Berthold Detection Systems).
In Solution Virus Capture Assay
ELISA microplates were coated overnight with 2 μg/ml rabbit anti-human Fab fragment in PBS at 4 °C. The capture antibodies (2G12 Fab variants) were mixed with 100-fold concentrated JR-FL pseudovirus at a final antibody concentration of 5 μg/ml. The resulting antibody/virus mix was incubated at 37 °C for 2 h and subsequently pelleted for 50 min at 16,000 × g at 4 °C. In the meantime, the coated plates were washed with PBS and blocked (3% nonfat dry milk) for 1 h at room temperature. The resulting antibody/virus pellets were resuspended in PBS, transferred onto the ELISA plates, and incubated for 1 h at 37 °C. Finally, plates were washed and overlaid with 1 × 104 TZM-bl cells in 100 μl/well. After 48 h, the luciferase activity was measured as described above. All experiments were performed in duplicate.
Antibody Modeling
The antibody models were generated with COOT (14) and PyMOL (15).
RESULTS AND DISCUSSION
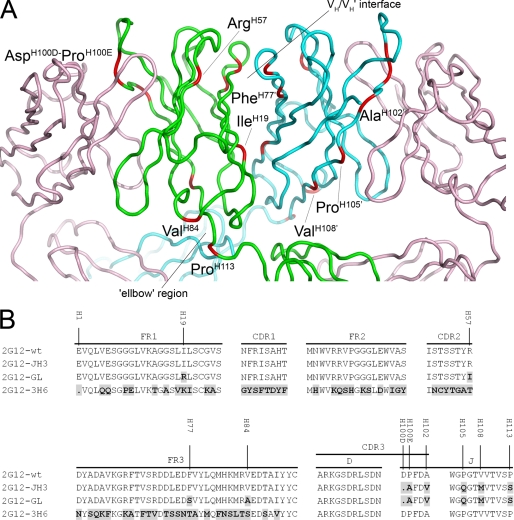
To investigate the mechanism of domain swapping in 2G12 further, particularly the roles of previously highlighted residues at the VH/VH′ dimer interface and elbow region, three IgG1 2G12 heavy chain mutants with multiple “germ line” amino acid substitutions in the VDJ region were created (Fig. 1A). An amino acid alignment of the VH domains of these 2G12 mutants with that of wild-type 2G12 (2G12-wt) is shown in Fig. 1B. In the first mutant, 2G12-JH3, the J region of 2G12-wt was replaced by a corresponding germ line region IGHJ3*01, resulting in five amino acid substitutions and one deletion (Fig. 1B). Two of these substitutions and the amino acid deletion occur in the complementarity-determining region (CDR) 3, whereas the remaining three substitutions are located in the J region. For the second mutant, 2G12-GL, four additional germ line amino acid substitutions are found in the VH region, including one substitution in the framework region (FR) 1, one in the CDR2, and two in the FR3. In a third construct, 2G12-3H6, the 2G12 VH region was completely exchanged by a different VH region derived from a previously described antibody (16, 17) to verify the responsibility of the 2G12 DJ region for domain swapping.
FIGURE 1.
2G12 Fab crystal structure A and amino acid alignment of the VDJ regions of 2G12 mutants compared with 2G12-wt B. A, the crystal structure of Fab 2G12 with the positions of the amino acid substitutions and deletions found in 2G12-GL shown in red and labeled. The light chains of the Fab dimer are shown in pink, with the heavy chains from Fab 1 and Fab 2 shown in green and blue, respectively. The VH/VH′ interface and the elbow region are also indicated (adapted from Protein Data Bank code 1op3) (6). B, differences in amino acid composition of the 2G12-JH3, 2G12-GL, and 2G12–3H6 mutants compared with 2G12-wt are shown in black with gray background. The amino acid numbering in A and B is based on the Kabat and Wu numbering system (19).
All modified 2G12 heavy chain genes and the corresponding 2G12-wt light chain genes (codon-optimized and synthesized by GeneArt) were stably transfected into protein-free cultivated Chinese hamster ovary dehydrofolate reductase-deficient suspension cells (ATCC CRL-9096). The protein A-purified IgG1 2G12 mutants exhibited electrophoretic characteristics similar to those of IgG 2G12-wt as revealed by SDS-PAGE (supplemental Fig. S1).
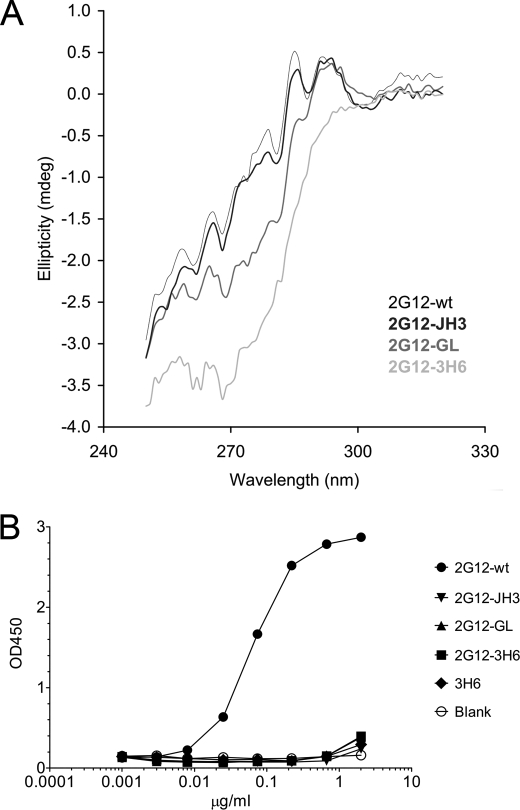
The IgG1 2G12-wt and IgG1 2G12 mutants were further analyzed by CD spectrometry. Measurements in the near-UV region (240–340 nm) are shown in Fig. 2A. IgG1 2G12-JH3 exhibited a spectrum identical to that of IgG1 2G12-wt, indicating that the exchange of the J region of IgG1 2G12-wt had no influence on the overall structure. Substitution of four additional amino acids in the IgG1 2G12-GL mutant was reflected by an apparent structural change. However, this difference may have derived from subtle changes in the local environment of particular aromatic residues, which are not necessarily associated with any major structural change (i.e. domain exchange). The spectrum of the third mutant, IgG1 2G12–3H6, revealed the largest difference from IgG1 2G12-wt. In this mutant, the whole VH region was exchanged, which appeared to be accompanied by major structural changes. To investigate potential structural differences between the IgG1 2G12-wt and the IgG1 2G12 mutants further, electron microscopy was carried out. We used freeze drying and heavy metal shadowing to visualize the frozen IgG samples in the electron microscope. Although the resolution of this preparation method was slightly lower compared with a previously described staining method (18), we were able to discriminate between I-shaped and Y-shaped conformations of the 2G12 samples (6, 18). The IgG1 2G12-wt, IgG1 2G12-JH3, and IgG1 2G12-GL displayed a successively decreasing predominance of the I-shaped conformation (∼90%, ∼75%, and ∼40%, respectively). In contrast, mutant 2G12-3H6 IgG1 existed predominantly in a Y-shaped conformation (∼90%). Taken together, these electron microscopy results indicate qualitative structural differences between the different IgG1 2G12 samples (supplemental Fig. S2).
FIGURE 2.
2G12 IgG CD measurements and 2G12 Fab specificity ELISA on recombinant gp160. A, the CD spectra in the near-UV range revealed no major structural differences between IgG1 2G12-wt and IgG1 2G12-JH3. IgG1 2G12-GL already exhibited a slight apparent structural change. In contrast, the spectrum of IgG1 2G12-3H6 suggested major structural changes compared with IgG1 2G12-wt. B, only 2G12-wt Fab was able to bind to recombinant gp160 MN.
We next examined the binding capacities of Fab 2G12 mutants (produced by papain digestion) to recombinant gp160 MN (Fig. 2B). Relative affinities (i.e. apparent affinities determined from the antibody concentration at half-maximal binding) of Fab 2G12-JH3, Fab 2G12-GL, and Fab 2G12-3H6 compared with Fab 2G12-wt were less than 1%. These findings are in good correlation with those of Calarese et al. (6), who discovered that most of their mutations to the primary combining site, secondary binding site, and VH/VH′ interface resulted in a dramatic loss of binding to gp120. Considering that the gp160 specificity ELISA might not represent an optimal environment for the 2G12 mutants, the same samples were analyzed further for binding activity to native gp120 (i.e. in solution virus capture assay) as well as neutralization potency on JR-FL pseudovirus. In contrast to 2G12-wt, the mutants were neither able to capture free virus nor exhibited any neutralization capacity (data not shown).
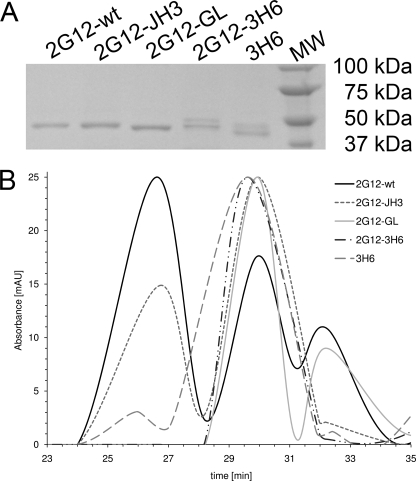
Finally, the Fab 2G12 variants were examined for their ability to form three-dimensional domain-swapped dimers. For this experiment, Fab 2G12 fractions were concentrated, visualized by SDS-PAGE (Fig. 3A), and separated by gel filtration. Fig. 3B shows the normalized spectrograms. Fab 2G12-wt eluted as two dominant peaks at 26.5 and 30 min, corresponding to dimeric and monomeric forms, respectively. Fab 2G12-JH3 also eluted as two peaks at 26.5 and 30 min. However, the proportion of dimer was significantly less than that for Fab 2G12-wt. In contrast, Fab 2G12-GL, Fab 2G12-3H6, and the Fab 3H6 control exhibited dominant peaks at 30 min, indicating monomer only.
FIGURE 3.
SDS-PAGE of 2G12 Fab variants A and gel filtration B. A, one band (approximately 53 kDa) was visible for 2G12-wt, 2G12-JH3, and 2G12-GL. An additional band (approximately 55 kDa) was visible for 2G12–3H6. This latter band derives from antibody 3H6, which contains an N-glycosylation site in the VH CDR2 (20) and correlates with the two bands observed for 3H6 Fab. B, Fab 2G12-wt eluted as both dimer and monomer. Fab 2G12-JH3 also eluted as both dimer and monomer, but with a significantly lower proportion of dimer than observed for Fab 2G12-wt. Fab 2G12-GL, Fab 2G12-3H6, and Fab 3H6 eluted as monomer only. Several protein standards with known molecular mass were also loaded onto the column to determine the size distribution of the peaks.
The fact that 2G12-JH3 still forms a dimer (Fig. 3B) indicates that the ProH113 to Ser substitution in the J region does not prevent VH domain exchange. The high resolution crystal structure of Fab 2G12 (Protein Data Bank code 1op3) (6) revealed that Pro N, Cα, Cβ at the elbow, or hinge region of the domain-exchanged dimer are within van der Waals (VDW) contact of ValH84 Cγ2 (Fig. 4A). These hydrophobic interactions were proposed to stabilize the conformation of the hinge region in which the VH domain pivots around ProH113 to facilitate dimerization. Modeling of the ProH113 to Ser substitution, as found in 2G12-JH3, by simply mutating the ProH133 side chain to that of the most probable rotamer of the Ser side chain using the program COOT (14) suggests that these interactions could be conserved in the mutant (Fig. 4A). Moreover, the ϕ and ψ angles of the modeled Ser lie within the preferred regions of the Ramachandran plot. Consequently, the conformation of the hinge as observed in the Fab 2G12 structure may be maintained in 2G12-JH3 via hydrophobic interactions between ValH84 and SerH113 that are analogous to those between ValH84 and ProH113.
FIGURE 4.
Structural analysis of sequence differences among 2G12-wt, 2G12-JH3, and 2G12-GL. A, in the high resolution crystal structure of Fab 2G12 (Protein Data Bank code 1op3) (6), ValH84 Cγ2 is within VDW contact of ProH113 N, Cα, Cβ, and ValH111 O at the elbow region of the domain-exchanged dimer. Modeling of Fab 2G12 with a ProH113 to Ser substitution (Ser side chain is shown in yellow) suggests that the VDW interactions mediated by ValH84 Cγ2 at the elbow region in 2G12-wt could be maintained in the 2G12-JH3 mutant. B, the hydrophobic side chain of IleH19 forms extensive VDW contacts with SerH7′, GlyH8′, IleH19′, LeuH20′, SerH21′, and TyrH79′ in the VH/VH′ interface of the domain-swapped dimer. C, ArgH57 forms a salt bridge with AspH72′ at the VH/VH′ interface of the domain-swapped dimer. This interaction not only likely plays an important role in stabilization of the VH/VH′ interface, but also likely plays an indirect role in 2G12 binding to clustered high mannose oligosaccharides on gp120.
On the other hand, the fact that 2G12-GL does not form a dimer (Fig. 3B) could be due to one or a combination of the four additional substitutions found in this mutant over 2G12-JH3. As mentioned above, in the Fab 2G12 structure, ValH84 Cγ2 was observed to form several VDW interactions with ProH113′ at the elbow region as well as with ValH111 O (6) (Fig. 4A). Modeling of the ValH84 to Ala substitution suggests that these interactions would be lost in 2G12-GL, and this may be enough to destabilize the hinge region and, hence, inhibit dimerization.
In regard to the IleH19 to Arg substitution, the region of the VH/VH′ interface around IleH19 was revealed to be very tightly packed in the Fab 2G12 structure (6) (Fig. 4B), with the hydrophobic side chain of IleH19 being within VDW contact of SerH7′, GlyH8′, IleH19′, LeuH20′, SerH21′, and TyrH79′. The binding interactions in the VH/VH′ interface would probably be decreased significantly by disruption of these hydrophobic contacts. Moreover, because of steric clashes, it is not possible to model any rotamers of the Arg side chain at this position using COOT without substantially affecting the location and rotamers of the surrounding side chains. The IleH19 to Arg substitution may, therefore, not even be tolerated within the dimer interface.
In relation to the PheH77 to Ser substitution, the aromatic side chain of PheH77 was observed to be within VDW contact of ThrH68′ and GlnH81′ in the Fab 2G12 structure (6). Modeling a Ser at this position suggests that these interactions would be lost in the 2G12-GL mutant.
The ArgH57 to Ile substitution would break a salt bridge with AspH72′ observed at the VH/VH′ dimer interface of Fab 2G12 (Fig. 4C) (6). In addition to the putative role of this salt bridge in stabilizing the VH/VH′ interface, it is also likely to be important for positioning of the neighboring TyrH56 which, in the complex with Manα1,2Man (6), forms pi-stacking interactions with TyrL94 at the VH/VL interface. These interactions in turn may be important for the positioning of GlyL93 O which H-bonds with the terminal mannose O6 in the Manα1,2Man containing arms of high mannose sugars. Notably, a TyrH56 to Ala substitution was reported to cause a dramatic loss in affinity of 2G12 for gp120 (6). Moreover, SerH54, ThrH55, and ArgH57 to Ala substitutions were similarly observed to lead to significantly decreased gp120 binding (6). These effects could similarly arise from changes in the conformation of the aromatic side chain of TyrH56 which prevent it from forming pi-stacking interactions with TyrL94.
Consequently, the unusual domain-exchanged configuration of 2G12 could be fostered by the single or combined effects of four amino acid side chains that either help to stabilize the elbow region (H113) or the VH/VH′ dimer interface. Moreover, a proline at H113 has been shown here to not be required for the domain swapping capability of 2G12. Rather, the hydrophobic contacts between the residue at this position and the side chain of residue His84 appear to be predominantly responsible for stabilization of the elbow region.
The lack of domain swapping capability observed for the mutant 2G12–3H6 clearly indicates that the 2G12-wt CDR3 and J region alone are not sufficient to promote VH domain exchange when the complete VH region of 2G12 is replaced by another antibody VH region. Taken together, the present study shows that the I-shaped conformation of 2G12 cannot be traced back to a single amino acid mutation but rather involves a complex interplay of amino acid replacements. It remains to be further investigated at which stage these mutations occurred in the evolution of this very unique antibody.
Supplementary Material
Acknowledgments
We thank Zara Fulton (Dept. of Molecular Biology, The Scripps Research Institute, La Jolla, CA) for providing the 2G12 models and for critically reading the manuscript and M. Löschel, R. Marvan, and K. Bauer for excellent technical support.
This work was funded by European Union Project 019052, “HIVAB,” and the Austrian Science Fund J2845-B13.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- VH
- variable heavy
- VL
- variable light
- CDR
- complementarity-determining region
- ELISA
- enzyme-linked immunosorbent assay
- FR
- framework
- PBS
- phosphate-buffered saline
- VDW
- van der Waals
- wt
- wild-type.
REFERENCES
- 1.Liu Y., Eisenberg D. (2002) Protein Sci. 11, 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett M. J., Sawaya M. R., Eisenberg D. (2006) Structure 14, 811–824 [DOI] [PubMed] [Google Scholar]
- 3.Gronenborn A. M. (2009) Curr. Opin. Struct. Biol. 19, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trkola A., Pomales A. B., Yuan H., Korber B., Maddon P. J., Allaway G. P., Katinger H., Barbas C. F., 3rd, Burton D. R., Ho D. D., Moore J. P. (1995) J. Virol. 69, 6609–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astronomo R. D., Lee H. K., Scanlan C. N., Pantophlet R., Huang C. Y., Wilson I. A., Blixt O., Dwek R. A., Wong C. H., Burton D. R. (2008) J. Virol. 82, 6359–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calarese D. A., Scanlan C. N., Zwick M. B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M. R., Stanfield R. L., Roux K. H., Kelly J. W., Rudd P. M., Dwek R. A., Katinger H., Burton D. R., Wilson I. A. (2003) Science 300, 2065–2071 [DOI] [PubMed] [Google Scholar]
- 7.Pantophlet R., Burton D. R. (2006) Annu. Rev. Immunol. 24, 739–769 [DOI] [PubMed] [Google Scholar]
- 8.Calarese D. A., Lee H. K., Huang C. Y., Best M. D., Astronomo R. D., Stanfield R. L., Katinger H., Burton D. R., Wong C. H., Wilson I. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwong P. D., Wilson I. A. (2009) Nat. Immunol. 10, 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reisinger H., Steinfellner W., Stern B., Katinger H., Kunert R. (2008) Appl. Microbiol. Biotechnol. 81, 701–710 [DOI] [PubMed] [Google Scholar]
- 11.Marsche G., Furtmüller P. G., Obinger C., Sattler W., Malle E. (2008) Cardiovasc. Res. 79, 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellenberger E., Kistler J. (1979) in Unconventional Electron Microscopy for Molecular Structure Determination (Hoppe S., Mason R. eds) pp. 49–79, F. Vieweg and Sohn, Braunschweig, Germany [Google Scholar]
- 13.Nelson J. D., Brunel F. M., Jensen R., Crooks E. T., Cardoso R. M., Wang M., Hessell A., Wilson I. A., Binley J. M., Dawson P. E., Burton D. R., Zwick M. B. (2007) J. Virol. 81, 4033–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 15.DeLano W. L. (2002) Curr. Opin. Struct. Biol. 12, 14–20 [DOI] [PubMed] [Google Scholar]
- 16.Gach J. S., Maurer M., Hahn R., Gasser B., Mattanovich D., Katinger H., Kunert R. (2007) J. Biotechnol. 128, 735–746 [DOI] [PubMed] [Google Scholar]
- 17.Bryson S., Julien J. P., Isenman D. E., Kunert R., Katinger H., Pai E. F. (2008) J. Mol. Biol. 382, 910–919 [DOI] [PubMed] [Google Scholar]
- 18.Roux K. H., Zhu P., Seavy M., Katinger H., Kunert R., Seamon V. (2004) Mol. Immunol. 41, 1001–1011 [DOI] [PubMed] [Google Scholar]
- 19.Kabat E. A., Wu T. T. (1991) J. Immunol. 147, 1709–1719 [PubMed] [Google Scholar]
- 20.Gach J. S., Quendler H., Weik R., Katinger H., Kunert R. (2007) AIDS Res. Hum. Retroviruses 23, 1405–1415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.