Abstract
PKD1 (polycystin-1), the disease-causing gene for ADPKD, is widely expressed in various cell types, including osteoblasts, where its function is unknown. Although global inactivation of Pkd1 in mice results in abnormal skeletal development, the presence of polycystic kidneys and perinatal lethality confound ascertaining the direct osteoblastic functions of PKD1 in adult bone. To determine the role of PKD1 in osteoblasts, we conditionally inactivated Pkd1 in postnatal mature osteoblasts by crossing Oc (osteocalcin)-Cre mice with floxed Pkd1 (Pkd1flox/m1Bei) mice to generate conditional heterozygous (Oc-Cre;Pkd1flox/+) and homozygous (Oc-Cre;Pkd1flox/m1Bei) Pkd1-deficient mice. Cre-mediated recombination (Pkd1Δflox) occurred exclusively in bone. Compared with control mice, the conditional deletion of Pkd1 from osteoblasts resulted in a gene dose-dependent reduction in bone mineral density, trabecular bone volume, and cortical thickness. In addition, mineral apposition rates and osteoblast-related gene expression, including Runx2-II (Runt-related transcription factor 2), osteocalcin, osteopontin, and bone sialoprotein, were reduced proportionate to the reduction of Pkd1 gene dose in bone of Oc-Cre;Pkd1flox/+ and Oc-Cre;Pkd1flox/m1Bei mice. Primary osteoblasts derived from Oc-Cre;Pkd1flox/m1Bei displayed impaired differentiation and suppressed activity of the phosphatidylinositdol 3-kinase-Akt-GSK3β-β-catenin signaling pathways. The conditional deletion of Pkd1 also resulted in increased adipogenesis in bone marrow and in osteoblast cultures. Thus, PKD1 directly functions in osteoblasts to regulate bone formation.
Introduction
PC1 (polycystin-1) is a highly conserved, receptor-like multidomain membrane protein widely expressed in various cell types and tissues (1, 2). Mutations of human PKD1 (polycystic kidney disease gene 1) cause autosomal dominant polycystic kidney disease (ADPKD)2 (3, 4). The genetics of ADPKD is complex, because it is widely held that inactivation of the normal copy of the PKD1 gene by a second somatic mutation in conjunction with the inherited mutation of the other allele is required for renal cyst formation, which occurs in only a subset of the dually affected tubules (5). Although primarily affecting the kidney, ADPKD is also a multisystem disorder (6, 7). Extrarenal manifestations include intracranial and aortic aneurysms and cystic disease of liver and pancreas (8–11). The biological functions of PC1 are poorly defined in some tissues that express PKD1 transcripts, such as bone. Indeed, the absence of clinically demonstrable skeletal abnormalities in patients with ADPKD initially delayed the investigation of PKD1 function in bone. The apparent lack of abnormalities in other tissues expressing PC1 may arise because of differences in the frequency of a second hit somatic mutation, the presence of other modifying factors that may compensate for lack of PC1 function in other organs (12), or failure to detect more subtle phenotypes. For example, lung was not thought to be affected by PKD1 mutations until computed tomography scans of lungs of ADPKD patients showed a 3-fold increase in the prevalence of bronchiectasis compared with controls (13).
Pkd1 is highly expressed in bone, and several mouse models with inactivating mutations of Pkd1 have skeletal abnormalities in the setting of polycystic kidney disease and embryonic lethality (6, 7, 14–16). Most recently, however, the heterozygous Pkd1m1Bei mouse, which has an inactivating mutation of Pkd1 and survives to adulthood without polycystic kidney disease, has been shown to develop osteopenia and impaired osteoblastic differentiation (17, 18), suggesting that Pkd1 may function in bone. Because homozygous PKD1/Pkd1 mutations in humans and mice are lethal, and most of the existing models are globally Pkd1-deficient, the significance of inactivation of Pkd1 in osteoblasts remains uncertain, and the bone changes might reflect an indirect effect due to loss of PKD1, in the kidney or other tissues.
In the current study, to determine if PKD1 in osteoblasts has a direct function in regulating postnatal skeletal functions, we used mouse genetic approaches to conditionally delete Pkd1 in osteoblasts. We demonstrate that conditional deletion of Pkd1 from osteoblasts using Oc-Cre results defective osteoblast function in vivo and in vitro, and osteopenia, indicating that PKD1 has a direct role to regulate osteoblast function and skeletal homeostasis.
EXPERIMENTAL PROCEDURES
Mice
We obtained the floxed Pkd1 mice from Dr. Gregory Germino at Johns Hopkins University (19) and Oc (osteocalcin)-Cre mice from Dr. Thomas Clemens at the University of Alabama (20). The Pkd1m1Bei heterozygous mice were available in our laboratory as described previously (18). These mice were bred and maintained on a C57BL/6J background. At first, we created double heterozygous Oc-Cre;Pkd1m1Bei/+ mice and homozygous Pkd1flox/flox mice. Then double heterozygous Oc-Cre;Pkd1m1Bei/+ mice were mated with homozygous Pkd1flox/flox mice to generate excised floxed Pkd1 heterozygous (Oc-Cre;Pkd1flox/+) and null mice (Oc-Cre;Pkd1flox/m1Bei or Pkd1Oc-cko) as well as Beier Pkd1 heterozygous mice (Pkd1m1Bei/flox) and Oc-Cre negative control mice (Pkd1flox/+, equivalent to wild type). These mice were used for phenotypic analysis. Animal experiments were performed, following review and approval by the University of Kansas Medical Center's Animal Care and Use Committee.
Genotyping PCR and Real-time PCR to Detect Mutations and Deletions
Genomic DNA was prepared from bone and other tissue specimens using standard procedures. PCR genotyping was performed using the following primers (19) (Fig. 1A): F1, 5′-CTT CTA TCG CCT TCT TGA CGA GTT C-3′; R1, 5′-AGG GCT TTT CTT GCT GGT CT-3′; R2, 5′-TCG TGT TCC CTT ACC AAC CCT C-3′. Pkd1 floxed (Pkd1flox) alleles were identified in 2% agarose gels as 670 bp bands (Fig. 1B). The Δ floxed Pkd1 (Pkd1Δflox) allele was detected as a 0.85 kb band in 1% agarose gels (Fig. 1B). The Pkd1m1Bei allele was genotyped using SYBR® Green (Bio-Rad) real-time PCR as described previously (18).
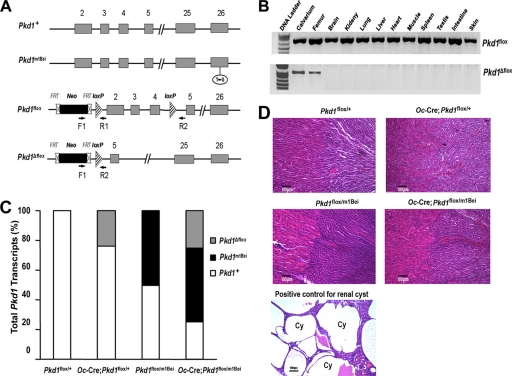
FIGURE 1.
Oc-Cre-mediated bone specific deletion of Pkd1 from the floxed Pkd1 allele (Pkd1flox). A, schematic illustration of wild-type (Pkd1+), mutant (Pkd1m1Bei), and floxed Pkd1 allele before (Pkd1flox) and after deletion (Pkd1Δflox) of the lox P cassette containing exons 2–4 via Cre-mediated recombination. B, genotype PCR analysis of different tissues that were harvested from 16-week-old Oc-Cre;Pkd1flox/m1Bei mice showed bone-specific deletion of the Pkd1 gene. Osteocalcin-Cre-mediated recombination of excised floxed Pkd1 (Pkd1Δflox) allele occurred exclusively in bone, whereas non-skeletal tissues retained the floxed Pkd1 allele (Pkd1flox). C, real-time RT-PCR analysis of total Pkd1 transcripts. Data are expressed as the percentage expression of wild-type (Pkd1+ and Pkd1flox), mutant (Pkd1m1Bei), and conditional deleted (Pkd1Δflox) Pkd1 alleles for each genotype from 5–6 tibias of 16-week-old mice. Expression of total Pkd1 transcripts was performed using Pkd1-allele-specific primers as described under “Experimental Procedures.” The normal Pkd1+ versus cyclophilin A is normalized to the mean ratio of five control mice, which has been set to 1. The percentage of conditional deleted and mutant transcripts was calculated from the relative levels of the normal Pkd1+ transcripts in different Pkd1 exons. D, histology of adult kidney. Hematoxylin-eosin-stained sections from 16-week-old mice failed to identify any cystic tubules in either cortical or medullary regions of kidney from Oc-Cre;Pkd1flox/+ or Pkd1Oc-cko mice, consistent with the absence of Oc-Cre expression in the kidney. In contrast, ablation of Pkd1 in the kidney caused massive cyst formation, which served as a positive control. Cy, cyst. Scale bars, 100 μm.
Bone Densitometric, Histomorphometric, and Microcomputed Tomography (μCT) Analysis
Bone mineral density (BMD) of femurs was assessed at 16 weeks of age using a LUNARPIXIMUS bone densitometer (Lunar Corp., Madison, WI). Calcein (Sigma) double labeling of bone and histomorphometric analyses of periosteal mineral apposition rate (MAR), mineralized surface per bone surface (MS/BS), and bone formation rate (BFR) in tibias were performed using the osteomeasure analysis system (Osteometrics). Goldner and Von Kossa staining were performed according to standard protocols (21, 22). The distal femoral metaphyses were also scanned using a Scanco μCT 40 (Scanco Medical AG, Brüttisellen, Switzerland). A three-dimensional image analysis was done to determine bone volume/total volume and cortical thickness as described previously (18, 21).
Detection of Bone Marrow Adipocytes in Long Bones by Oil Red O Lipid Staining
Whole intact femurs with encapsulated marrow were dissected from 18-week-old mice, fixed for 48 h in phosphate-buffered paraformaldehyde, decalcified in 14% EDTA, and then embedded in tissue freezing medium. Cryosectioning was performed on a Leica CM1900 cryostat (Leica, Nussloch, Germany) equipped with a CryoJane frozen sectioning kit (Instrumedics, Hackensack, NJ). 10-μm thick sections were then stained with Oil Red O for bone marrow adipocytes as described before (23). Briefly, the sections were rinsed in 60% isopropyl alcohol; stained for 20 min in 0.5% Oil Red O, isopropyl alcohol solution; differentiated in 60% isopropyl alcohol; rinsed in tap water; and mounted in glycerin jelly. Sections were examined with a Leica DM LB microscope equipped with an Optronics digital camera.
Detection of Bone Marrow Fat in Long Bones by μCT
Whole intact tibiae with encapsulated marrow were dissected from 18-week-old mice, fixed for 48 h in phosphate-buffered paraformaldehyde, decalcified in 14% EDTA, and stained for 2 h in 2% aqueous osmium tetroxide (OsO4). Bones were rinsed in water for 48 h and then scanned at 6 μm resolution using a Scanco μCT 40, 45 keV, and 177 μA. Quantification of fat volume, density, and distribution throughout the marrow was registered to low contrast decalcified bone.
Real-time RT-PCR
For quantitative real-time RT-PCR, 2.0 μg of total RNA isolated from either the long bone of 16-week-old mice or 10-day cultured primary osteoblasts in differentiation medium was reverse transcribed as described previously (24). PCRs contained 100 ng of template (cDNA or RNA), 300 nm each forward and reverse primers, and 1× iQTM SYBR® Green Supermix (Bio-Rad) in 50 μl. The threshold cycle of tested gene product from the indicated genotype was normalized to the threshold cycle for cyclophilin A. Expression of total Pkd1 transcripts was performed using the following Pkd1 allele-specific primers: in exon 26, forward primer of normal Pkd1+ transcript (5′-CTG GTG ACC TAT GTG GTC AT-3′), forward primer of mutant Pkd1m1Bei transcript (5′-CTG GTG ACC TAT GTG GTC AG-3′), and common reverse primer (5′-AGC CGG TCT TAA CAA GTA TTT C-3′); in exons 2–4, forward primer of normal Pkd1+ transcript (5′-ATA GGG CTC CTG GTG AAC CT-3′) and reverse primer (5′-CCA CAG TTG CAC TCA AAT GG-3′). The normal Pkd1+ versus cyclophilin A is normalized to the mean ratio of five control mice, which has been set to 1. The percentage of conditional deleted and mutant transcripts was calculated from the relative levels of the normal Pkd1+ transcripts in different Pkd1 exons (25).
Serum Biochemistry
Serum osteocalcin levels were measured using a mouse osteocalcin enzyme immunoassay kit (Biomedical Technologies Inc., Stoughton, MA). Serum urea nitrogen was determined using a serum urea nitrogen diagnostic kit from Pointe Scientific, Inc. Serum calcium was measured by the colorimetric cresolphthalein binding method, and phosphorus was measured by the phosphomolybdate-ascorbic acid method (Stanbio Laboratory, TX). Serum TRAP (tartrate-resistant acid phosphatase) was assayed with the enzyme-linked immunosorbent assay-based SBA Sciences mouseTRAPTM assay (Immunodiagnostic Systems, Fountain Hills, AZ).
Primary Osteoblast Culture for Proliferation, Differentiation, and Western Blot Analysis
Primary osteoblasts from newborn mouse calvarias were cultured in α-minimum essential medium containing 10% fetal bovine serum and 1% penicillin/streptomycin as described previously (24). Cell proliferation was detected by bromodeoxyuridine incorporation assays as the manufacturer describes (QIA58, Calbiochem). To induce differentiation, primary osteoblasts were plated at a density of 1 × 105 cells/well in a 6-well plate and grown for period of up to 21 days in α-minimum essential medium containing 10% fetal bovine serum supplemented with 5 mm β-glycerophosphate and 25 μg/ml ascorbic acid. Alkaline phosphatase activity and alizarin red-S histochemical staining for mineralization were performed as described previously (24). Total DNA content was measured with a PicoGreen® double-stranded DNA quantitation reagent and kit (Molecular Probes, Inc., Eugene, OR).
To examine the amounts of cytoplasmic Akt, GSK, and β-catenin, the cells were prepared using 1× passive lysis buffer for 30 min at 4 °C (Promega, Madison, WI) and centrifuged at 100,000 × g for 45 min at 4 °C. Protein concentrations of the supernatant were determined with a Bio-Rad protein assay kit (Bio-Rad). Equal quantities of protein were subjected to NuPAGETM 4–12% BisTris gel (Invitrogen) and were analyzed with standard Western blot protocols (horseradish peroxidase-conjugated secondary antibodies from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and ECL from Amersham Biosciences). Antibodies against phospho-Akt (Ser-473), Akt, phospho-GSK (Ser-9), and GSK were from Cell Signaling Technology (Beverly, MA). Anti-β-catenin (sc-7199) and anti-β-actin (sc-47778) antibodies were from Santa Cruz Biotechnology, Inc.
Transient Transfection
Both MC3T3-E1 and primary osteoblasts were cultured in α-minimum essential medium containing 10% fetal bovine serum and 1% penicillin/streptomycin. To examine if PC1 regulates Runx2-P1 promoter activity by coupling with PI3K-Akt signaling, 1 × 106 MC3T3-E1 cells were transfected with either control expression vector (sIg℘) or gain-of-function PC1 C-tail construct (PC1-AT) along with the Runx2-P1 luciferase reporter (p0.42Runx2-P1-Luc) construct by electroporation using Cell Line Nucleofector Kit R according to the manufacturer's protocol (Amaxa Inc., Gaithersburg, MD). A total of 10.2 μg of plasmid DNA was used for each electroporation, with 3.6 μg of PC1 C-tail construct, 2.4 μg of p0.42Runx2-P1-Luc reporter, the indicated amounts of a dominant negative Akt (dn-Akt) construct in combination with empty vector (3.6 μg), and 0.6 μg of Renilla luciferase-null (RL-null) as internal control plasmid. Promoter activity was assessed by measuring luciferase activity 48 h after transfection in the presence or absence of a PI3K inhibitor (0.1–10 μm; LY294002) and a dn-Akt construct (1.2–3.6 μg) as described previously (17, 18).
To explore potential abnormalities of the WNT pathway in excised floxed Pkd1 null mice, control (Pkd1flox/+) and excised floxed Pkd1 null (Pkd1Oc-cko) osteoblasts were transiently cotransfected with either pTOPFLASH or pFOPFLASH along with Renilla luciferase-null (RL-null; Promega, Madison, WI) as an internal control as described above. Promoter activity will be assessed by measuring luciferase activity 48 h after transfection in the presence or absence of 100 ng/ml recombinant Wnt3a treatment for the last 8 h.
Statistics
We evaluated differences between groups by one-way analysis of variance. All values are expressed as means ± S.D. All computations were performed using GraphPad Prism5 (GraphPad Software, Inc., La Jolla, CA).
RESULTS
Oc-Cre-mediated Bone-specific Deletion of Pkd1
The four genotypes from the breeding strategy (Oc-Cre;Pkd1flox/m1Bei or Pkd1Oc-cKO, Oc-Cre;Pkd1flox/+, Pkd1flox/m1Bei, and Pkd1flox/+) were born at the expected Mendelian frequency, and all exhibited survival indistinguishable from that of wild-type mice. The normal survival of conditional Pkd1Oc-cKO null mice (Oc-Cre;Pkd1flox/m1Bei) contrasts with perinatal lethality of homozygous Pkd1m1Bei/m1Bei mice (17). Oc-Cre expression is limited to cells of the osteoblast lineage (late osteoblasts > osteocytes) with onset of expression just before birth and persisting throughout the mature osteoblast lineage (20). To confirm that the Pkd1 floxed allele was selectively deleted in bone, we performed PCR analysis using a combination of primers that specifically detect floxed Pkd1 alleles (Pkd1flox) and the excised floxed Pkd1 alleles (Pkd1Δflox) in Oc-Cre;Pkd1flox/+ or Oc-Cre;Pkd1flox/m1Bei mice (Fig. 1A). We demonstrated that Oc-Cre-mediated floxed recombination occurred exclusively in tissues that contain osteoblastic cells, whereas non-skeletal tissues retained the intact floxed Pkd1 alleles (Pkd1flox) (Fig. 1B). The Pkd1m1Bei mutation, which functions as a null allele, was used in combination with the floxed Pkd1 allele (Pkd1flox) to increase the net efficiency of Pkd1 inactivation by Cre-recombinase to reduce functional Pkd1 expression. Therefore, we examined the percentage of Pkd1 conditional deleted and the Pkd1m1Bei mutant alleles in bone. The level of conditional deleted Pkd1Δflox alleles and the presence of the Pkd1m1Bei mutation from the femurs of these four genotypes of mice were assessed by real time PCR (Fig. 1C). Both Pkd1flox/m1Bei and Oc-Cre;Pkd1flox/m1Bei mice expressed 50% of the Pkd1m1Bei mutant allele, whereas Oc-Cre;Pkd1flox/+ and Oc-Cre;Pkd1flox/m1Bei mice exhibited ∼25% excision of the floxed exons 2–4 from Pkd1, indicating that Oc-Cre-mediated bone-specific deletion of the floxed Pkd1 allele is incomplete (Fig. 1C). The combined effect of Pkd1m1Bei and (Pkd1Δflox) in Oc-Cre;Pkd1flox/m1Bei resulted in a net reduction of Pkd1 expression by ∼75% in bone (Fig. 1C). Real-time RT-PCR to assess the level of expression of the residual functional Pkd1 transcript confirmed the progressive reduction of functional Pkd1 message in conditional mutant mice (i.e. Pkd1flox/+ (100%), Oc-Cre;Pkd1flox/+ (76%), Pkd1flox/m1Bei (50%), and Oc-Cre;Pkd1flox/m1Bei (25%) mice) (data not shown). In addition, Oc-Cre;Pkd1flox/m1Bei mice heterozygous for the conditional deleted Pkd1Δflox allele and mutant Pkd1m1Bei allele demonstrated no cyst formation in the kidney, consistent with the bone-specific inactivation of Pkd1 (Fig. 1D). In contrast, positive control mice lacking Pkd1 in the kidney have massive cyst formation in the kidney (Fig. 1D).
Additive Effects of Global Mutant (Pkd1m1Bei) and Conditional Deleted (Pkd1Δflox) Pkd1 Alleles Suggest a Direct Role for PKD1 in Bone
At 16 weeks of age, the gross appearance and body weight of single global and conditional heterozygous (Pkd1flox/m1Bei or Oc-Cre;Pkd1flox/+) and control (Pkd1flox/+) mice were not significantly different. The global Pkd1 heterozygous mice (Pkd1flox/m1Bei) and heterozygous conditional deleted Pkd1 mice (Oc-Cre;Pkd1flox/+), however, were osteopenic, as evidenced by respective 9 and 7% reduction BMD in both male and female adult mice (Fig. 2A). The phenotype of the Pkd1Oc-cKO mice was more severe. The body weight of both male and female Pkd1Oc-cKO mice was reduced by ∼16 and 12% (data not shown) compared with the control mice (Pkd1flox/+). In addition, Pkd1Oc-cKO mice had greater loss in BMD, with respective reductions in BMD of 14 and 13% reduction in male and female adult mice (Fig. 2A). The abnormalities in BMD in the various groups, although present at 6 weeks of age, segregated by gene dose by 16 weeks of age (Fig. 2B).
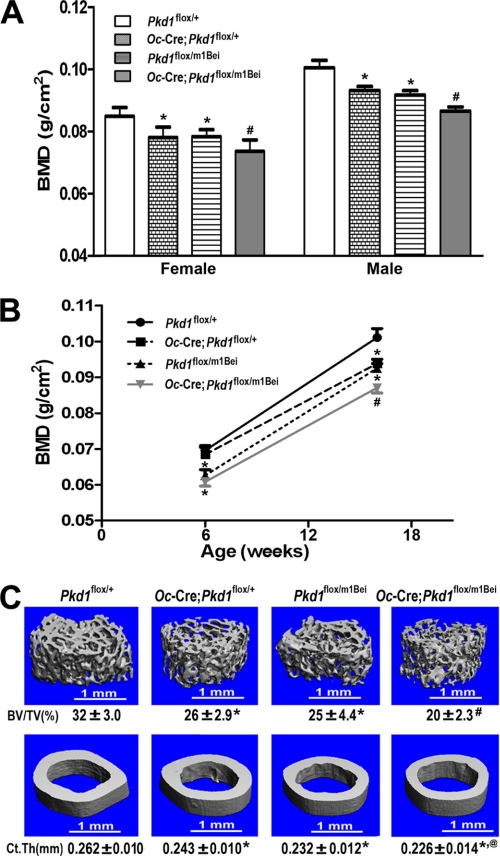
FIGURE 2.
Cre-mediated somatic loss of Pkd1 results in loss of bone mass. A, effects of Pkd1Δflox allele on BMD at 16 weeks of age. Similar to Beier Pkd1 heterozygous mice (Pkd1m1Bei/+), there was ∼7–9% reduction in both male and female of BMD in single excised floxed Pkd1 heterozygous mice (Oc-Cre;Pkd1flox/+) compared with age-matched control mice (Pkd1flox/+), and an even greater reduction (13–14%) in double heterozygous Oc-Cre;Pkd1flox/m1Bei (Pkd1Oc-cko) mice, indicating an additive effect of global mutant and conditional deleted Pkd1 alleles on loss of bone mass. B, age-dependent effects of Pkd1Δflox allele on BMD. Double heterozygous Pkd1Oc-cko mice displayed a significant decrease in femur BMD compared with Pkd1m1Bei/+ mice until 16 weeks of age but not at 6 weeks of age, indicating an age-dependent effect of Pkd1Δflox allele on bone mass. C, effects of Pkd1Δflox allele on bone structure of femurs and midshaft diaphyses. μCT analysis of the distal femoral metaphyses and midshaft diaphyses revealed that double heterozygous Pkd1Oc-cko mice had greater loss in both trabecular and cortical bone than single Oc-Cre;Pkd1flox/+ and Pkd1m1Bei/+ heterozygous mice, consistent with additive effects of global mutant and conditional deleted Pkd1 alleles on bone structure and a direct role of Pkd1 in bone. Data represent the mean ± S.D. from 8–10 individual mice. *, significant difference from control (Pkd1flox/+); @, significant difference from single heterozygous Oc-Cre;Pkd1flox/+; #, significant difference from single heterozygous Oc-Cre;Pkd1flox/+ and Pkd1flox/m1Bei mice at p < 0.05, respectively.
μCT analysis revealed that the reduction in bone mass in heterozygous Pkd1-deficient mice (either Pkd1flox/m1Bei or Oc-Cre;Pkd1flox/+) was caused by a reduction in trabecular bone volume (24.3 and 25.5%, respectively) and cortical bone thickness (9.7 and 10.8%, respectively) (Fig. 2C). Pkd1cKO/m1Bei had greater loss in both trabecular (44.5%) and cortical bone (21.0%) than did single heterozygous mice (Fig. 2C).
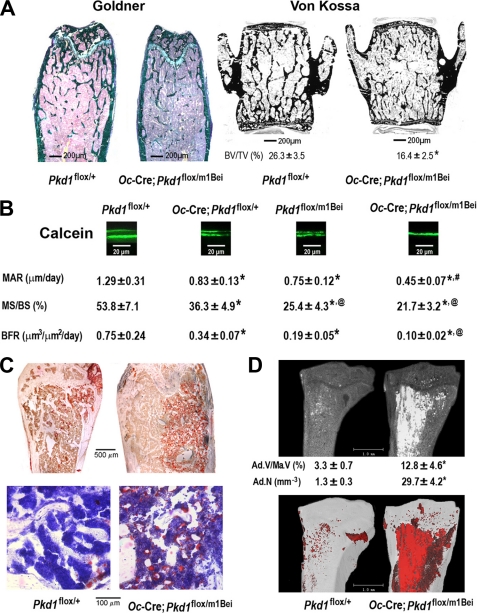
Consistent with a low bone mass phenotype by BMD and μCT analysis, both Goldner staining in distal femur and Von Kossa staining in vertebrae confirmed marked reductions in bone volume and cortical thickness in Pkd1Oc-cKO null mice (Fig. 3A). In addition, we found that bone loss was associated with a significant Pkd1 gene dose-dependent decrease in periosteal MAR, MS/BS, and BFR. In this regard, MAR, MS/BS, and BFR were reduced by ∼36%, ∼33%, and ∼57% in heterozygous Pkd1flox/m1Bei and Oc-Cre;Pkd1flox/+ mice and ∼65, ∼60, and ∼87% in Pkd1Oc-cKO null mice compared with age-matched controls, respectively (Fig. 3B).
FIGURE 3.
Histological analysis of Pkd1Oc-cKO mice in bone. A, Goldner and von Kossa staining of non-decalcified bone. Representative images of distal femur and lumbar vertebrate sections displayed markedly reductions in trabecular bone volume from 16-week-old Pkd1Oc-cko mice compared with age-matched control mice. Scale bars, 200 μm. B, bone histomorphometric analyses. There was a significant reduction in periosteal MAR, MS/BS, and BFR in single Oc-Cre;Pkd1flox/+ and Pkd1m1Bei/+ heterozygous mice compared with age-matched control Pkd1flox/+ mice and an even greater decrement in double heterozygous Pkd1Oc-cko mice, indicating an additive effect of both global and conditional Pkd1 deficiency to impair osteoblast-mediated bone formation. Representative images of the distal tibia-fibula junction sections from 16-week-old mice for each genotype showed progressive reductions in the distance between the two calcein labels (scale bars, 20 μm). C, Oil Red O staining of decalcified femur sections. Representative images of femoral bone marrow showed that the numbers of adipocytes and fat droplets were greater in 18-week-old Pkd1Oc-cko mice compared with age-matched control mice. Scale bars, 100 and 500 μm. D, OsO4 staining of decalcified tibias by μCT analyses. Qualitatively, the images of osmium staining (white or red areas) were much higher in the proximal tibia from 18-week-old Pkd1Oc-cko mice compared with age-matched control mice. Quantifications of fat cell number and volume were also performed as described under “Experimental Procedures.” Ad.V/Ma.V (%), adipocyte volume/marrow volume; Ad.N (mm−3), adipocyte number (mm−3). Scale bars, 1.0 mm. Data are mean ± S.D. from 3–5 individual mice. *, significant difference from control (Pkd1flox/+); @, significant difference from single heterozygous Oc-Cre;Pkd1flox/+; #, significant difference from single heterozygous Oc-Cre;Pkd1flox/+ and Pkd1flox/m1Bei mice at p < 0.05, respectively.
To investigate the effects of Pkd1 deficiency on gene expression profiles in bone, we examined by real-time RT-PCR the expression levels of a panel of osteoblast lineage-, osteoclast-, and chondrocyte-related mRNAs from the femurs of 16-week-old control, heterozygous Pkd1-deficient (Oc-Cre;Pkd1flox/+ and Pkd1m1Bei/+), and Oc-Cre;Pkd1flox/m1Bei mice (Table 1). Bone derived from heterozygous Oc-Cre;Pkd1flox/+ and Pkd1m1Bei/+ mice had measurable reductions in the osteoblast lineage gene transcripts, including Runx2-II, total Runx2, osteocalcin, osteopontin, Bsp (bone sialoprotein), Opg (osteoprotegerin), RankL (Rank ligand), Dmp1 (dentin matrix protein 1), and Phex (phosphate-regulating gene with homologies to endopeptidases on the X chromosome) mRNA levels compared with control mice. Significantly greater reductions of Runx2-II, total Runx2, osteocalcin, osteopontin, Bsp, RankL, and Dmp1 were observed in Pkd1Oc-cKO null mice. In this regard, the Opg/RankL expression ratio was increased in a gene dose-dependent manner (Table 1). Consistent with a ratio of Opg/RankL that favors the reduced osteoclastogenesis, bone expression of Trap and Mmp9 (matrix metalloproteinase 9), markers of bone resorption, were also reduced in heterozygous Pkd1-deficient mice and to a greater extent in Pkd1Oc-cKO null mice (Table 1). Transcripts of chondrocyte-related genes did not differ between heterozygous Pkd1-deficient and Pkd1Oc-cKO null mice (Table 1).
TABLE 1.
Gene expression profiles in 16-week-old mice
Data are mean ± S.D. from 5–6 tibias of 6-week-old individual mice and expressed as the -fold changes relative to the housekeeping gene cyclophilin A subsequently normalized to control (Pkd1flox/+) mice.
| Gene | Accession no. | Oc-Cre;Pkd1flox/+ | Pkd1flox/m1Bei | Oc-Cre;Pkd1flox/m1Bei | p value |
|---|---|---|---|---|---|
| Osteoblast lineage | |||||
| Runx2-II | NM_009820 | 0.73 ± 0.04a | 0.71 ± 0.08a | 0.45 ± 0.07a,b | 0.0007 |
| Runx2-I | D14636 | 1.14 ± 0.32 | 1.16 ± 0.14 | 1.15 ± 0.18 | 0.5961 |
| Runx2 | NM_009820 | 0.73 ± 0.09a | 0.71 ± 0.16a | 0.51 ± 0.07a,b | 0.0009 |
| Osteocalcin | NM_007541 | 0.82 ± 0.12a | 0.80 ± 0.11a | 0.46 ± 0.08a,b | <0.0001 |
| Osteopontin | AF515708 | 0.73 ± 0.11a | 0.69 ± 0.11a | 0.44 ± 0.03a,b | 0.0027 |
| Bsp | NM_008318 | 0.73 ± 0.07a | 0.71 ± 0.08a | 0.52 ± 0.06a,b | <0.0001 |
| Opg | MMU94331 | 0.71 ± 0.15a | 0.70 ± 0.11a | 0.68 ± 0.20a | 0.0295 |
| Rank ligand | NM_011613 | 0.70 ± 0.12a | 0.57 ± 0.07a | 0.39 ± 0.07a,b | <0.0001 |
| Mmp13 | NM_008607 | 0.97 ± 0.16 | 0.65 ± 0.05a | 0.43 ± 0.11a,b | <0.0001 |
| Dmp1 | MMU242625 | 0.71 ± 0.06a | 0.64 ± 0.12a | 0.40 ± 0.05a,b | <0.0001 |
| Phex | NM_011077 | 0.72 ± 0.12a | 0.67 ± 0.12a | 0.58 ± 0.14a | 0.0002 |
| Osteoclast | |||||
| Trap | NM_007388 | 0.68 ± 0.11a | 0.63 ± 0.13a | 0.36 ± 0.09a,b | <0.0001 |
| Mmp9 | NM_013599 | 0.95 ± 0.16 | 0.69 ± 0.08a | 0.63 ± 0.07a | 0.0001 |
| Chondrocyte | |||||
| Collagen II | NM_031163 | 0.99 ± 0.13 | 1.03 ± 0.26 | 1.13 ± 0.23 | 0.5764 |
| VegfA | NM_009505 | 1.07 ± 0.26 | 1.06 ± 0.21 | 0.97 ± 0.31 | 0.8905 |
| Adipocyte | |||||
| PPARγ | NM_009505 | 1.08 ± 0.15 | 1.20 ± 0.19 | 1.40 ± 0.13a | 0.0025 |
| aP2 | NM_024406 | 1.13 ± 0.16 | 1.55 ± 0.23a | 1.95 ± 0.26a,b | <0.0001 |
| Lpl | NM_008509 | 1.38 ± 0.25a | 1.42 ± 0.27a | 2.01 ± 0.29a,b | <0.0001 |
| Others | |||||
| IFNγ | NM_008337 | 2.42 ± 0.79a | 2.70 ± 1.21a | 3.57 ± 1.51a | 0.0039 |
| TNFα | NM_013693 | 1.18 ± 0.32 | 1.23 ± 0.69 | 1.13 ± 0.39 | 0.8371 |
| TRAIL | NM_009425 | 1.36 ± 0.51 | 1.33 ± 0.48 | 1.29 ± 0.45 | 0.4500 |
| Alox15 | NM_009660 | 1.93 ± 0.38a | 2.22 ± 0.89a | 2.40 ± 0.95a | 0.0106 |
a Significant difference from control (Pkd1flox/+) at p < 0.05.
b Significant difference from single heterozygous Oc-Cre;Pkd1flox/+ and Pkd1flox/m1Bei mice at p < 0.05.
Changes in gene expression in bone correlated with alterations in serum biomarkers. In this regard, further evidence for osteoblast dysfunction includes a reduction in osteocalcin in serum from 16-week-old heterozygous Oc-Cre;Pkd1flox/+ and Pkd1m1Bei/+ mice (Table 2). Serum levels of TRAP, a marker of bone resorption, were also reduced in heterozygous Pkd1-deficient mice compared with control littermates (Table 2). As with other parameters, Pkd1Oc-cKO null mice had greater reductions in bone formation and resorption markers, indicating additive effects of inactivation of both Pkd1 alleles in bone homeostasis. Collectively, these findings suggest that Pkd1-mediated bone loss results from low bone formation rates rather than increased bone resorption (Fig. 2, A–C, and Table 2).
TABLE 2.
Biochemistry analysis of serum in 16-week-old mice
Data are mean ± S.D. from nine individual mice. Osteocalcin is produced by osteoblasts, and TRAP is produced by osteoclasts.
| Genotype | Pkd1flox/+ | Oc-Cre;Pkd1flox/+ | Pkd1flox/m1Bei | Oc-Cre;Pkd1flox/m1Bei |
|---|---|---|---|---|
| Serum urea nitrogen (mg/dl) | 19 ± 4.9 | 20 ± 4.8 | 22 ± 4.3 | 22 ± 5.2 |
| Calcium (mg/dl) | 8.9 ± 0.27 | 8.7 ± 0.38 | 8.6 ± 0.34 | 8.5 ± 0.37 |
| Phosphorus (mg/dl) | 8.3 ± 0.95 | 8.5 ± 1.17 | 8.6 ± 1.50 | 8.8 ± 0.76 |
| Osteocalcin (ng/ml) | 87 ± 21.3 | 74 ± 13.8a | 64 ± 18.6a | 56 ± 12.6a,b |
| TRAP (units/liter) | 12.6 ± 4.54 | 8.0 ± 2.94a | 6.5 ± 1.61a | 5.1 ± 0.94a,b |
a Significant difference from control (Pkd1flox/+) at p < 0.05.
b Significant difference from single heterozygous Oc-Cre;Pkd1flox/+ and Pkd1flox/m1Bei mice at p < 0.05.
PPARγ (peroxisome proliferator-activated receptor γ), an adipocyte transcription factor, and adipocyte markers, including Lpl (lipoprotein lipase) and aP2 (adipocyte fatty acid-binding protein 2) were increased femurs of Pkd1-deficient mice in a Pkd1 gene dosage-related manner (Table 1). Consistent with increased adipogenic markers, bone marrow exhibited an increased percentage of fat cells in Pkd1Oc-cKO mice, as evidenced by a higher number of adipocytes and volume of fat droplets in decalcified femurs and tibias stained with Oil Red O and OsO4 (Fig. 3C). In addition, the inflammatory cytokine IFNγ (interferon-γ) level, but not TNFα (tumor necrosis factor-α) and TRAIL (TNF-related apoptosis-inducing ligand) expression levels, was significantly elevated in Pkd1-deficient mice (Table 1). Moreover, Alox15 (arachidonate 15-lipoxygenase) was also markedly increased in conditional Pkd1-deficient mice.
Effect of Conditional Deletion of Pkd1 on Osteoblastic Function ex Vivo
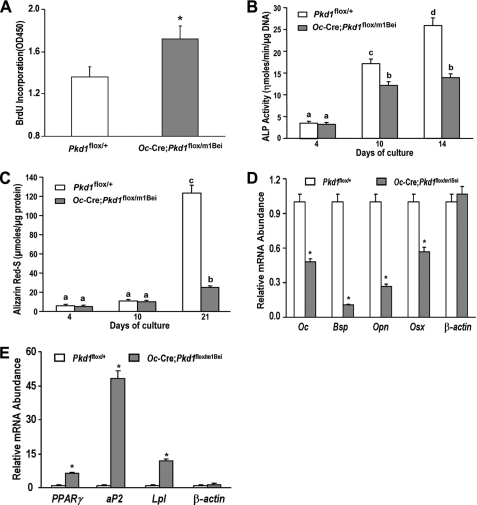
To determine the impact of conditional deleted Pkd1 on osteoblast function ex vivo, we examined cell proliferation and osteoblastic differentiation and gene expression profiles in primary osteoblast cultures derived from control and Pkd1Oc-cKO null mice. Consistent with defects in bone formation, we found that Pkd1Oc-cKO null osteoblasts had a higher bromodeoxyuridine incorporation than control osteoblasts, indicating a greater proliferation rate in Pkd1Oc-cKO null osteoblasts (Fig. 4A). In addition, conditional null osteoblasts displayed impaired osteoblastic differentiation and maturation, as evidenced by lower alkaline phosphatase activity, diminished calcium deposition in extracellular matrix, and reduced osteoblastic differentiation markers compared with controls (Fig. 4, B–D). In agreement with increased adipogenic activity in vivo, the cultured primary calvarial cells under osteogenic conditions exhibited a marked increase of adipocyte markers, including PPARγ, aP2, and Lpl (Fig. 4E), suggesting impairment of osteogenesis and enhancement of adipogenesis in Pkd1Oc-cKO null osteoblast cultures.
FIGURE 4.
Pkd1Oc-cKO osteoblasts have a developmental defect ex vivo. A, bromodeoxyuridine (BrdU) incorporation. Primary cultured Pkd1Oc-cKO osteoblasts exhibited a higher bromodeoxyuridine incorporation than control Pkd1flox/+ osteoblasts for 6 h, indicating increased proliferation in the Pkd1Oc-cKO osteoblasts. B, alkaline phosphatase (ALP) activity. Primary cultured Pkd1Oc-cKO osteoblasts displayed time-dependent increments in alkaline phosphatase activities for 14 days of culture, but the alkaline phosphatase activity was significantly lower at different time points compared with control Pkd1flox/+ osteoblasts. C, quantification of mineralization. Alizarin Red-S was extracted with 10% cetylpyridinium chloride and quantified as described under “Experimental Procedures.” Primary cultured Pkd1Oc-cKO osteoblasts had time-dependent increments in Alizarin Red-S accumulation for 21 days of culture, but the accumulation was significantly lower at different time points compared with control Pkd1flox/+ osteoblasts. D and E, gene expression profiles by real-time RT-PCR. 10-day cultured Pkd1Oc-cKO osteoblasts in osteogenic differentiation medium showed a significant attenuation in osteogenesis compared with control osteoblasts, as evidenced by a significant reduction in osteoblastic markers, including osteocalcin (Oc), bone sialoprotein (Bsp), osteopontin (Opn), and osterix (Osx). However, a marked increase of adipocyte markers, such as PPARγ, aP2, and Lpl, was observed from the Pkd1Oc-cKO osteoblasts under the same differentiation medium when compared with control osteoblasts. Data are mean ± S.D. from triple three independent experiments. *, significant difference from control (Pkd1flox/+) mice at p < 0.05. Values sharing the same superscript in B and C are not significantly different at p < 0.05.
Effect of Conditional Deletion of Pkd1 on PI3K-Akt-GSK-β-Catenin Signaling Pathway in Osteoblasts
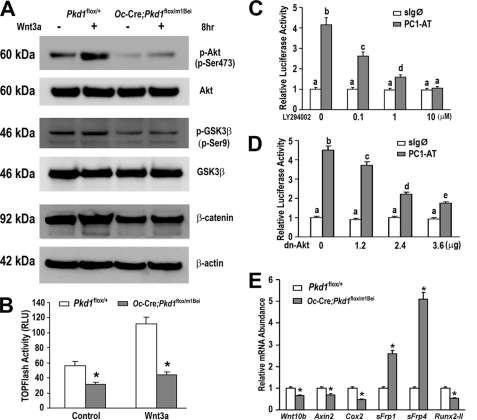
PC1 encoded by Pkd1 is coupled to multiple signal transduction pathways, including activation of canonical WNT/β-catenin and Akt-dependent pathways. There is evidence that Akt-dependent serine 9 phosphorylation of GSK3β prevents phosphorylation and degradation of β-catenin (26–29). Because the PI3K/Akt pathway and WNT signaling play a role in osteoblastic development (30, 31), we determined the level of Akt phosphorylation in Pkd1Oc-cKO derived osteoblasts. Basal phospho-Akt relative to total Akt expression and phosphorylation of serine 9 of GSK3β were reduced in Pkd1Oc-cKO null osteoblasts compared with controls. To determine if potential cross-talk between the PC1/PI3K/Akt pathway and the WNT/GSK3β/β-catenin pathways have functional consequences (28), we next examined the response of osteoblasts derived from Pkd1Oc-cKO mice to Wnt3a. Pkd1Oc-cKO-derived osteoblasts had a reduced response to Wnt3a-mediated phosphorylation of Akt when compared with control cells (Fig. 5A). In contrast, the addition of Wnt3a resulted in an increase in phosphorylation of GSK3β in control cells, leading to the inactivation of GSK3β (Fig. 5A). To determine if inhibition of GSK3β activated β-catenin (32), we assessed the accumulation of free β-catenin in the cytoplasm (33). Consistent with reduction of GSK3β phosphorylation, we found that the basal level of cytosolic β-catenin was lower in Pkd1Oc-cKO null osteoblasts and exhibited no increase following Wnt3a treatment, whereas control cells increased cytoplasmic β-catenin levels following Wnt3a stimulation (Fig. 5A).
FIGURE 5.
Signaling pathways in Pkd1Oc-cKO osteoblasts. A, Western blot analysis. Comparison of Akt, GSK3β, and β-catenin expressions in control and Pkd1Oc-cKO osteoblasts treated with or without Wnt3a (100 ng/ml) for the indicated times. Phosphorylated Akt at Ser-473 (panel 1) coincides with phosphorylation of GSK3β at Ser-9 (panel 3), reflecting its inactivation by Akt and followed by accumulation of cytoplasmic β-catenin, as detected by Western blot. Total Akt (panel 4), GSK3β (panel 4), and β-actin (panel 6) were used as loading controls for phospho-Akt, phospho-GSK3β, and β-catenin in the cytoplasm, respectively. Pkd1Oc-cKO osteoblasts exhibited suppressed activity of the PI3K-Akt-GSK3β-β-catenin signaling pathway under basal and Wnt3a-stimulated conditions. B, TCF/LEF-dependent transcriptional activation as assessed by pTOPFLASH activity. Pkd1Oc-cKO osteoblasts display lower levels of basal TCF/LEF-dependent reporter activity (TOPFLASH) when compared with control Pkd1flox/+ osteoblasts. Wnt3a (100 ng/ml) induced reporter activity by more than 2.0-fold above control in Pkd1flox/+ osteoblasts, whereas Wnt3a induced reporter activity by only 1.4-fold above control in Pkd1Oc-cKO osteoblasts. C and D, PC1-mediated regulation of Runx2-II-P1 prompter activity via the PI3K/Akt pathway. Wild-type MC3T3-E1 cells were transiently transfected either with control expression vector (sIg℘) or gain-of-function PC1 C-tail construct (PC1-AT) along with the Runx2-II-P1 luciferase reporter (p0.42Runx2-P1-Luc) construct in the presence or absence of PI3K inhibitor LY294002 or dn-Akt construct. PC1-mediated activation of Runx2-II-P1 promoter activity was dose-dependently diminished by either PI3K inhibitor LY294002 or dn-Akt construct. E, gene expression profiles by real-time RT-PCR. 10-day cultured Pkd1Oc-cKO osteoblasts in differentiation medium showed a significant attenuation in WNT/β-catenin signaling compared with control osteoblasts, as evidenced by a significant down-regulation of WNT/β-catenin-targeting genes, including Wnt10b, Axin2, Cox2, and Runx2-II, and up-regulation of its antagonist genes, such as sFrp1 and sFrp4. Data are mean ± S.D. from triple independent experiments. *, significant difference from control (Pkd1flox/+) mice at p < 0.05. Values sharing the same superscript in C and D are not significantly different at p < 0.05.
To examine the effect of Pkd1 inactivation on Wnt/β-catenin transcriptional activity, we examined TOPFlash activity in Pkd1Oc-cKO null osteoblasts. We observed a significant reduction of basal TOPFLASH activity in primary osteoblasts derived from Pkd1Oc-cko mice compared with the controls. In addition, Wnt3a-induced TOPFLASH activity was more than 2-fold above basal level in the control osteoblasts, whereas Wnt3a-induced TOPFLASH activity was only 1.4-fold above basal level in the Pkd1Oc-cKO null mice (Fig. 5B), indicating that loss of PC1 significantly attenuates responsiveness of the Wnt/β-catenin pathway in osteoblasts.
To examine the role of PC1-dependent Akt activation on transcriptional control of osteoblast development, we examined Runx2-II promoter activity. In previous studies, we have shown that PC1 is coupled to Runx2-II expression, a master regulator in osteoblast function, and that transfection of the C-terminal region of PC1 (PC1-AT) was sufficient to activate the Runx2-II P1 promoter/reporter construct (17). As shown in Fig. 5, C and D, we found that either the PI3K inhibitor LY294002 or cotransfection with a dn-Akt construct resulted in a dose-dependent inhibition of PC1-AT-mediated increase in Runx2-II P1 promoter activity.
Examination of Wnt-related gene expression in Pkd1Oc-cKO null osteoblasts further supports impairment of WNT/β-catenin signaling. In this regard, we found evidence for down-regulation of Wnt10b, Axin2, Cox2, and Runx2-II and up-regulation of the negative regulators sFrp1 and sFrp4 (Fig. 5E).
DISCUSSION
PC1 is expressed in cells within the osteoblast lineage (18), and skeletal abnormalities have been reported in Pkd1 mutant mouse models (14, 15, 17, 18), but from these generalized loss-of-function observations it was not clear if the observed skeletal abnormalities were an indirect consequence of loss of PC1 in multiple tissues or a direct effect of loss of PC1 in osteoblasts. In the present studies, we have addressed this question by using Oc-Cre to conditionally inactivate Pkd1 in mature osteoblasts postnatally. First, we have shown that the heterozygous conditional reduction of Pkd1 in osteoblasts in Oc-Cre;Pkd1flox/+ results in an osteopenic bone phenotype indistinguishable from the global Pkd1flox/m1Bei mice. The fact that Oc-Cre;Pkd1flox/+ mice had a ∼25% reduction in Pkd1 expression, whereas Pkd1flox/m1Bei mice had a 50% reduction but had identical effects on bone, suggests that loss of Pkd1 function in osteoblasts is responsible for the observed reduction in bone mass in both models. Moreover, the additive effects on the severity of osteopenia in combined conditional deletion of Pkd1 in osteoblasts superimposed on the inactivated Pkd1m1Bei allele in Oc-Cre;Pkd1flox/m1Bei (or Pkd1Oc-cKO) mice, which resulted in ∼2-fold greater (∼14%) reduction in BMD associated with a 75% overall reduction in Pkd1 expression in bone, compared with a 50% reduction in other tissues, indicates a dose-dependent function of Pkd1 in mature osteoblasts.
We purposely did not create Oc-Cre;Pkd1flox/floxmice, due to the relative inefficiency of the Oc-Cre, which, based on the 25% reduction in Pkd1 transcripts observed in Oc-Cre;Pkd1flox/+ mice, may have only reduced expression in Oc-Cre;Pkd1flox/flox mice to levels similar to the heterozygous Pkd1flox/m1Bei mice (e.g. 50%). Thus, to achieve greater reduction in Pkd1 expression in osteoblasts (e.g. ∼75%), we created Oc-Cre;Pkd1flox/m1Bei mice. It is of note that the magnitude of bone loss in Pkd1Oc-cKO mice is comparable with the 16% reduction in BMD found in Lrp5 null mice (34), a receptor known to have important anabolic osteoblast-mediated functions in bone through activation of canonical Wnt signaling pathways, and exceeds bone loss observed in oophorectomized mice (which is typically <10%) (35). Because Pkd1Oc-cKO had no demonstrable extraskeletal phenotypes and loss of Pkd1 was greatest in bone, these findings are most consistent with a direct role of PC1 to regulate osteoblast function. Evidence for impaired osteoblastic function in Pkd1Oc-cko null mice is evident from both in vivo and ex vivo analysis. Bone from Pkd1Oc-cko null mice displayed decreased MAR, MS/BS, BFR, and reduced expression of osteoblastic markers, including the Runx2-II isoform, which regulates osteoblast development and function.
Ex vivo assessment of primary osteoblasts isolated confirmed an intrinsic impairment of osteoblast maturation as well as identified increased proliferation, which are typically inversely related (36). A feature of ADPKD renal cells is increased proliferation rate as well as impaired differentiation of epithelial cells (37); our findings suggest that this phenotypic switch may also occur in osteoblasts. Indeed, the Pkd1Oc-cKO null osteoblasts showed a greater proliferation rate as well as impaired osteoblastic differentiation and maturation. Because others have shown that the response of the renal epithelium to acquired loss of Pkd1 is determined by the developmental state of the organ (38), our data showing a defect in osteoblast maturation in remodeling bone raises the possibility that Pkd1 may also play a role in the embryogenesis of bone. Osteoblasts and adipocytes undergo renewal from bone marrow-derived precursors, and the ratio of osteoblasts and adipocytes appears to be reciprocally controlled in response to physiological stimuli and aging (39). We also found an inverse dose relationship between Pkd1 expression and adipogenesis marker expression in the Pkd1Oc-cKO osteoblast cultures compared with the control. In agreement with this in vitro data, our in vivo study also showed strong evidence for lipid droplet accumulation in the bone marrow of Pkd1Oc-cKO mice compared with the control mice via Oil Red O and osmium staining. This phenomenon was also supported by an enhanced expression of inflammatory mediators, such as IFNγ and Alox15 (40, 41), which, produced by adipose tissues, strongly suppress osteoblastogenesis. The findings of increased fat cells in bone marrow of Pkd1Oc-cKO mice and impaired development of osteoblast cultures derived from Pkd1Oc-cKO mice suggest that Pkd1 deficiency may lead to impaired differentiation of mesenchymal stem cells into osteoblasts and enhanced differentiation into adipocytes.
Because Oc-Cre-mediated deletion of Pkd1 in osteoblasts would carry forward to the terminal differentiated osteocytes, we cannot exclude a possible role of the osteocyte in the bone phenotype in Pkd1Oc-cko null mice. In addition to being expressed throughout the osteoblastic lineage during embryogenesis and in postnatal bone, Pkd1 is also expressed in chondrocytes (6). Consequently, the more severe skeletal abnormalities in global Pkd1 null compared with Pkd1Oc-cKO mice could be due to PC1 regulation of osteoblast differentiation or chondrocyte function during development or to systemic effects caused by the presence of polycystic kidneys (6, 7). Additional studies that ablate Pkd1 earlier in the osteoblast lineage during embryogenesis or later in osteocytes, as well in mature and immature chondrocytes, will be needed to establish a direct role of PC1 in skeletal development and to establish the respective contributions of PKD1 function in preosteoblasts, osteoblasts, osteocytes, and chondrocytes.
We previously demonstrated that PC1 selectively regulates Runx2-II P1 promoter activity in osteoblasts through an intracellular calcium pathway linked to nuclear factor I family and AP-1 transcription factors (17). The PC1 regulates a variety of signal transduction pathways in renal epithelial cells, including Akt1 and GSK3β (26–28), which are also important regulators of bone mass (42, 43). The present studies indicate that Pkd1 also regulates PI3K-Akt-GSK3β signaling in osteoblasts and that this pathway is upstream of Runx2. The decline in Akt-GSK3β-β-catenin signaling in osteoblasts from Pkd1Oc-cko null mice and the finding that PC1-PI3K-Akt signaling regulates Runx2-II P1 promoter activity suggest that this may represent another PKD1-dependent pathway regulating osteoblast function. The interactions between WNT- and PKD1-dependent signaling pathways, however, are complex, and further studies will be needed to define the mechanism whereby loss of polycystin 1 results in decreased β-catenin activity.
The broader role of PC1 in regulating bone physiology is not revealed by our studies. There is no known ligand for PC. Polycystin potentially functions as a mechanosensor, a chemosensor, or as a sensor of cell-cell or cell-matrix interactions (44, 45). PC1 colocalizes to the primary cilium (46), which is also present in osteoblasts, osteocytes, and chondrocytes (47, 48). Also, cleavage of PC1 at the G protein-coupled receptor proteolytic site upstream of the first TM segment may be required for bioactivity (49), suggesting that PC1 may be constitutively active. Although the precise activator in bone remains to be defined, PC1 presence or its ability to sense environmental cues during bone remodeling may allow it to function as a “hub” or a common connection point that permits cells in the osteoblast lineage to sense diverse environmental signals during skeletal development and translate these into multiple signaling pathways required for maintenance of bone mass.
In contrast to the role of PKD1 in mouse bone, a PC1-dependent bone phenotype has not been reported in humans with ADPKD, who have superimposed renal osteodystrophy. It may yet be possible to detect small, clinically unapparent reductions of bone density in humans heterozygous for PKD1 mutations prior to developing kidney failure. In addition, there are some very interesting but rare families with early onset of polycystic kidney disease in newborns that are associated with severe skeletal malformations, similar to those observed in mice with homozygous Pkd1 mutations, suggesting that broadly disrupting both alleles in osteoblasts/osteocytes (analogous to the Pkd1 null mouse as opposed to random second hits in ADPKD) might cause clinically apparent bone disease in humans (12, 50).
In conclusion, the finding that selective reduction of Pkd1 in osteoblasts results in osteopenia defines a new signaling paradigm in bone that has the potential to expand our understanding of how bone senses environmental signals maintaining bone mass as well as regulating osteoblast growth and development. The further study of polycystin function in bone could provide a variety of new insights, ranging from new mechanosensing mechanisms potentially involving primary cilium to identifying a molecular target for the development of pharmacological approaches to increased bone mass in osteopenic disorders.
Acknowledgments
We are particularly grateful to Dr. Gregory Germino at Johns Hopkins University for providing the floxed Pkd1 mice.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AR049712 and R21-AR056794.
- ADPKD
- autosomal dominant polycystic kidney disease
- BMD
- bone mineral density
- μCT
- microcomputed tomography
- MAR
- mineral apposition rate
- MS/BS
- mineralized surface per bone surface
- BFR
- bone formation rate
- RT
- reverse transcription
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PI3K
- phosphatidylinositol 3-kinase.
REFERENCES
- 1.Xu H., Shen J., Walker C. L., Kleymenova E. (2001) DNA Seq. 12, 361–366 [DOI] [PubMed] [Google Scholar]
- 2.Chauvet V., Qian F., Boute N., Cai Y., Phakdeekitacharoen B., Onuchic L. F., Attié-Bitach T., Guicharnaud L., Devuyst O., Germino G. G., Gubler M. C. (2002) Am. J. Pathol. 160, 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson P. D. (2004) N. Engl. J. Med. 350, 151–164 [DOI] [PubMed] [Google Scholar]
- 4.Gabow P. A. (1993) N. Engl. J. Med. 329, 332–342 [DOI] [PubMed] [Google Scholar]
- 5.Torra R., Badenas C., San Millán J. L., Pérez-Oller L., Estivill X., Darnell A. (1999) Am. J. Hum. Genet. 65, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu W., Shen X., Pavlova A., Lakkis M., Ward C. J., Pritchard L., Harris P. C., Genest D. R., Perez-Atayde A. R., Zhou J. (2001) Hum. Mol. Genet. 10, 2385–2396 [DOI] [PubMed] [Google Scholar]
- 7.Boulter C., Mulroy S., Webb S., Fleming S., Brindle K., Sandford R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12174–12179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peczkowska M., Januszewicz A., Grzeszczak W., Moczulski D., Janaszek-Sitkowska H., Kabat M., Biederman A., Hendzel P., Prejbisz A., Cendrowska-Demkow I., Zieliñski T., Januszewicz M. (2004) Blood Press 13, 283–286 [DOI] [PubMed] [Google Scholar]
- 9.van Dijk M. A., Chang P. C., Peters D. J., Breuning M. H. (1995) J. Am. Soc. Nephrol. 6, 1670–1673 [DOI] [PubMed] [Google Scholar]
- 10.Hassane S., Claij N., Lantinga-van Leeuwen I. S., Van Munsteren J. C., Van Lent N., Hanemaaijer R., Breuning M. H., Peters D. J., DeRuiter M. C. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 2177–2183 [DOI] [PubMed] [Google Scholar]
- 11.Housset C. (2005) Gastroenterol. Clin. Biol. 29, 861–869 [DOI] [PubMed] [Google Scholar]
- 12.Watnick T. J., Torres V. E., Gandolph M. A., Qian F., Onuchic L. F., Klinger K. W., Landes G., Germino G. G. (1998) Mol. Cell 2, 247–251 [DOI] [PubMed] [Google Scholar]
- 13.Driscoll J. A., Bhalla S., Liapis H., Ibricevic A., Brody S. L. (2008) Chest 133, 1181–1188 [DOI] [PubMed] [Google Scholar]
- 14.Kolpakova-Hart E., McBratney-Owen B., Hou B., Fukai N., Nicolae C., Zhou J., Olsen B. R. (2008) Dev. Biol. 321, 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolpakova-Hart E., Nicolae C., Zhou J., Olsen B. R. (2008) Matrix Biol. 27, 505–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou B., Kolpakova-Hart E., Fukai N., Wu K., Olsen B. R. (2009) Bone 44, 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao Z., Zhang S., Magenheimer B. S., Luo J., Quarles L. D. (2008) J. Biol. Chem. 283, 12624–12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao Z., Zhang S., Mahlios J., Zhou G., Magenheimer B. S., Guo D., Dallas S. L., Maser R., Calvet J. P., Bonewald L., Quarles L. D. (2006) J. Biol. Chem. 281, 30884–30895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piontek K. B., Huso D. L., Grinberg A., Liu L., Bedja D., Zhao H., Gabrielson K., Qian F., Mei C., Westphal H., Germino G. G. (2004) J. Am. Soc. Nephrol. 15, 3035–3043 [DOI] [PubMed] [Google Scholar]
- 20.Zhang M., Xuan S., Bouxsein M. L., von Stechow D., Akeno N., Faugere M. C., Malluche H., Zhao G., Rosen C. J., Efstratiadis A., Clemens T. L. (2002) J. Biol. Chem. 277, 44005–44012 [DOI] [PubMed] [Google Scholar]
- 21.Xiao Z., Awad H. A., Liu S., Mahlios J., Zhang S., Guilak F., Mayo M. S., Quarles L. D. (2005) Dev. Biol. 283, 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glass D. A., 2nd, Bialek P., Ahn J. D., Starbuck M., Patel M. S., Clevers H., Taketo M. M., Long F., McMahon A. P., Lang R. A., Karsenty G. (2005) Dev. Cell 8, 751–764 [DOI] [PubMed] [Google Scholar]
- 23.David V., Martin A., Lafage-Proust M. H., Malaval L., Peyroche S., Jones D. B., Vico L., Guignandon A. (2007) Endocrinology 148, 2553–2562 [DOI] [PubMed] [Google Scholar]
- 24.Xiao Z. S., Hjelmeland A. B., Quarles L. D. (2004) J. Biol. Chem. 279, 20307–20313 [DOI] [PubMed] [Google Scholar]
- 25.Lantinga-van Leeuwen I. S., Leonhard W. N., van de Wal A., Breuning M. H., Verbeek S., de Heer E., Peters D. J. (2006) Genesis 44, 225–232 [DOI] [PubMed] [Google Scholar]
- 26.Boca M., D'Amato L., Distefano G., Polishchuk R. S., Germino G. G., Boletta A. (2007) Mol. Biol. Cell 18, 4050–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boca M., Distefano G., Qian F., Bhunia A. K., Germino G. G., Boletta A. (2006) J. Am. Soc. Nephrol. 17, 637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim E., Arnould T., Sellin L. K., Benzing T., Fan M. J., Grüning W., Sokol S. Y., Drummond I., Walz G. (1999) J. Biol. Chem. 274, 4947–4953 [DOI] [PubMed] [Google Scholar]
- 29.Raucci A., Bellosta P., Grassi R., Basilico C., Mansukhani A. (2008) J. Cell. Physiol. 215, 442–451 [DOI] [PubMed] [Google Scholar]
- 30.Westendorf J. J., Kahler R. A., Schroeder T. M. (2004) Gene 341, 19–39 [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee A., Rotwein P. (2009) J. Cell Sci. 122, 716–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aoki K., Taketo M. M. (2008) Methods Mol. Biol. 468, 307–331 [DOI] [PubMed] [Google Scholar]
- 33.Constantinou T., Baumann F., Lacher M. D., Saurer S., Friis R., Dharmarajan A. (2008) J. Mol. Signal 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwaniec U. T., Wronski T. J., Liu J., Rivera M. F., Arzaga R. R., Hansen G., Brommage R. (2007) J. Bone Miner. Res. 22, 394–402 [DOI] [PubMed] [Google Scholar]
- 35.Kim D., Cho S. W., Her S. J., Yang J. Y., Kim S. W., Kim S. Y., Shin C. S. (2006) Stem Cells 24, 1798–1805 [DOI] [PubMed] [Google Scholar]
- 36.Lian J. B., Stein G. S. (1995) Iowa Orthop. J. 15, 118–140 [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi T., Wallace D. P., Magenheimer B. S., Hempson S. J., Grantham J. J., Calvet J. P. (2004) J. Biol. Chem. 279, 40419–40430 [DOI] [PubMed] [Google Scholar]
- 38.Piontek K., Menezes L. F., Garcia-Gonzalez M. A., Huso D. L., Germino G. G. (2007) Nat. Med. 13, 1490–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nuttall M. E., Gimble J. M. (2000) Bone 27, 177–184 [DOI] [PubMed] [Google Scholar]
- 40.Rocha V. Z., Folco E. J., Sukhova G., Shimizu K., Gotsman I., Vernon A. H., Libby P. (2008) Circ. Res. 103, 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein R. F., Allard J., Avnur Z., Nikolcheva T., Rotstein D., Carlos A. S., Shea M., Waters R. V., Belknap J. K., Peltz G., Orwoll E. S. (2004) Science 303, 229–232 [DOI] [PubMed] [Google Scholar]
- 42.Kawamura N., Kugimiya F., Oshima Y., Ohba S., Ikeda T., Saito T., Shinoda Y., Kawasaki Y., Ogata N., Hoshi K., Akiyama T., Chen W. S., Hay N., Tobe K., Kadowaki T., Azuma Y., Tanaka S., Nakamura K., Chung U. I., Kawaguchi H. (2007) PLoS One 2, e1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kugimiya F., Kawaguchi H., Ohba S., Kawamura N., Hirata M., Chikuda H., Azuma Y., Woodgett J. R., Nakamura K., Chung U. I. (2007) PLoS One 2, e837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weimbs T. (2007) Am. J. Physiol. Renal Physiol. 293, F1423–F1432 [DOI] [PubMed] [Google Scholar]
- 45.Wilson P. D., Geng L., Li X., Burrow C. R. (1999) Lab. Invest. 79, 1311–1323 [PubMed] [Google Scholar]
- 46.Li Q., Montalbetti N., Wu Y., Ramos A., Raychowdhury M. K., Chen X. Z., Cantiello H. F. (2006) J. Biol. Chem. 281, 37566–37575 [DOI] [PubMed] [Google Scholar]
- 47.Jensen C. G., Poole C. A., McGlashan S. R., Marko M., Issa Z. I., Vujcich K. V., Bowser S. S. (2004) Cell Biol. Int. 28, 101–110 [DOI] [PubMed] [Google Scholar]
- 48.Takaoki M., Murakami N., Gyotoku J. (2004) Biol. Sci. Space 18, 181–182 [PubMed] [Google Scholar]
- 49.Qian F., Boletta A., Bhunia A. K., Xu H., Liu L., Ahrabi A. K., Watnick T. J., Zhou F., Germino G. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16981–16986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turco A. E., Padovani E. M., Chiaffoni G. P., Peissel B., Rossetti S., Marcolongo A., Gammaro L., Maschio G., Pignatti P. F. (1993) J. Med. Genet. 30, 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]