Abstract
The formation of SiO2-based cell walls by diatoms (a large group of unicellular microalgae) is a well established model system for the study of molecular mechanisms of biological mineral morphogenesis (biomineralization). Diatom biomineralization involves highly phosphorylated proteins (silaffins and silacidins), analogous to other biomineralization systems, which also depend on diverse sets of phosphoproteins (e.g. mammalian teeth and bone, mollusk shells, and sponge silica). The phosphate moieties on biomineralization proteins play an essential role in mineral formation, yet the kinases catalyzing the phosphorylation of these proteins have remained poorly characterized. Recent functional genomics studies on the diatom Thalassiosira pseudonana have revealed >100 proteins potentially involved in diatom silica formation. Here we have characterized the biochemical properties and biological function of one of these proteins, tpSTK1. Multiple tpSTK1-like proteins are encoded in diatom genomes, all of which exhibit low but significant sequence similarity to kinases from other organisms. We show that tpSTK1 has serine/threonine kinase activity capable of phosphorylating silaffins but not silacidins. Cell biological and biochemical analysis demonstrated that tpSTK1 is an abundant component of the lumen of the endoplasmic reticulum. The present study provides the first molecular structure of a kinase that appears to catalyze phosphorylation of biomineral forming proteins in vivo.
Introduction
Numerous organisms ranging from prokaryotes to mammals produce inorganic materials (biominerals) with species specific structures and properties to serve as endo- or exoskeletons. Because of their intricate morphologies and extraordinary physical properties, biominerals are remarkable examples of biological morphogenesis. Additionally, because of the structural intricacies and exceptional mechanical properties of biominerals, the processes that enable their formation are regarded as paradigms for developing new routes for inorganic materials synthesis (1–7). Insight into the molecular mechanisms that control biomineral formation (biomineralization) is currently emerging. Biomineral-associated organic macromolecules have been identified that are intimately involved in the mineral biogenesis process and typically include phosphoproteins. The presence of phosphate residues at multiple serine and threonine residues is required for functionality (8–10). Additionally, highly phosphorylated proteins play a role in extracellular adhesion processes (11, 12).
Despite the importance of extracellular phosphoproteins, the kinases that catalyze their phosphorylation are only poorly characterized. Such kinases need to be situated within the secretory pathway (e.g. endoplasmic reticulum and Golgi apparatus) rather than the cytosol, as biomineralization proteins become co-translationally imported into the ER2 and transported through the Golgi apparatus before reaching the mineral-forming compartment (i.e. a specialized extracellular space or a specific intracellular vesicle) (13–16). In membrane fractions from mammalian cells, ER- and Golgi apparatus-associated casein kinase-like activities have been identified that are believed to be involved in the biogenesis of secretory phosphoproteins including proteins involved in biomineralization of bone and teeth (e.g. osteopontin, bone sialoprotein, and dentin sialophosphoprotein) (17–26). However, to date the primary structures of these kinases have remained elusive. Furthermore, there is a complete lack of knowledge about the kinases that phosphorylate biomineralization proteins in non-mammalian organisms.
One of the best studied model systems for biomineralization is the formation of the SiO2 (silica)-based cell walls in diatoms, a large group of unicellular photosynthetic eukaryotes. Diatom biosilica formation involves (among other yet unidentified components) long-chain polyamines and two families of phosphoproteins, termed silaffins and silacidins (27, 28). Silaffins and silacidins are rich in serine residues (∼20–40%), with many or even all of them being phosphorylated (9, 29–31). In vitro data indicate that silica formation depends on an organic matrix established by electrostatic interactions between the positively charged long-chain polyamine molecules and the numerous negatively charged phosphate residues of silaffins and silacidins (27). Thus, the kinases that catalyze the attachment of phosphate residues onto the polypeptide backbones of silaffins and silacidins are essential components of the biosilica-forming machinery in diatoms. Characterization of the structures and properties of these kinases is therefore required to fully understand the mechanism of silica biomineralization and its regulation in diatoms.
The genome sequences from the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum have revealed the presence of 64 and 117 proteins containing putative kinase domains, respectively (32, 33). Recent functional genomics studies of T. pseudonana revealed a set of putative kinase genes that are potentially involved in silica biogenesis (34, 35). However, no experimental data have been obtained regarding the enzymatic activities, substrate specificities, and intracellular localization for any of these putative kinases. One of the putative kinases, encoded by GenBank™ accession number EED92887.1 (for simplicity denoted hereafter as tpstk1), exhibits a cell cycle-dependent mRNA expression pattern similar to the silaffin encoding gene tpsil3 (34). This observation prompted our idea that the encoded putative kinase might be involved in the phosphorylation of silaffins. Below we describe the biochemical characterization of the protein encoded by tpstk1 and experiments to determine its role in the phosphorylation of silaffins and silacidins in vivo.
EXPERIMENTAL PROCEDURES
Materials
Antimycin A, dephosphorylated casein, chymotrypsin, cytochrome c, histones I and II, Igepal, inositol diphosphate, myelin basic protein, phosphoserine, phosphotyrosine, and phosphothreonine were purchased from Sigma-Aldrich. ATP, isopropyl β-d-thiogalactopyranoside, guanidinium hydrochloride, NADH, phenylmethylsulfonyl fluoride, and sucrose were purchased from EMD Biosciences (Gibbstown, NJ). Recombinant silaffins were purified as described previously (36).
Culture Conditions
T. pseudonana clone CCMP1335 was grown in an artificial seawater medium according to the North East Pacific Culture Collection at 18 °C under constant light at 10,000–15,000 lux.
Determination of the tpstk1 Gene Sequence
Total RNA isolation was performed as described by Poulsen and Kröger (37) followed by isolation of mRNA using oligo(dT)25-coupled magnetic beads (Dynabeads, Invitrogen) as described in Fischer et al. (38). The 3′-end of the tpstk1 cDNA was amplified using gene-specific sense primers 5′-TGCAGCAGGGCAGTGCATCC-3′ for the first PCR and 5′-GCATCC AAGCAGTACATTCC-3′ for the nested PCR and antisense primers 5′-CAGCCGCCGAATTCCCAG(T)18-3′ for the first PCR and 5′-GCCGCCG AATTCCCAGTTT-3′ for the nested PCR. The cDNA coupled to magnetic beads was prepared for 5′-RACE by adding a poly(C) tail to the 5′-end with terminal deoxynucleotidyltransferase according to the manufacturer's instructions (Fermentas, Glen Burnie, MD). The 5′-end of the tpstk1 cDNA was amplified using gene-specific antisense primers 5′-GTAGTGATACTGTCTCCATC-3′ for the first PCR and 5′-CCTCACTGGTGACGTAGTGA-3′ for the second PCR, and sense primers 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′ for the first PCR and 5′-GGCCACGCGTCGACTAGTAC-3′ for the second PCR. The 544- and 500-bp DNA fragments produced from 3′- and 5′-RACE PCR, respectively, were ligated into pGEMT (Promega Corp., Madison, WI) and sequenced. The full-length tpstk1 cDNA was amplified by PCR (40 cycles: 94 °C for 20 s, 58 °C for 20 s, and 72 °C for 90 s) using sense primer 5′-ATGAGAGTCATACGCAATGTTTC-3′ and antisense primer 5′-CTAAAAGCTGATAGGACCGATG-3′.
Cloning, Expression, Isolation, and Refolding of Recombinant tpSTK1
The coding region of tpstk1 was amplified from T. pseudonana genomic DNA by PCR using sense primer 5′- GCAGGCCTATGAGAGTCATACGCAATGTTTCTC-3′ and antisense primer 5′-CCAAGCTTCTAATGGTGATGGTGATGGTGAAAGCTGATAGGACCGATGCC-3′, which introduced a StuI restriction site (bold), a HindIII site (italic), and a C-terminal hexahistidine tag (underlined). The PCR product obtained was ligated into the StuI/HindIII sites of pPROEX-HTb (Invitrogen) and introduced into Escherichia coli DH5α. By this strategy a gene was generated that encoded the full-length tpSTK1 protein fused to an N-terminal hexahistidine tag followed by a spacer region (DYDIPT), an rTEV protease cleavage site, and a C-terminal hexahistidine tag. This fusion protein was termed recombinant tpSTK1.
For expression of recombinant tpSTK1 a freshly transformed E. coli DH5α clone was inoculated into LB medium containing 100 μg/ml ampicillin (LBamp), grown overnight at 37 °C, diluted 50-fold with fresh LBamp medium, and then grown to an absorbance (A) of 0.6 (at λ = 600 nm). Expression of recombinant tpSTK1 was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mm. After an additional 3 h of shaking at 37 °C, cells were harvested by centrifugation, washed once with 1% (w/v) NaCl, and resuspended in lysis buffer A (50 mm Tris-HCl, pH 8.0, 25% (w/v) sucrose, 1 mm EDTA, 10 mm DTT). Lysozyme, DNase I, and MgCl2 were added at final concentrations of 0.5 mg/ml, 19 μg/ml, and 2.0 mm, respectively. Subsequently, an equal volume of lysis buffer B (50 mm Tris-HCl, pH 8.0, 0.1 m NaCl, 10 mm DTT) was added, and the cells were incubated at room temperature for 1 h. EDTA was added to a final concentration of 6.7 μm, and the cells were frozen in liquid nitrogen. The cell pellet was thawed for 30 min at 37 °C, MgCl2 was added to a final concentration of 4.1 mm, and the cells were incubated at room temperature for 1 h. EDTA (6.7 μm) was added, and the lysate was centrifuged at 23,000 × g at 4 °C for 30 min to pellet inclusion bodies. The pellet was resuspended in 10 ml of wash buffer I (50 mm Tris-HCl, pH 8.0, 0.1 m NaCl, 1 mm EDTA, 10 mm DTT) and then centrifuged at 23,000 × g at 4 °C for 30 min. The pellet was resuspended with 10 ml of wash buffer II (50 mm Tris-HCl, pH 8.0, 0.1 m NaCl, 1 mm EDTA, 1 mm DTT) and centrifuged at 23,000 × g at 4 °C for 30 min. The final pellet was dissolved in purification buffer (50 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 1 mm DTT, 20 mm imidazole, 8 m urea) for 30 min and applied to a 1-ml HisTrap-FF column (GE Healthcare) that had been pre-equilibrated with purification buffer at 4 °C for 1 h at 0.5 ml/min. The column was washed with 25 ml of purification buffer before refolding of the protein was performed on the column with a linear gradient (0.5 ml/min, 50 min) from 100% purification buffer to 100% refolding buffer (50 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 20 mm imidazole) at 4 °C. After washing with 25 ml of refolding buffer, the protein was eluted with a linear gradient (0.5 ml/min, 20 min) from 100% refolding buffer to 100% elution buffer (1 m imidazole, 50 mm Tris-HCl, pH 8.0, 0.5 m NaCl). Fractions containing recombinant tpSTK1 were dialyzed against 50 mm Tris-HCl, pH 8.0, 0.1 m NaCl, 10% (v/v) glycerol, and 1 mm phenylmethylsulfonyl fluoride. The dialysate was concentrated to ∼2 mg/ml recombinant tpSTK1 using a 3000 Da molecular mass cut-off Centricon filter (Millipore, Billerica, MA).
Preparation of Anti-tpSTK1 IgG
Recombinant tpSTK1 (5 mg) was purified by preparative SDS-PAGE and used for immunization of rabbits by a commercial vendor (Lampire Biological Laboratories). The antiserum obtained 50 days after immunization was purified using an affinity matrix that carried recombinant tpSTK1. IgGs from the preimmune serum were purified using a protein G column (GE Healthcare) according to the manufacturer's instructions.
Semiquantitative Reverse Transcription-PCR from Synchronized T. pseudonana Cells
T. pseudonana cells were synchronized as described by Hildebrand et al. (39). At the indicated times equal culture volumes were harvested for total RNA isolation and cDNA synthesis as described by Poulsen and Kröger (37). The tpstk1 gene was amplified using sense primer 5′-AAGCTTTCAGCTCAACTTATACTTGCTCCC-3′ and antisense primer 5′-GAGCAGGCCTTCACATCACAACGCACCGTC-3′; the tpsil3 gene was amplified using sense primer 5′-GGACACCAAGAGTGGAAAG-3′ and antisense primer 5′-TCAAGCGCTCATGGAGTGG-3′; the tpfcp9 gene was amplified using sense primer 5′-TTCGAGGATGAGCTCGGTG-3′ and antisense primer 5′-GAACATCACGCATGAAGGC-3′. PCR was performed with 25 cycles of amplification (94 °C for 20 s, 60 °C for 20 s, and 72 °C for 40 s).
Cloning, Expression, and Isolation of a rSilNH10-rSic Fusion Protein
The rSilN gene was amplified from pET16b/ProY by PCR using the sense primer 5′-CATGCCATGGCTGCCCAAAGCATTGCTGAC-3′ and the antisense primer 5′-CGCGGGATCCGTGATGGTGATGGTGATGGTGATGGTGATGCAAGATACGAAGTTCTTCCTC-3′, which introduced an NcoI restriction site (bold), a BamHI site (italic), and a C-terminal decahistidine tag (underlined). The 310-bp PCR product obtained was ligated into the NcoI/BamHI sites of pET28a (Novagen, Gibbstown, NJ) and introduced into E. coli DH5α yielding plasmid pET28a/rSilNH10.
A DNA molecule corresponding to nucleotides 154–730 of the silacidin gene (31) was amplified from T. pseudonana genomic DNA by PCR using the sense primer 5′-CGCGGGATCCATGATCAACAAAGTCCGCCGTCTC-3′ and the antisense primer 5′-GCCGCTCGAGCTATGCATCAAACATCATCAAATCTTCACT-3′, which introduced a BamHI site (bold) and an XhoI site (italic). The resulting 596-bp product, which encodes amino acids 52–235 of the silacidin precursor polypeptide, was ligated into pJET1.2 (Fermentas) yielding plasmid pJET1.2/rSic. After the rSic gene sequence was confirmed by DNA sequencing, plasmid pJET1.2/rSic was digested with BamHI and XhoI, and the 596-bp product was ligated into the BamHI and XhoI sites of pET28a/rSilNH10 yielding expression plasmid pET28a/rSilNH10-rSic. The expression plasmid was introduced into E. coli DH5α, and the composition and correct orientation of the rSilNH10-rSic fusion gene was verified by restriction site mapping.
For expression of rSilNH10-rSic, an E. coli BL21(DE3) clone harboring pET28a/rSilNH10-rSic was inoculated into LB medium containing 100 μg/ml kanamycin (LBkan), grown overnight at 37 °C, diluted 50-fold with fresh LBkan medium, and grown to A = 0.6 (at λ = 600 nm). Expression of the recombinant fusion protein was induced by the addition of isopropyl β-d-thiogalactopyranoside to 1 mm. After growing for an additional 3 h at 37 °C, cells were harvested by centrifugation, washed once with 1% (w/v) NaCl, and resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 1 m NaCl, 10 mm imidazole, 6 m guanidinium hydrochloride). Fusion protein rSilNH10-rSic was purified via immobilized affinity chromatography as described previously for rSilN (36).
Cyanogen Bromide Cleavage of rSilNH10-rSic
Cyanogen bromide (Acros Organics, Morris Plains, NJ) dissolved in 70% (v/v) formic acid was added to lyophilized rSilNH10-rSic at 1 mg of cyanogen bromide/0.1 mg of dry protein. The reaction mixture was incubated at room temperature in the dark for 24 h. Excess cyanogen bromide was removed by a steady stream of N2 gas, and the residue was dissolved in 0.2 ml of H2O and lyophilized. The dry sample was resuspended in 1 m Tris, neutralized with NaOH, and incubated with nickel-nitrilotriacetic acid-agarose (Qiagen, Valencia, CA) in binding buffer (50 mm Tris-HCl, pH 8.0, 1 m NaCl, 10 mm imidazole). The unbound material contained pure rSic (see supplemental Fig. S2), which was dialyzed exhaustively against 10 mm ammonium acetate. The dialysate was dried by lyophilization, and the residue was resuspended in H2O.
Kinase Assays
Protein substrates were incubated with 10 μg of recombinant tpSTK1 or membrane fraction (10 μg of total protein) in kinase activity buffer (50 mm Tris-HCl, pH 8.0, 40 mm MgCl2, 10 μm ATP) for 1 h at 37 °C. To test for divalent metal ion requirement, MgCl2 was replaced by an equal concentration of CaCl2, MnCl2, or ZnCl2. For detection, an equal volume of luminescent kinase assay reagent (Kinase-Glo® kit, Promega Corp.) was added to the reaction and incubated at room temperature for 10 min. Luminescence was measured using a Synergy 2 plate reader (Biotek, Winooski, VT).
Reversed Phase High Pressure Liquid Chromatography (RP-HPLC)
Recombinant silaffins before and after phosphorylation with recombinant tpSTK1 were separated using RP-HPLC on a C18 column by applying a linear acetonitrile gradient of 0–25% buffer B for 35 min (buffer A: 0.1% (v/v) trifluoroacetic acid in H2O; buffer B: 0.085% (v/v) trifluoroacetic acid in acetonitrile).
Chemical Analysis of Phosphorylated Silaffins
Phosphate content was determined according to a standard protocol (40). Phosphoamino acid analysis was performed as described previously (30).
Expression of Green Fluorescent Protein (GFP) Fusion Proteins
To construct a vector for expression of C-terminal GFP fusion proteins, the egfp gene was amplified by PCR from a plasmid (a kind gift from Dr. Kirk Apt, Martek Biosciences) using sense primer 5′-ATCGCATGCGGTACCGGCGGAATGGTGAGCAAGGGCGAGG-3′ (SphI site underlined; KpnI site in bold) and antisense primer 5′-GAATGCGGCCGCTTACTTGTAACAGCTCGTCCATG-3′ (NotI site underlined). The PCR product was ligated into the EcoRV and NotI sites of vector pTpfcp (41) in which the KpnI site had been destroyed. The resulting plasmid, pTpfcp/ctGFP, consisted of the regulatory sequences of the fcp gene flanking the egfp gene, which carries a 5′ extension containing three unique restriction sites (EcoRV, SphI, and KpnI) allowing for the in-frame insertion of genes.
To generate a gene encoding STK1–24GFPDDEL, a DNA fragment encoding the tpSTK1 signal peptide was amplified by PCR using sense primer 5′-ACCAAAATGAGAGTCATACGCAATG-3′ and antisense primer 5′-ATTCGGTACCGGAGGTGACGGACACAAGAAA-3′. The resulting 88-bp PCR product was ligated into the EcoRV site of pTpfcp/ctGFP generating pTpfcp/STK11–24GFP. Plasmid pTpfcp/STK11–24GFP was then used as a template for amplification of a STK11–24GFP-encoding gene using sense primer 5′-ACCAAAATGAGAGTCATACGCAATG-3′ and antisense primer 5′-GATTCGCGGCCGCTTACAGCTCGTCATC-3′, which introduced a NotI restriction site (bold) and a segment encoding the tetrapeptide DDEL (underlined). The resulting 820-bp product was digested with NotI and ligated into the EcoRV and NotI site of the previously described vector, pTpfcp (41), generating pTpfcp/STK11–24GFPDDEL.
To generate a gene encoding BiP1–27GFP, the signal peptide encoding part of the BiP gene (nucleotides 1–81) was amplified by PCR from T. pseudonana genomic DNA using sense primer 5′-GGTCGATATCAAAATGGCGTTCAAACGGCGGTTCC-3′ and antisense primer 5′-GATCGGTACCGGCGAGATTTGTAGCGATGTG-3′, which introduced an EcoRV restriction site (bold) and a KpnI restriction site (underlined). The resulting 104-bp PCR product was ligated into the EcoRV and KpnI sites of pTpfcp/ctGFP. The resulting plasmid was termed pTpfcp/BIP1–27GFP.
To generate a gene encoding BiP1–39GFPDDEL, the part of the BiP gene encoding amino acids 1–39 was amplified by PCR from T. pseudonana genomic DNA using sense primer 5′- ATCATAATCATGGCGTTCAAACGGCGGTTC-3′ and antisense primer 5′-GATCGCGGCCGCTTGTGTGCTCGTCGCTATTGTG-3′, which introduced a NotI restriction site (bold). The resulting 140-bp PCR product was digested with NotI and ligated into the EcoRV and NotI sites of the previously described vector pTpNR (41). The resulting plasmid was termed pTpNR/BIP1–39. The egfp gene was amplified with sense primer 5′-GAATCGCGGCCGCATGGTGAGCAAGGGCGAGGA-3′ and antisense primer 5′-GATTCGCGGCCGCTTACAGCTCGTCATCCTTGTACAGCTCGCCATGCC-3′, which introduced NotI restriction sites (bold) and a sequence encoding the tetrapeptide DDEL (underlined). The resulting 758-bp PCR product was digested with NotI and ligated into the NotI site of pTpNR/BIP1–39. Transformant E. coli clones were screened for the correct orientation of the eGFPDDEL-encoding part resulting in the final plasmid, pTpNR/BIP1–39eGFPDDEL.
To generate a gene encoding GFPDDEL, the egfp gene was amplified with the sense primer 5′-ACCAAAAATGGTGAGCAAGGGCGAGGAC-3′ and antisense primer 5′-GATTCGCGGCCGCTTACAGCTCGTCATCCTTGTACAGCTCGTCCATGCC-3′, which introduced a NotI restriction site (bold) and a sequence encoding the tetrapeptide DDEL (underlined). The resulting 750-bp PCR product was digested with NotI and ligated into the EcoRV and NotI sites of the previously described vector pTpfcp (41) generating pTpfcp/GFPDDEL. The sequences of all PCR products were verified by DNA sequencing. Plasmids were introduced into T. pseudonana cells by microparticle bombardment as described previously (41).
Fluorescence Microscopy
T. pseudonana cells expressing GFP fusion proteins were imaged using a Zeiss LSM510 confocal microscope as described previously (42).
Fractionation of Diatom Membranes
One liter of T. pseudonana cells (∼106 cells/ml) was harvested by centrifugation (3000 × g, 10 min), washed with ice cold lysis buffer (50 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.2 mm DTT, 10 mm NaF, 1 mm Na3VO4, 250 mm sucrose, 1× protease inhibitor mixture (Roche Applied Science)), and resuspended in 3 ml of lysis buffer. The cells were equally distributed into three 2-ml tubes, 0.5 ml of glass beads (0.25–0.30 mm) was added to each tube, and tubes were vortexed for 30 s. The glass beads were allowed to settle, and the supernatants were removed from each tube, combined, and then centrifuged at 16,000 × g for 2 min at 4 °C (supernatant = fraction S0). The pellets were resuspended in 1.0 ml of lysis buffer and treated with glass beads as described above. After three lysis/centrifugation cycles, the S0 fraction was centrifuged at 16,000 × g for 15 min at 4 °C. The pellet (P1) was resuspended in lysis buffer, and the supernatant (S1) was centrifuged at 100,000 × g for 1 h at 4 °C yielding fractions P2 (pellet) and S2 (supernatant).
For fractionation of membranes by sucrose density gradient centrifugation, the P2 fraction was brought to 20% (w/v) sucrose in lysis buffer and layered atop a stepwise sucrose gradient (sucrose concentrations in % (w/v): 22.5, 25.0, 27.5, 30.0, 32.5, 35.0 in lysis buffer). After centrifugation at 100,000 × g for 24 h at 4 °C, each sucrose layer was collected (top layer, F1; bottom layer, F7), diluted 5-fold with lysis buffer, and centrifuged at 100,000 × g for 1 h at 4 °C. Each membrane pellet was resuspended in 1.0 ml of lysis buffer and immediately used for enzyme assays or protease accessibility tests. To solubilize membranes, fractions were incubated with 1% (v/v) Igepal in lysis buffer for 1 h at 4 °C with rotation and then centrifuged at 100,000 × g for 1 h at 4 °C.
Quantification of Chlorophyll and Protein
Samples were extracted with acetone (80% (v/v) final concentration), and the chlorophyll content was determined as described previously (43). The solid residue remaining after acetone extraction was washed three times with 100% (v/v) acetone and dried. The dried residue was resuspended in 1% (w/v) SDS and incubated at 95 °C for 5 min. Protein concentrations were determined using the BCA assay (44).
Marker Enzyme Assays
Assays for inositol diphosphatase, antimycin-insensitive NADH-cytochrome c reductase, and cytochrome c oxidase were performed as described previously (45). The vanadate-sensitive adenosine triphosphatase assay was performed as described in Galbraith et al. (46). Membrane fractions containing 10 μg of total protein were used in each assay.
Protease Accessibility Assay
Intact and solubilized membrane fractions in lysis buffer (50 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, 0.2 mm DTT, 10 mm NaF, 1 mm Na3VO4, 250 mm sucrose) containing 10 μg of total protein were incubated in the absence or presence of chymotrypsin (1.0 μg/μl) for 1 h at 0 °C. After the addition of 5 mm phenylmethylsulfonyl fluoride, an equal volume of SDS loading buffer was added and the mixture was incubated at 95 °C for 15 min before analysis by SDS-PAGE and Western blot.
Antibody Inhibition Assays
Sucrose gradient membrane fractions F4 and F5 (intact and solubilized) or recombinant tpSTK1 was incubated at 4 °C for 2 h with the indicated concentrations of anti-tpSTK1 IgG, or IgG from preimmune serum, or in the absence of IgG. Subsequently, kinase assays were performed as described above.
Precipitation of Secreted Proteins
T. pseudonana cells (density ∼106 cells/ml) were pelleted by centrifugation at 3000 × g for 10 min. Proteins in the supernatant were precipitated by the addition of trichloroacetic acid (final concentration 8.5% w/w) and sodium deoxycholate (final concentration 0.9% (w/w)). After incubation for 15 min at 25 °C, the sample was centrifuged at 16,000 × g for 10 min at 4 °C. The precipitate was air-dried, dissolved in SDS sample buffer, and incubated at 95 °C for 5 min before SDS-PAGE.
RESULTS
Primary Structure and Cell Cycle-specific Expression of tpSTK1
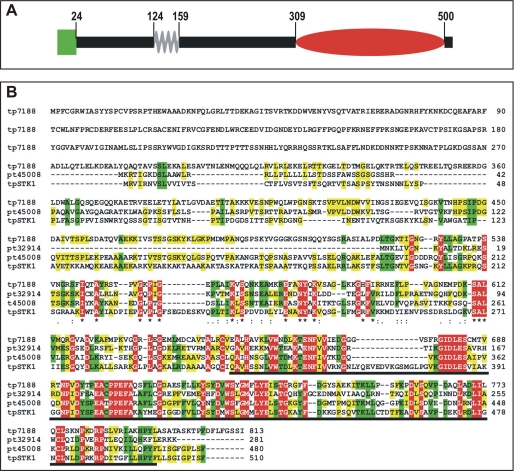
To determine the translation start and stop codons of the tpstk1 gene, 5′- and 3′-RACE PCRs were performed. Reverse transcription-PCR covering the entire gene confirmed the absence of introns as predicted by the tpstk1 gene model in the T. pseudonana data base (32). The tpstk1 gene encodes a protein, tpSTK1, of 510 amino acids with a predicted isoelectric point of 8.95. Sequence analysis using the BLAST algorithm revealed that the C-terminal domain (amino acids 309–500) exhibits homology (E-value: e-16) to the catalytic domain of Ser/Thr kinases, and the N-terminal half contains a predicted coiled-coil domain (amino acids 124–159). Amino acids 1–24 are predicted by sequence analysis programs (47–49) to be either a signal peptide for co-translational import into the ER or a membrane anchor domain. No other potential membrane-spanning domains were identified (Fig. 1A).
FIGURE 1.
Primary structures of tpSTK1 and related proteins. A, schematic structure of tpSTK1. The green box represents the N-terminal signal peptide or transmembrane domain, the gray zig-zag line indicates the predicted coiled-coil domain, and the red oval shows the putative Ser/Thr kinase domain. Numbers indicate the amino acid positions at the borders of the domains. B, alignment of tpSTK1 with predicted protein tp7188 (GenBank™ accession number ACI64649.1) from T. pseudonana and predicted proteins pt45008 and pt32914 (GenBank™ accession numbers EEC49800.1 and EEC50886.1) from P. tricornutum. The alignment was performed using the ClustalW2 program (62). Amino acids that are conserved among all (red), three (green), and two (yellow) sequences are highlighted. The predicted kinase domain of tpSTK1 is underlined.
A BLAST search in the NCBI data base revealed significant sequence alignments to more than 50 proven or predicted Ser/Thr kinases from a broad range of organisms (including humans). The alignment to kinases from non-diatom organisms had relatively high e-values (between 2e-10 and 8e-7), whereas three predicted diatom kinases, tp7188 (GenBank™ accession number ACI64649.1) from T. pseudonana and pt45008 (GenBank™ accession number EEC49800.1) and pt32914 (GenBank™ accession number EEC50886.1) from P. tricornutum, exhibited much lower e-values (3e-86, 1e-71, and 5e-44, respectively) and thus much higher sequence similarity.
tpSTK1 and the three tpSTK1-like predicted diatom kinases each contain a kinase domain in the C-terminal part of the protein. The kinase domains exhibit 26.0–28.8% amino acid sequence conservation among all four proteins, and the positions of 44.4–48.0% of the amino acid residues within the kinase domains are conserved among at least three of the predicted diatom kinases (Fig. 1B).
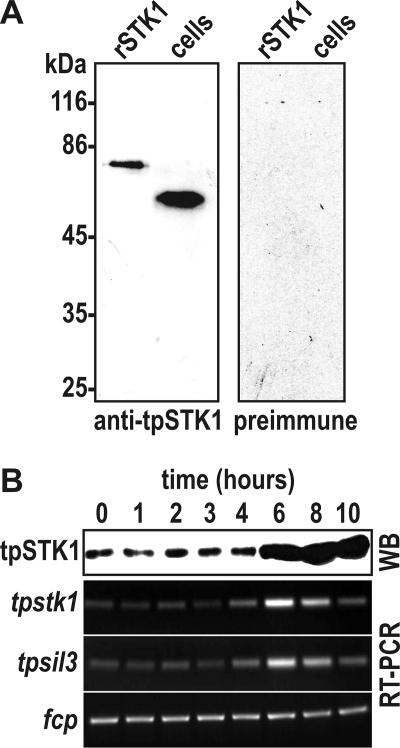
Previously, Hildebrand and co-workers (34) have demonstrated that the expression of tpstk1 mRNA is strongly up-regulated during formation of the valve silica after mitosis, thus closely following the expression pattern of the silaffin-encoding gene, tpsil3. To investigate expression of the tpSTK1 protein during the cell cycle, a polyclonal antiserum was produced against a recombinant version of the protein. From the antiserum anti-tpSTK1 IgG antibodies were obtained by dual affinity chromatography on a recombinant tpSTK1-loaded column followed by protein G-agarose. The anti-tpSTK1 IgGs specifically detected a protein of apparent molecular mass of ∼60 kDa in a T. pseudonana whole cell lysate (Fig. 2A).
FIGURE 2.
Cell cycle-specific expression of native tpSTK1. A, Western blots probed with anti-tpSTK1 IgG (0.2 μg/ml) and preimmune IgG (0.2 μg/ml) that were isolated from the same rabbit. Lanes labeled rSTK1 contained 100 ng of purified recombinant tpSTK1; lanes labeled cells contained lysates from 1.3 × 106 T. pseudonana cells. B, time course of tpSTK1 protein and tpstk1 mRNA expression in synchronized T. pseudonana cells. The times refer to hours after addition of silicic acid to silicon-starved cells. Formation of valve silica is at maximum between 4 and 6 h (39). For each time point, SDS-lysed cells from equal culture volumes were used for Western blot analysis (WB) and cDNA synthesis. The Western blot was probed with 0.2 μg/ml anti-tpSTK1 IgG. Reverse transcription (RT)-PCRs were performed from identical amounts of cDNA using primers specific for the indicated genes: tpsil3, gene encoding silaffin tpSil3; fcp, gene encoding a fucoxanthin chlorophyll-associated protein that is constitutively expressed in T. pseudonana under constant illumination (A. Scheffel and N. Kröger, unpublished communication).
To investigate expression of the tpSTK1 protein during the cell cycle, T. pseudonana cells were synchronized according to an established protocol (39). Semiquantitative reverse transcription-PCR demonstrated that the highest level of tpstk1 mRNA expression was reached at 6 h, which corresponds to the time point of formation of the valve part of the biosilica (Fig. 2B). This is in agreement with a previous quantitative reverse transcription-PCR study on synchronized T. pseudonana cells (34). Western blot analysis of whole cell lysates from the synchronized culture also detected the strongest signal of tpSTK1 at 6 h, demonstrating that biosynthesis of the tpSTK1 protein closely followed expression of the mRNA. The high level of the tpSTK1 protein was maintained at 8 and 10 h, whereas the tpstk1 mRNA level decreased over the same period of time.
Kinase Activity of Recombinant tpSTK1
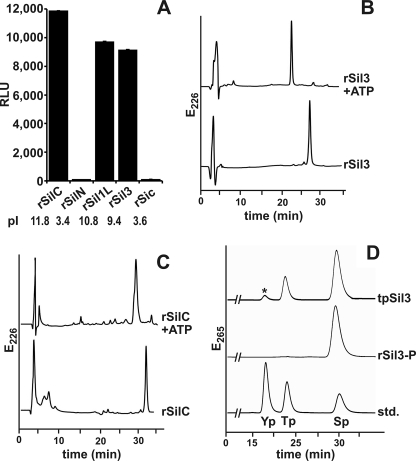
To investigate whether tpSTK1 is a protein kinase, a recombinant tpstk1 gene was constructed that encoded a histidine-tagged derivative of tpSTK1. Recombinant tpSTK1 was expressed in E. coli, solubilized from inclusion bodies, and purified to homogeneity by immobilized ion affinity chromatography (see supplemental Fig. S1). Luminescence-based kinase assays were performed using recombinant silaffins (36) rSil1L (11.8 kDa), rSil3 (22.1 kDa), rSilC (17.6 kDa), and rSilN (9.9 kDa) and recombinant silacidin rSic (19.9 kDa; see supplemental Fig. S2) as substrates. Substantial kinase activities were detected when substrates rSil1L, rSil3, and rSil1C were employed, whereas kinase activities with substrates rSilN and rSic were negligible (Fig. 3A).
FIGURE 3.
Phosphorylation activity of recombinant tpSTK1 with recombinant silaffins (rSilN, rSilC, rSil3, and rSil1L) and recombinant silacidin (rSic) as substrates. A, luminescence-based kinase assay with 0.10 mg/ml substrate protein. Each measurement was repeated at least three times. ⫟, indicates S.D. RLU, relative luminescence units. B and C, RP-HPLC analysis after incubation of the indicated silaffin with tpSTK1 in the presence (+ATP) and absence of ATP. D, phosphoamino acid analysis by anion exchange HPLC of tpSTK1-phosphorylated rSil3 (rSil3-P) and native silaffin tpSil3. The peak labeled with an asterisk contains oxygen-sulfated serine, which was artificially generated during tpSil3 hydrolysis (N. Poulsen and N. Kröger, unpublished result). As a reference the chromatogram of a mixture of the standard amino acids, phosphotyrosine (Yp), phosphothreonine (Tp), and phosphoserine (Sp), is shown.
To ensure that kinase activities determined by the assay were due to phosphorylation of the substrates rather than ATP hydrolysis (note that the assay method is based on quantifying a decrease in ATP concentration), RP-HPLC analysis of the substrates was performed before and after incubation with recombinant tpSTK1. After incubation with recombinant tpSTK1 in the presence of ATP, the substrates rSil3 and rSilC exhibited significant downshifts in their RP-HPLC elution times (Fig. 3, B and C), consistent with a decrease in hydrophobicity due to phosphorylation. Indeed, chemical analysis detected 2.8–3.1 and 0.8–1.0 mol of phosphate/mol of protein for recombinant tpSTK1-treated rSil3 and rSilC, respectively (see supplemental Fig. S3 and Table S1). Phosphoamino acid analysis of RP-HPLC-purified phosphorylated rSil3 detected phosphoserine (Fig. 3D), consistent with the prediction that tpSTK1 is a Ser/Thr kinase. Phosphoserine is also present in the corresponding native silaffin, tpSil3, isolated from T. pseudonana (Fig. 3D).
Further analysis of the substrate specificity of recombinant tpSTK1 demonstrated a preference of the kinase for protein substrates with isoelectric points of >7 (see supplemental Fig. S4A), which is consistent with its inability to phosphorylate rSilN and rSic, both very acidic proteins (see Fig. 3A). Other characteristics of recombinant tpSTK1 are the inability to use GTP instead of ATP (see supplemental Fig. S4B) and the use of Mg2+ or Mn2+ but not Zn2+ or Ca2+ as metal co-factors (see supplemental Fig. S4C).
Intracellular Location of tpSTK1
Current sequence analysis tools do not allow unequivocal prediction of whether the N-terminal amino acids 1–24 in tpSTK1 represent a transient targeting peptide for co-translational ER import or a permanent membrane anchor, as the structural characteristics of both sequence elements are very similar (49). Distinguishing between the two possibilities is important because only a signal peptide would mediate entry into the secretory pathway, thus enabling tpSTK1 to encounter silaffins and other proteins involved in silica biomineralization in vivo.
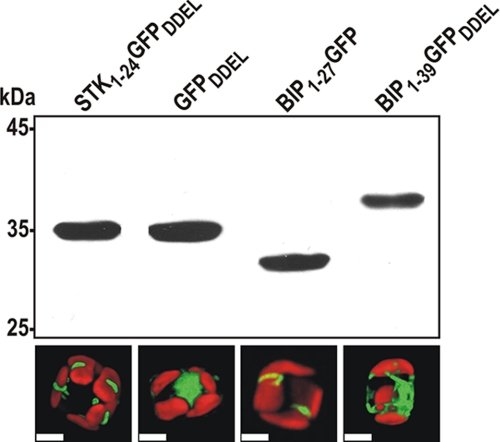
To investigate whether amino acids 1–24 of tpSTK1 act as a signal peptide for import into the ER, fusion protein STK1–24 GFPDDEL was expressed in T. pseudonana. The fusion protein contained amino acids 1–24 of tpSTK1 (STK1–24) at the N terminus fused to GFP and the C-terminal tetrapeptide DDEL, which is the ER retention signal from the T. pseudonana BiP protein.3 If STK1–24 acted as a signal peptide, the fusion protein was expected to localize in the ER and the signal peptide be removed by the endogenous signal peptidase. Western blot analysis revealed that the apparent molecular mass of the fusion protein in T. pseudonana was identical to that of T. pseudonana-expressed GFPDDEL (i.e. GFP fused to the DDEL tetrapeptide) (Fig. 4). This result suggested that the STK1–24 part was cleaved off from the STK1–24GFPDDEL fusion protein during maturation in T. pseudonana. The GFPDDEL protein derived from the processing of STK1–24GFPDDEL was located in a clamp-like structure associated with the chloroplast. In contrast, the GFPDDEL protein, which was expressed without an N-terminal extension, exhibited a cytosolic localization pattern as expected (Fig. 4). The chloroplast-associated structure containing the STK1–24GFPDDEL-derived GFPDDEL protein may be a subcompartment of the ER, as diatom chloroplasts are tightly surrounded by ER membranes (50). This assumption is supported by the results from expression in T. pseudonana of BiP1–27GFP, a GFP fusion protein carrying the signal peptide for import of the ER-resident protein BiP, because this protein is located in a seemingly similar chloroplast associated compartment (Fig. 4). Western blot analysis confirmed the removal of the N-terminal signal peptide from the BIP1–27GFP fusion protein, as the resulting protein, GFP, was smaller in apparent molecular mass than GFP carrying the DDEL tetrapeptide (Fig. 4). As an additional control, BiP1–39GFPDDEL was expressed in T. pseudonana. This GFP fusion protein contained amino acids 1–39 and the C-terminal ER retention signal of BiP. Removal of the signal peptide during intracellular maturation was expected to generate a GFP fusion protein that contained 12 and 16 amino acids more than the proteins derived from STK1–24 GFPDDEL and BiP1–27GFP, respectively. Indeed, Western blot analysis confirmed that the BiP1–39GFPDDEL had the highest molecular mass of all of the GFP fusion proteins that were analyzed (Fig. 4). Interestingly, the BiP1–39GFPDDEL-derived protein exhibited an ER-like localization pattern (Fig. 4), suggesting that the additional amino acids derived from mature BiP have a strong influence on location within the ER. Altogether, these data indicate that amino acids 1–24 of tpSTK1 function as a cleavable signal peptide for import into the ER and not as a permanent membrane anchor domain.
FIGURE 4.
Intracellular processing and localization of GFP fusion proteins. For names and composition of the GFP fusion proteins refer to “Results”. Top, Western blot with anti-GFP antibodies (0.5 μg/ml). In each lane, whole cell lysates of T. pseudonana expressing the indicated GFP fusion protein were loaded. After removal of the N-terminal signal peptide from BIP1–39GFPDDEL 12 amino acids derived from the mature BiP sequence remain attached to the N terminus of GFP. Bottom, corresponding confocal fluorescence micrographs of individual T. pseudonana cells expressing the GFP fusion proteins indicated in the top panel. The red color is caused by autofluorescence of the chloroplasts. Scale bars = 2 μm.
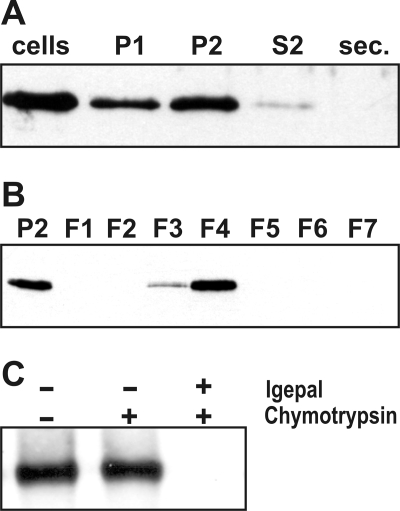
Multiple attempts to use GFP tagging to determine the intracellular location of the full-length tpSTK1 protein by fluorescence microscopy failed, as no GFP-fluorescent transformants were obtained. Therefore, a biochemical approach was pursued to localize tpSTK1. T. pseudonana cells were gently lysed with glass beads, and the lysate was separated into three fractions by differential centrifugation: P1 (16,000 × g pellet), P2 (100,000 × g pellet), and S2 (100,000 × g supernatant). S2 is the cytosolic fraction; P1 contains non-lysed cells, cell walls, and ∼80% of the chloroplasts (based on chlorophyll content); and P2 contains plastid-depleted cellular membranes (Table 1). Western blot analysis using anti-tpSTK1 IgG revealed that tpSTK1 is present predominantly in the P2 fraction, is virtually absent from the cytosol, and is not secreted into the medium (Fig. 5A).
TABLE 1.
Enzyme activities and protein and chlorophyll content in subcellular fractions of T. pseudonana
The listed numbers represent averages from three independent experiments. The number for the highest activity of a particular enzyme is shown in bold. IDPase, inositol diphosphatase; CCRase, cytochrome c reductase; COx, cytochrome c oxidase; vsATPase, vanadate-sensitive ATPase; Chl, chlorophyll; ND, not determined.
| Fraction | IDPase | CCRase | COx | vsATPase | Protein | Chl |
|---|---|---|---|---|---|---|
| nmol Pi/s | nmol/min | nmol/min | nmol Pi/s | mg | mg | |
| S2 | ND | ND | ND | ND | 1.31 | 0.04 |
| P1 | ND | ND | ND | ND | 0.79 | 2.45 |
| P2 | ND | ND | ND | ND | 1.20 | 0.65 |
| F1 + F2 | 0.04 | 0.07 | 0.04 | 0.01 | 0.41 | 0.11 |
| F3 | 0.05 | 0.33 | 0.03 | 0.01 | 0.14 | 0.06 |
| F4 | 0.34 | 1.74 | 0.03 | 0.01 | 0.21 | 0.03 |
| F5 | 1.44 | 0.04 | 0.11 | 0.04 | 0.19 | 0.02 |
| F6 + F7 | 0.06 | 0.08 | 0.81 | 1.41 | 0.34 | 0.40 |
FIGURE 5.
Intracellular location of native tpSTK1 in T. pseudonana wild type cells. The Western blots were probed with anti-tpSTK1 IgG (0.2 μg/ml). A, distribution of native tpSTK1 in subcellular fractions and the culture medium. Equal aliquots of whole cell lysate (cells), cell culture medium (sec.), and subcellular fractions (P1, P2, S2) were subjected to SDS-PAGE (P1 contains significant amounts of intact cells). B, distribution of native tpSTK1 in membrane fractions from density sucrose gradient centrifugation. Equal aliquots of subcellular fraction P2 and sucrose gradient fractions F1–F7 were loaded. C, protease accessibility test. Equal amounts of membranes from fraction F4 were incubated in the absence (−) or presence (+) of the detergent Igepal and the protease chymotrypsin.
The P2 membranes were fractionated by sucrose density gradient centrifugation yielding fractions F1–F7. Each fraction was analyzed for the presence of tpSTK1, total protein, and chlorophyll content, and the activities of marker enzymes for the ER (NADH-cytochrome c reductase), mitochondria (cytochrome c oxidase), Golgi apparatus (inositol diphosphatase), and plasma membrane (vanadate-sensitive ATPase). According to the marker enzyme assays, fraction F4 contains the majority of the ER membranes (71% of total cytochrome c reductase activity), most of the Golgi membranes are present in fraction F5 (74% of total inositol diphosphatase activity), and combined fractions F6 and F7 contain the majority of mitochondria (79% of total cytochrome c oxidase activity) and plasma membrane (96% of total vanadate-sensitive ATPase activity) (Table 1). Western blot analysis of the same gradient fractions using anti-tpSTK1 antibodies detected tpSTK1 mainly in fraction F4, and to a smaller extent also in fraction F3, but not in any other fraction (Fig. 5B). The fact that the distribution of tpSTK1 within the sucrose gradient perfectly paralleled the distribution of ER membranes strongly suggests that the kinase is associated with ER membranes in vivo.
To determine whether tpSTK1 is localized on the cytosolic or the luminal side of membranes, protease accessibility experiments were performed with the membranes of fraction F4. Treatment of the membranes with chymotrypsin resulted in degradation of several major protein components (see supplemental Fig. S5) but did not disrupt the activity of the ER luminal enzyme cytochrome c reductase (see Supplement for Table S2). However, cytochrome c reductase activity was completely disrupted when the membranes were detergent-solubilized prior to chymotrypsin treatment (see supplemental Table S2). These results indicated that the surfaces of the isolated ER membranes in fraction F4 were correctly oriented (“inside-in”). Western blot analysis of chymotrypsin-treated intact and detergent-solubilized F4 membranes revealed that tpSTK1 was inaccessible to protease digestion in intact membranes (Fig. 5C), demonstrating that it is located in the membrane lumen.
Kinase Activity of Native tpSTK1
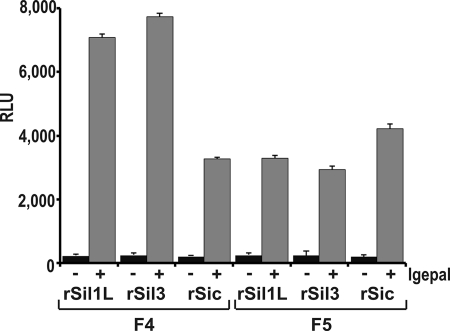
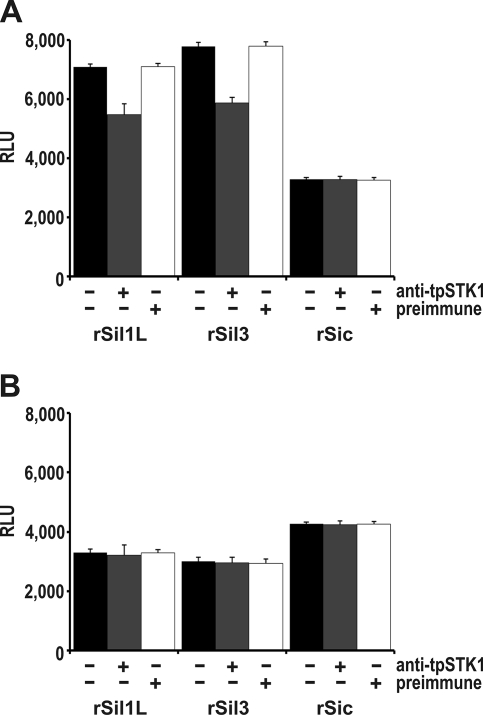
To investigate the phosphorylation of silaffins and silacidins by protein kinases associated with the ER and Golgi apparatus, kinase assays were performed with membranes from sucrose gradient fractions F5 (contains Golgi membranes) and F4 (contains ER membranes) using recombinant silaffins rSil3 and rSil1L and recombinant silacidin rSic as substrates. Kinase activity with either substrate was very low with intact membranes and at least 10-fold higher with detergent-solubilized membranes (Fig. 6). The kinase activities observed with silaffin substrates were substantially higher with solubilized membranes from fraction F4 than from fraction F5, whereas the extent of rSic phosphorylation was higher with solubilized membranes from fraction F5. To determine the contribution of native tpSTK1 to the observed phosphorylation activities, kinase assays were performed in the presence of anti-tpSTK1 IgG. The tpSTK1:IgG ratio used in the assays was sufficient to completely inhibit the activity of recombinant tpSTK1 (see supplemental Figs. S6 and S7). In the presence of the antibodies the kinase activities of F4 and F5 membranes with substrate rSic remained virtually unchanged (Fig. 7). With the silaffin substrates the kinase activity of F5 membranes did not decrease in the presence of the antibodies (Fig. 7B), whereas the kinase activity of F4 membranes decreased by 24% for rSil3 and 29% for rSil1L (Fig. 7A). Using the same concentration of preimmune IgG instead of anti-tpSTK1 IgG did not inhibit the activities of the kinases present in solubilized membrane preparations from either fraction (Fig. 7). These data indicated that native tpSTK1 was responsible for 24–29% of the silaffin kinase activity in the ER lumen and that it was unable to phosphorylate silacidin.
FIGURE 6.
Phosphorylation of silaffins rSil3 and rSil1L and silacidin rSic in subcellular fractions of T. pseudonana containing ER (F4) and Golgi (F5) membranes. For each assay equal amounts of membranes (10 μg of total protein) were used. The concentration of each substrate protein was 0.1 mg/ml. Kinase activities of intact membranes (−) and membranes solubilized with 1.0% (v/v) Igepal (+) are shown. ⫟, indicates S.D. RLU, relative luminescence units.
FIGURE 7.
Inhibition of tpSTK1 activity in subcellular membrane fractions of T. pseudonana. Phosphorylation of silaffins (rSil3 and rSil1L) and silacidin (rSic) in detergent-solubilized membranes of ER-containing fraction F4 (A) and Golgi-containing fraction F5 (B) in the presence (+) or absence (−) of equal concentrations of anti-tpSTK1 IgG or preimmune IgG. Each measurement was repeated at least three times. ⫟, indicates S.D. RLU, relative luminescence units.
DISCUSSION
The data presented here demonstrate that the serine/threonine kinase tpSTK1 contains a functional N-terminal signal peptide for import into the ER and exhibits the biochemical characteristics of a protein resident in the lumen of the ER. The preferred substrates of tpSTK1 in vitro are proteins with basic isoelectric points including recombinant silaffins and excluding the acidic recombinant silacidin. As silaffins pass through the ER on their pathway to the silica deposition vesicle (i.e. the organelle of silica formation) (27), and because tpSTK1 is a relatively abundant protein in the ER (∼0.5% of total ER protein; see supplemental Fig. S6), it is reasonable to assume that silaffins are phosphorylated by tpSTK1 in vivo. Therefore, we have denoted tpSTK1 as a silaffin kinase. It is expected that additional kinases are required for the biogenesis of native silaffins in vivo based on the following observations. (i) tpSTK1 catalyzed the attachment of only 1–3 phosphate residues/silaffin molecule, whereas native silaffins from T. pseudonana contain between 35 and 50 phosphate residues/molecule (29) (it is unknown what fraction of the phosphate residues in native silaffins is attached to glycan moieties). (ii) Recombinant tpSTK1 was only able to phosphorylate the serine residues of rSil3, yet the corresponding native silaffin tpSil3 contains both phosphorylated serine and threonine residues. (iii) The lumina of the ER and Golgi apparatus contain kinases with substantial activity for rSil3 and rSil1L phosphorylation, despite the inhibition/absence of functional tpSTK1.
An additional candidate silaffin kinase in T. pseudonana is the predicted protein tp7188 (GenBank™ accession number ACI64649.1), which exhibits 28.4% amino acid sequence identity with the entire tpSTK1 sequence and 39.1% identity with the kinase domain of tpSTK1 (see Fig. 1B). Predicted protein tp7188 does not contain an N-terminal signal peptide,4 yet it contains a predicted internal membrane-spanning domain in the N-terminal part (amino acids 183–199), which could potentially act as a signal-anchor sequence for insertion into the ER membrane. If tp7188 is inserted as a type II membrane protein, the C-terminal part containing the putative Ser/Thr domain would face the lumen of the ER (51).
Although intensive research has been conducted on kinases operating in the cytosol, protein phosphorylation within the secretory pathway is not well understood. The first primary structure of a kinase operating in the secretory pathway, the Golgi-localized cadherin phosphorylating Ser/Thr kinase Fj from Drosophila melanogaster, has only recently been identified (25). The tyrosine kinase Fyn, which previously was believed to be active only in the cytosol, has also been shown to be located in the lumen of the ER and is implicated in phosphorylation of the ER-resident chaperone Grp94 (26). These highly specific kinases are different from the casein kinase-like activities in the mammalian ER and Golgi apparatus that catalyze the phosphorylation of a large variety of secretory phosphoproteins including proteins involved in biomineralization of bone and teeth (e.g. osteopontin, dentin sialophosphoprotein, and bone sialoprotein) (18, 19, 52). To date no information is available on the molecular structures of these kinases. The silaffin kinase tpSTK1 described in the present work is, therefore, the first structurally characterized kinase likely to be involved in biomineralization. The closest sequence homologues of tpSTK1 are the three predicted kinases, pt45008 (GenBank™ accession number EEC49800.1) and pt32914 (GenBank™ accession number EEC50886.1) from P. tricornutum and tp7188 (GenBank™ accession number ACI64649.1) from T. pseudonana. Preliminary analysis of emerging genome sequencing data from Fragilariopsis cylindrus revealed five predicted kinases that exhibited higher sequence similarity to tpSTK1 and the tpSTK1-like diatom kinases (e-values: 8e-45 to 5e-70) than to any other Ser/Thr kinases from diatoms or non-diatom organisms.4 These data suggest that tpSTK1 and its diatom homologues might represent a novel family of kinases involved in unconventional cellular pathways like silica biomineralization. Using these diatom kinase sequences, future extensive bioinformatics analysis may enable us to identify related kinases with unknown functions in other biomineral-forming organisms.
The experimental finding that tpSTK1 is an ER-resident protein was unexpected, as the kinase does not contain a C-terminal ER retrieval signal (C-terminal DDEL in diatoms, (H/K)DEL in other organisms). This suggests either that tpSTK1 binds to other ER-resident proteins that carry an ER retrieval signal (e.g. BiP and GRP94) or that tpSTK1 may contain an as yet unknown ER retention motif. There are precedents for both of these scenarios (53–56). The exact location of tpSTK1 within the ER is currently unknown. The signal peptides of tpSTK1 and BiP targeted GFP to a plastid-associated clamp-like structure (see Fig. 6B), which probably represents an ER subcompartment, as diatom plastids are surrounded by rough ER (50). However, it is possible that mature tpSTK1 is not exclusively located in the clamp-like structure, because the GFP fusion protein containing 16 additional amino acid residues of the mature BiP protein was not confined to this structure but rather exhibited a widespread distribution within the ER. A similar structure, termed “blob”-like structure (BLS) and located at the periphery of plastids, was observed by Kroth and co-workers (57, 58) in the diatom P. tricornutum. BLS is an accumulation site for GFP-tagged nuclear encoded plastid proteins, which contain an N-terminal signal peptide for ER import but lack a functional plastid import sequence (import of nuclear encoded proteins into diatom plastids proceeds through the ER, requiring a bipartite targeting sequence (50, 59–61)). We speculated that the clamp-like structure in T. pseudonana and BLS in P. tricornutum may be analogous ER subcompartments through which ER-imported proteins transit before reaching their final destination.
Future research will be needed to address the exact localization of tpSTK1 by immunolocalization techniques (immunofluorescence microscopy and immunogold electron microscopy). Furthermore, using membrane-permeable chemical cross-linking reagents in combination with “pulldown” methods and mass spectrometric analysis may allow identification of the entire set of in vivo substrates of the novel kinase tpSTK1.
Supplementary Material
Acknowledgments
We are grateful to D. Hipp for help with establishing the kinase assay and A. Scheffel for critically reading the manuscript.
This work was supported by the Georgia Institute of Technology, Grant DMR 0845939 from the National Science Foundation (to N. K.), Cottrell Scholar Award 7492 from the Research Corporation for Science Advancement (to N. K.), and a U. S. Department of Education Graduate Assistance in Areas of National Need (GAANN) fellowship (to V. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1 and S2.
N. Poulsen and N. Kröger, unpublished observation.
N. Kröger, unpublished observation.
- ER
- endoplasmic reticulum
- RACE
- rapid amplification of cDNA ends
- DTT
- dithiothreitol
- RP
- reversed phase
- HPLC
- high pressure liquid chromatography
- r
- recombinant
- BLS
- blob-like structure
- GFP
- green fluorescent protein
- BiP
- binding protein.
REFERENCES
- 1.Mann S. (1993) Nature 365, 499–505 [Google Scholar]
- 2.Sanchez C., Arribart H., Guille M. M. (2005) Nat. Mater. 4, 277–288 [DOI] [PubMed] [Google Scholar]
- 3.Mayer G. (2005) Science 310, 1144–1147 [DOI] [PubMed] [Google Scholar]
- 4.Hamm C. E., Merkel R., Springer O., Jurkojc P., Maier C., Prechtel K., Smetacek V. (2003) Nature 421, 841–843 [DOI] [PubMed] [Google Scholar]
- 5.Wilt F. H. (2005) Dev. Biol. 280, 15–25 [DOI] [PubMed] [Google Scholar]
- 6.Belcher A. M., Hansma P. K., Stucky G. D., Morse D. E. (1998) Acta Mater. 46, 733–736 [Google Scholar]
- 7.Crookes-Goodson W. J., Slocik J. M., Naik R. R. (2008) Chem. Soc. Rev. 37, 2403–2412 [DOI] [PubMed] [Google Scholar]
- 8.Boskey A. L., Maresca M., Ullrich W., Doty S. B., Butler W. T., Prince C. W. (1993) Bone Miner. 22, 147–159 [DOI] [PubMed] [Google Scholar]
- 9.Kröger N., Lorenz S., Brunner E., Sumper M. (2002) Science 298, 584–586 [DOI] [PubMed] [Google Scholar]
- 10.He G., Ramachandran A., Dahl T., George S., Schultz D., Cookson D., Veis A., George A. (2005) J. Biol. Chem. 280, 33109–33114 [DOI] [PubMed] [Google Scholar]
- 11.Waite J. H., Qin X. (2001) Biochemistry 40, 2887–2893 [DOI] [PubMed] [Google Scholar]
- 12.Zhao H., Sun C., Stewart R. J., Waite J. H. (2005) J. Biol. Chem. 280, 42938–42944 [DOI] [PubMed] [Google Scholar]
- 13.Lowenstam H. A., Weiner S. (1989) On Biomineralization, Oxford University Press U.S.A., Oxford, NY [Google Scholar]
- 14.Simkiss K., Wilbur K. M. (1989) Biomineralization: Cell Biology and Mineral Deposition, Academic Press Inc., San Diego [Google Scholar]
- 15.Kröger N., Sumper M. (1998) Protist 149, 213–219 [DOI] [PubMed] [Google Scholar]
- 16.Bäuerlein E. (2003) Angew. Chem. Int. Ed. Engl. 42, 614–641 [DOI] [PubMed] [Google Scholar]
- 17.Bingham E. W., Farrell H. M., Jr. (1974) J. Biol. Chem. 249, 3647–3651 [PubMed] [Google Scholar]
- 18.Veis A., Sfeir C., Wu C. B. (1997) Crit. Rev. Oral Biol. Med. 8, 360–379 [DOI] [PubMed] [Google Scholar]
- 19.Lasa M., Chang P. L., Prince C. W., Pinna L. A. (1997) Biochem. Biophys. Res. Commun. 240, 602–605 [DOI] [PubMed] [Google Scholar]
- 20.Lasa M., Marin O., Pinna L. A. (1997) Eur. J. Biochem. 243, 719–725 [DOI] [PubMed] [Google Scholar]
- 21.Pasqualini E., Caillol N., Valette A., Lloubes R., Verine A., Lombardo D. (2000) Biochem. J. 345, 121–128 [PMC free article] [PubMed] [Google Scholar]
- 22.Brunati A. M., Contri A., Muenchbach M., James P., Marin O., Pinna L. A. (2000) FEBS Lett. 471, 151–155 [DOI] [PubMed] [Google Scholar]
- 23.Drzymala L., Castle A., Cheung J. C., Bennick A. (2000) Biochemistry 39, 2023–2031 [DOI] [PubMed] [Google Scholar]
- 24.Lee S. N., Hwang J. R., Lindberg I. (2006) J. Biol. Chem. 281, 3312–3320 [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa H. O., Takeuchi H., Haltiwanger R. S., Irvine K. D. (2008) Science 321, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frasson M., Vitadello M., Brunati A. M., La Rocca N., Tibaldi E., Pinna L. A., Gorza L., Donella-Deana A. (2009) Biochim. Biophys. Acta 1793, 239–252 [DOI] [PubMed] [Google Scholar]
- 27.Kröger N., Poulsen N. (2008) Annu. Rev. Genet. 42, 83–107 [DOI] [PubMed] [Google Scholar]
- 28.Sumper M., Brunner E. (2008) Chembiochem 9, 1187–1194 [DOI] [PubMed] [Google Scholar]
- 29.Poulsen N., Kröger N. (2004) J. Biol. Chem. 279, 42993–42999 [DOI] [PubMed] [Google Scholar]
- 30.Poulsen N., Sumper M., Kröger N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12075–12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wenzl S., Hett R., Richthammer P., Sumper M. (2008) Angew. Chem. Int. Ed. Engl. 47, 1729–1732 [DOI] [PubMed] [Google Scholar]
- 32.Armbrust E. V., Berges J. A., Bowler C., Green B. R., Martinez D., Putnam N. H., Zhou S., Allen A. E., Apt K. E., Bechner M., Brzezinski M. A., Chaal B. K., Chiovitti A., Davis A. K., Demarest M. S., Detter J. C., Glavina T., Goodstein D., Hadi M. Z., Hellsten U., Hildebrand M., Jenkins B. D., Jurka J., Kapitonov V. V., Kröger N., Lau W. W., Lane T. W., Larimer F. W., Lippmeier J. C., Lucas S., Medina M., Montsant A., Obornik M., Parker M. S., Palenik B., Pazour G. J., Richardson P. M., Rynearson T. A., Saito M. A., Schwartz D. C., Thamatrakoln K., Valentin K., Vardi A., Wilkerson F. P., Rokhsar D. S. (2004) Science 306, 79–86 [DOI] [PubMed] [Google Scholar]
- 33.Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K., Kuo A., Maheswari U., Martens C., Maumus F., Otillar R. P., Rayko E., Salamov A., Vandepoele K., Beszteri B., Gruber A., Heijde M., Katinka M., Mock T., Valentin K., Verret F., Berges J. A., Brownlee C., Cadoret J. P., Chiovitti A., Choi C. J., Coesel S., De Martino A., Detter J. C., Durkin C., Falciatore A., Fournet J., Haruta M., Huysman M. J., Jenkins B. D., Jiroutova K., Jorgensen R. E., Joubert Y., Kaplan A., Kröger N., Kroth P. G., La Roche J., Lindquist E., Lommer M., Martin-Jézéquel V., Lopez P. J., Lucas S., Mangogna M., McGinnis K., Medlin L. K., Montsant A., Oudot-Le Secq M. P., Napoli C., Obornik M., Parker M. S., Petit J. L., Porcel B. M., Poulsen N., Robison M., Rychlewski L., Rynearson T. A., Schmutz J., Shapiro H., Siaut M., Stanley M., Sussman M. R., Taylor A. R., Vardi A., von Dassow P., Vyverman W., Willis A., Wyrwicz L. S., Rokhsar D. S., Weissenbach J., Armbrust E. V., Green B. R., Van de Peer Y., Grigoriev I. V. (2008) Nature 456, 239–24418923393 [Google Scholar]
- 34.Frigeri L. G., Radabaugh T. R., Haynes P. A., Hildebrand M. (2006) Mol. Cell. Proteomics 5, 182–193 [DOI] [PubMed] [Google Scholar]
- 35.Mock T., Samanta M. P., Iverson V., Berthiaume C., Robison M., Holtermann K., Durkin C., Bondurant S. S., Richmond K., Rodesch M., Kallas T., Huttlin E. L., Cerrina F., Sussman M. R., Armbrust E. V. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1579–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kröger N., Dickerson M. B., Ahmad G., Cai Y., Haluska M. S., Sandhage K. H., Poulsen N., Sheppard V. C. (2006) Angew. Chem. Int. Ed. Engl. 45, 7239–7243 [DOI] [PubMed] [Google Scholar]
- 37.Poulsen N., Kröger N. (2005) FEBS J. 272, 3413–3423 [DOI] [PubMed] [Google Scholar]
- 38.Fischer H., Robl I., Sumper M., Kröger N. (1999) J. Phycol. 35, 113–120 [Google Scholar]
- 39.Hildebrand M., Frigeri L. G., Davis A. K. (2007) J. Phycol. 43, 730–740 [Google Scholar]
- 40.Buss J. E., Stull J. T. (1983) Methods Enzymol. 99, 7–14 [DOI] [PubMed] [Google Scholar]
- 41.Poulsen N., Chesley P. M., Kröger N. (2006) J. Phycol. 42, 1059–1065 [Google Scholar]
- 42.Poulsen N., Berne C., Spain J., Kröger N. (2007) Angew. Chem. Int. Ed. Engl. 46, 1843–1846 [DOI] [PubMed] [Google Scholar]
- 43.Arnon D. I. (1949) Plant Physiol. 24, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 45.Packer L., Fleischer S. (1997) Biomembranes, pp. 214–219, Academic Press Inc., San Diego [Google Scholar]
- 46.Galbraith D. W., Bohnert H. J., Bourque D. P. (1995) Methods in Plant Cell Biology, pp. 137–142, Academic Press Inc., San Diego [Google Scholar]
- 47.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 48.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 49.Elofsson A., von Heijne G. (2007) Annu. Rev. Biochem. 76, 125–140 [DOI] [PubMed] [Google Scholar]
- 50.Kroth P. G. (2002) Int. Rev. Cytol. 221, 191–255 [DOI] [PubMed] [Google Scholar]
- 51.Goder V., Spiess M. (2001) FEBS Lett. 504, 87–93 [DOI] [PubMed] [Google Scholar]
- 52.George A., Veis A. (2008) Chem. Rev. 108, 4670–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arber S., Krause K. H., Caroni P. (1992) J. Cell Biol. 116, 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monnat J., Neuhaus E. M., Pop M. S., Ferrari D. M., Kramer B., Soldati T. (2000) Mol. Biol. Cell 11, 3469–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pimpl P., Denecke J. (2000) Plant Cell 12, 1517–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfeffer S. R. (2007) Annu. Rev. Biochem. 76, 629–645 [DOI] [PubMed] [Google Scholar]
- 57.Gruber A., Vugrinec S., Hempel F., Gould S. B., Maier U. G., Kroth P. G. (2007) Plant Mol. Biol. 64, 519–530 [DOI] [PubMed] [Google Scholar]
- 58.Kilian O., Kroth P. G. (2005) Plant J. 41, 175–183 [DOI] [PubMed] [Google Scholar]
- 59.Bhaya D., Grossman A. R. (1991) Mol. Gen. Genet. 229, 400–404 [DOI] [PubMed] [Google Scholar]
- 60.Lang M., Apt K. E., Kroth P. G. (1998) J. Biol. Chem. 273, 30973–30978 [DOI] [PubMed] [Google Scholar]
- 61.Apt K. E., Zaslavkaia L., Lippmeier J. C., Lang M., Kilian O., Wetherbee R., Grossman A. R., Kroth P. G. (2002) J. Cell Sci. 115, 4061–4069 [DOI] [PubMed] [Google Scholar]
- 62.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.