Abstract
Mutation of the tumor suppressor adenomatous polyposis coli (APC) is a key early event in the development of most colorectal tumors. APC promotes degradation of β-catenin and thereby negatively regulates Wnt signaling, whereas mutated APCs present in colorectal tumor cells are defective in this activity. APC also stimulates the activity of the guanine nucleotide exchange factor Asef and regulates cell morphology and migration. Truncated mutant APCs constitutively activate Asef and induce aberrant migration of colorectal tumor cells. Furthermore, we have recently found that Asef and APC function downstream of hepatocyte growth factor and phosphatidylinositol 3-kinase. We show here that Asef is required for basic fibroblast growth factor- and vascular endothelial growth factor-induced endothelial cell migration. We further demonstrate that Asef is required for basic fibroblast growth factor- and vascular endothelial growth factor-induced microvessel formation. Furthermore, we show that the growth as well as vascularity of subcutaneously implanted tumors are markedly impaired in Asef−/− mice compared with wild-type mice. Thus, Asef plays a critical role in tumor angiogenesis and may be a promising target for cancer chemotherapy.
Introduction
Mutational inactivation of the tumor suppressor adenomatous polyposis coli (APC)2 is responsible for sporadic and familial colorectal tumors (1, 2). Most somatic mutations in APC occur at its central region called the mutation cluster region, and mutated APCs typically generate truncated gene products. The most thoroughly studied function of APC is its ability to induce degradation of β-catenin, a key Wnt signaling effector (3–6). Truncated mutant APCs identified in colorectal tumors are defective in this activity, suggesting that the tumor-suppressive function of APC is dependent on its ability to negatively regulate Wnt signaling.
APC has also been shown to interact with various cellular proteins other than β-catenin and to regulate cytoskeletal networks (7–15). For example, APC binds via its armadillo repeat domain to the NH2-terminal regions of Asef and Asef2, guanine-nucleotide exchange factors (GEFs) specific for Rac1 and Cdc42 (9, 11, 15–16). APC enhances their GEF activity and thereby regulates cell morphology, adhesion, and migration. In the absence of APC, the activity of Asef is inhibited by the intramolecular interaction between its Dbl homology (DH) and Src homology 3 (SH3) domains (17, 18). However, a mutant form of Asef lacking the NH2-terminal region exhibits strong GEF activity even in the absence of APC. Thus, APC may activate the GEF activity of Asef by relieving the intramolecular negative regulation. Furthermore, truncated mutant APCs present in colorectal tumor cells activate Asef constitutively and cause decreased cell-cell adhesion and increased aberrant migration. Hence, the activation of Asef by truncated mutant APC may be important for colorectal tumorigenesis.
Our previous study revealed that activated Asef-induced changes in morphological and migratory properties are similar to those induced by some growth factors. These findings have encouraged us to examine the possibility that Asef could function as a downstream component of signal transduction pathways linking growth factors to morphological changes and cell migration. Indeed, we have recently found that Asef and APC function downstream of hepatocyte growth factor and that Asef is required for hepatocyte growth factor-induced cell migration (19). In the present study, we examined whether Asef functions downstream of bFGF and VEGF, which play critical roles in angiogenesis, a process important for embryonic development and pathogenesis including tumorigenesis (20). These factors regulate vascular endothelial cell migration and proliferation, which are essential to angiogenesis. Here we show that Asef is required for bFGF- and VEGF-induced microvessel formation. Furthermore, we demonstrate that Asef plays a critical role in tumor angiogenesis.
EXPERIMENTAL PROCEDURES
Construction of Targeting Vector
A genomic clone of Asef containing exon 2, which encodes a part of the APC-binding region (amino acids 73–126), was isolated from a C57BL/6 mouse genomic library (Incyte Genomics, Inc.). The 5′ end of the targeting vector contained a 7.1-kb genomic sequence ending with codon 73 of Asef, which was fused in-frame with a neomycin resistance gene (neo) under the control of the MC1 (herpes simplex virus-thymidine kinase) promoter and poly(A) signal. The 3′ end contained a 1.4-kb genomic sequence. The diphtheria toxin A cassette was ligated at the 3′ end of the construct for negative selection.
Generation of Asef-deficient Mice
TT2 embryonic stem cells were electroporated with 100 μg of the NotI-linearized targeting construct and cultured under conditions of G418 selection. The selected embryonic stem (ES) cell clones were screened by PCR and Southern blotting for correct homologous recombination. The karyotype was verified, and several correctly targeted ES cell clones were used to generate germ line chimeric mice by blastocyst aggregation. The resulting male chimeras were mated with wild-type C57BL/6J females to obtain heterozygous (Asef+/−) mice, which were intercrossed to produce homozygous (Asef−/−) mutants. Homozygous mice obtained from two independent ES cell clones were used for phenotype analysis. Genotyping of mice was performed by PCR using the following primers: wild-type Asef forward primer, 5′-CACATTGGTAAATGGCTTTTAACTGGACTC-3′; mutant Asef forward primer, 5′-CTTCCATGGAGATAACTTCGTATAGCATAC-3′; wild-type and mutant Asef reverse primer, 5′-CATACACAACATGTGTCAATTATAAATTAC-3′. The 943-bp and 693-bp fragments represent the Asef wild-type and targeted alleles, respectively.
Genotyping of ES Cells and Mice
Genomic DNA was isolated from ES cells or mouse tail biopsies. Extracts were treated with proteinase K, followed by phenol-chloroform extraction and ethanol precipitation. Purified DNA was digested with DraI, separated on 0.8% agarose gels, transferred to a positive charged membrane, and hybridized with an external (Probe P1) or internal (Probe P2) probe (Fig. 1A).
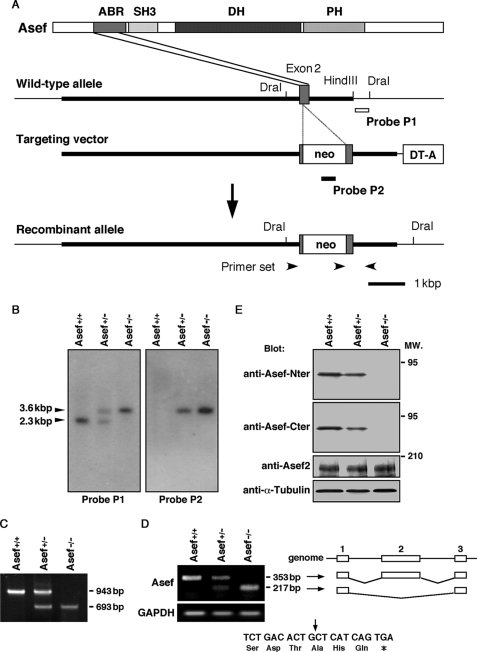
FIGURE 1.
Targeted disruption of the mouse Asef gene. A, construct containing a portion of the wild-type allele, targeting construct, and recombinant locus are shown. Asef was inactivated by the insertion of a neomycin cassette (neo) into codon 73 of Asef. The diphtheria toxin A cassette (DT-A) was placed outside the 3′ homologous region for negative selection. The probe P1 used for detection of homologous recombinants is represented by the open bar, and the probe P2 used for detection of a single insertion of the neomycine gene is represented by the filled bar. The primer set used for detection of recombinants is also represented by black arrowheads. ABR, APC-binding region; PH, Pleckstrin homology domain. B, the mutated Asef locus was identified by Southern blot analysis of genomic DNA digested with DraI and hybridized to internal (Probe P2) and external (Probe P1) genomic fragments. The 2.3-kbp fragment represents the wild-type allele, and the 3.6-kbp fragment represents the homologous recombinant allele. C, shown is a PCR analysis of genomic tail DNA from wild-type (Asef+/+), heterozygous (Asef+/−), and homozygous (Asef−/−) mice with three primers indicated in A. The 943 and 693-bp fragments represent the Asef wild-type and targeted alleles, respectively. D, analysis of Asef mRNA expression is shown. Reverse transcription-PCR analysis was performed on total RNA isolated from brains of Asef+/+, Asef+/−, and Asef−/− mice using primers located in exons 1 and 3. The 353-bp fragment represents the full-length transcript, whereas the 217-bp fragment lacks exon 2. Schematic representation of the structures of the 353- and 217-bp fragments are shown in the right panel. Lines indicate introns, and boxes indicate exons. The 217-bp fragment is derived from the direct splicing of exons 1–3, which introduces a stop codon 3 amino acids downstream in exon 3 and generates a truncated peptide of 104 amino acids (bottom; the arrow indicates the out-of-frame junction). Expression of GAPDH was examined as an internal control. E, an immunoblotting analysis of mutant mice is shown. Lysates prepared from the brain of adult Asef+/+, Asef+/−, and Asef−/− mice were analyzed by immunoblotting with antibodies against the amino-terminal (Nter) or carboxy-terminal (Cter) regions of Asef or Asef2. Anti-α-tubulin antibody was used as a control. MW., molecular weight.
Reverse Transcription-PCR
Total RNAs from organs and macrophages were isolated with TRIzol reagent (Invitrogen), and cDNA was generated from 1 μg RNA with oligo(dT) primers and ReverTra Ace reverse transcriptase (Toyobo). Asef transcripts were analyzed by PCR using the following primers: exon 1-secific primer, 5′-TCTCCAGAGTCTCCGCATCTTC-3′ (forward) and exon 3-specific primer 5′-GTGGCATCCATCACTTCGATG-3′ (reverse); exon 2-specific primer, 5′-GAGGAGGTGGAGAGCAACTG-3′ (forward) and exon 5-specific primer, 5′-GCGGTAGATGTCCTCGATGTTC-3′ (reverse). The 353-bp and 484-bp fragments represent the full-length transcripts, respectively. The exon 1-specific primer corresponds to a sequence in exon 1 of ARHGEF4, which is one of several splice variants of Asef. The primers for the analysis of mouse GAPDH were as follows: 5′-TGTGTCCGTCGTGGATCTGA-3′ (forward) and 5′-TTGCTGTTGAAGTCGCAGGAG-3′ (reverse).
Cell Culture and Transfection
B16-F10 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 4.5 g/liter dextrose. Human and mouse aortic endothelial cells (HAECs and MAECs) were cultured in EGM-2 medium (Cambrex) supplemented with bFGF, epidermal growth factor (EGF), VEGF, IGF-1, and 2% fetal bovine serum (FBS). Plasmids and siRNAs were transfected into cells using Lipofectamine 2000 (Invitrogen). Adenovirus infection was performed as described previously (10).
Expression Vectors and Antibodies
Hemagglutinin (HA)-tagged Asef was generated as described previously (9). Myc-tagged APC was subcloned into pCS2. The adenoviruses encoding FLAG-β-catenin S33Y and LacZ were constructed using the pAdenoX™ expression system (Clontech) according to the manufacturer's directions. Mouse monoclonal antibody (mAb) to APC was raised against a peptide containing amino acids 119–250 of APC. Rabbit polyclonal antibodies to Asef were prepared by immunizing rabbits with a peptide containing amino-terminal (amino acids 1–113) or carboxy-terminal (amino acids 485–605) region of Asef, respectively. Mouse mAb to Myc-tag (9E10), rabbit polyclonal antibodies to Cdc42, and goat polyclonal antibodies to Ki67 and VEGF were obtained from Santa Cruz Biotechnology. Mouse mAb to Rac1 was from BD Transduction Laboratories. Rat mAb to F4/80 and mouse mAb to α-tubulin were from Serotec and Oncogene Research, respectively. Rat mAb against CD31 and HA-tag (3F10) were from Pharmingen and Roche, respectively.
siRNA-mediated Gene Silencing
Stealth small interfering RNA (siRNA) duplexes against Asef and APC were purchased from Invitrogen. The sequences of the regions in the human Asef and APC cDNAs targeted for siRNAs were: Asef, 5′-CAAGGATGTTGAAGCCGCCTTGCAT-3′; APC-1, 5′-TCGTCTGATTCAGATTCCATCCTTT-3′; and APC-2, 5′-CCCACCTAATCTCAGTCCCACTATA-3′. HAECs were transfected with siRNA (1 μm) using Lipofectamine 2000 (Invitrogen). After 24 h of siRNA transfection, the medium was changed, and the cells were cultured for further 48 h and used for cell migration or tube formation assays. Validated stealth negative control RNA interference duplex with low GC content (Invitrogen) was used as a control.
Immunoprecipitation, Immunoblotting, and Immunostaining
Immunoprecipitation, immunoblotting, and immunostaining analyses were performed as described previously (10).
Isolation of Endothelial Cells
MAECs were isolated from Asef+/+ and Asef−/− mice as described previously (21) and maintained in EGM-2 medium (Cambrex) supplemented with bFGF, EGF, VEGF, IGF-1, and 2% FBS. At passages used for experiments, almost all cells were of endothelial origin as assessed by 1,1′-dioctadecyl-1-3,3,3′,3′-tetramethylindocarbocyanine-labeled acetylated low density lipoprotein uptake (Biomedical Technologies).
Cell Migration Assays
Cell migration assays were performed using Transwell migration chambers (Costar) as described (10). Both sides of the filter membrane were coated with 10 μg/ml collagen type I (Koken) overnight. Cells (HAECs, 1.6 × 104 cells/well; MAECs, 2.0 × 104 cells/well) were allowed to migrate to the underside of the top chamber for 3.5 or 4.5 h. Medium containing bFGF (25 ng/ml), VEGF (10 ng/ml) or 0.5% bovine serum albumin was added to the lower chamber. Cell migration was determined by counting the cells that had migrated to the lower side of the polycarbonate filters.
In Vitro Tube Formation Assays
Matrigel (200 μl/well) was plated evenly in a 24-well culture plate and incubated at 37 °C for at least 1 h to allow gel formation. MAECs were harvested after trypsin treatment, resuspended in serum-free EBM-2, plated onto the layer of Matrigel at a density of 5.0 × 104 cells/well, and followed by the addition of 2% FBS, 12 μg/ml bFGF, or 12 μg/ml VEGF. After 24 h of incubation, the formation of the capillary/tube-like networks were photographed and examined by using a phase-contrast microscope (22). The quantification was performed by measuring the number and length of tubes and the number of branch points in ×100 fields. Statistic analysis was performed using Student's t test. p value of < 0.05 was considered statistically significant.
GTPase Activation Assays
MAECs and HAECs were treated with bFGF (25 ng/ml for 2 h) or VEGF (10 ng/ml for 2 h) after overnight starvation and lysed in buffer containing 2% IGEPAL, 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, and 0.3 m NaCl. To detect the active GTP-bound form of GTPases in the cell lysates, the supernatants were mixed with 50 μg of recombinant GST-PAK-CRIB bound to glutathione-Sepharose beads for 30 min. The beads and the proteins bound to the fusion protein were washed two times with wash buffer containing 25 mm Tris-HCl (pH 7.5), 30 mm MgCl2, and 40 mm NaCl, and then the bound proteins were analyzed by SDS-PAGE followed by immunoblotting analysis.
Aortic Ring Assays
Thoracic aortas were removed from wild-type and Asef−/− mice after cervical dislocation. Extraneous fat and other tissues were removed, and the aortas were sliced transversely into 1-mm-thick rings. The rings were placed in the wells coated with 200 μl of Matrigel, sealed with an overlay of 300 μl of Matrigel and covered with 500 μl of EGM-2 (Cambrex) containing bFGF, EGF, VEGF, IGF-I, and 2% fetal calf serum. After 8 days of culture, microvessel outgrowth was visualized by phase contrast microscopy, and angiogenesis was quantitated by counting the numbers of capillary sprouts per ring.
In Vivo Angiogenesis Assay
Eight-week-old mice were injected subcutaneously around the abdominal region with 500 μl of growth factor-reduced Matrigel (BD Biosciences) supplemented with 400 ng/ml bFGF (BD Biosciences) and 50 mg/ml heparin. Mice were sacrificed 5 days after Matrigel injection, and neovascularization was analyzed by hematoxylin and eosin staining and immunohistological examination using anti-CD31 antibody. Infiltration of microvessels into the Matrigel was visualized by anti-CD31 staining and quantitated using ImageJ software.
Tumorigenesis Assays
Tumorigenicity of murine B16-F10 melanoma cells was assayed by injecting 1 × 106 cells subcutaneously into the flank of wild-type and Asef−/− mice. At several time points after tumor implantation, the size of the tumors was measured using calipers. The tumor volume was calculated according to the formula (V = π/6 × [L × W2]), where V is volume, L is length, and W is width (length is greater than width).
Isolation of Peritoneal Macrophages
Macrophages were isolated from peritoneal exudate cells as described (23). Peritoneal exudate cells were obtained 4 days after intraperitoneal injection of 4% Brewer thioglycollate medium (3 ml/mouse) and plated onto appropriate plates in RPMI 1640 medium containing 10% FBS. Macrophages were allowed to adhere for 1.5 h and then washed to remove nonadherent cells.
Histology, Immunohistochemistry, and Statistical Analysis
Plugs and tissues were embedded in paraffin or snap frozen in OCT compound (Tissue-Tek). Paraffin-embedded sections were stained with hematoxylin and eosin, anti-CD31 antibody, or anti-F4/80 antibody using the ABC staining kit (Vector Laboratories). Frozen sections were subjected to immunofluorescent staining with anti-CD31 antibody, anti-F4/80 antibody, or anti-VEGF antibody, or immunohistochemical staining with anti-Asef antibody using the ABC staining kit. Statistic analysis was performed using Student's t test. p value of < 0.05 was considered statistically significant.
Proliferation and Apoptosis Assays
Cryosections from Matrigel plugs were double-stained with anti-CD31 mAb and anti-Ki67 polyclonal antibodies or TUNEL reagents. TUNEL staining was performed using an apoptosis detection kit (TaKaRa) according to the manufacturer's instructions.
RESULTS
Generation of Asef-deficient Mice
To study the biological function of Asef, Asef-deficient mice were generated through conventional gene targeting in ES cells in which the neomycin cassette was inserted into exon 2 encoding the APC-binding region of Asef (Fig. 1A). Homologous recombinant ES cell clones were selected by PCR and Southern blot analysis and were used to generate germ line chimeric mice by blastocyst aggregation. Male chimeras from two independent ES cell clones were used for mating with C57BL/6J females to obtain heterozygous mice. To produce the mice used in subsequent analysis, mice carrying the mutant allele were backcrossed for six generations into C57BL/6J genetic background. Heterozygous offspring were crossed for homozygosity, and DNA from tail biopsy was taken to detect the mutated allele by PCR and Southern blot analysis (Fig. 1, B and C). When we performed reverse transcription-PCR analysis of RNA derived from mouse brain using exon 1- and 3-specific primers, we detected a fragment with an expected length (353-bp) in Asef+/+ and Asef+/− samples, but not Asef−/− samples (Fig. 1D). Unexpectedly, we detected a 217-bp fragment in Asef−/− and Asef+/− samples. Sequence analysis revealed that it is derived from the splicing of exon 1 to exon 3, which results in the generation of a truncated peptide of 104 amino acids (Fig. 1D). Moreover, immunoblotting analyses using antibody specific for the amino- or carboxy-terminal region of Asef revealed that ablation of the Asef locus leads to a concomitant loss of Asef protein expression (Fig. 1E). In addition, we found that the expression of the Asef-related Asef2 protein was not up-regulated in Asef−/− mice (Fig. 1E). Genotyping of offspring from Asef+/− intercrosses showed that there is no reduced viability of the Asef−/− mice and that mice of all three genotypes are present at the expected Mendelian frequency (Asef+/+, 27.5%; Asef+/−, 46.1%; Asef−/−, 26.4%; n = 557 animals). Hetero- and homozygous mutant mice were fertile and were indistinguishable from their wild-type littermates based on growth rates and external appearance. Similarly, pathological examination of a number of organs including the intestine did not show significant abnormalities except for modest impairment of retinal angiogenesis in neonatal mice (data not shown). Absence of Asef therefore appears to be compatible with normal physiologic functioning of mice.
Asef and APC Are Required for bFGF- and VEGF-induced Endothelial Cell Migration and Tube Formation
To study the physiological significance of Asef in angiogenesis, we cultured aortic endothelial cells (MAECs) generated from wild-type (Asef+/+) and Asef−/− mice, respectively. Immunostaining with anti-Asef antibody confirmed that Asef is expressed in MAECs from wild-type mice, but not Asef−/− mice (Fig. 2A). We found that the morphology and proliferation of Asef−/− MAECs were not altered compared with those of wild-type cells (Fig. 2B). However, when we compared their migratory behavior in the presence of bFGF or VEGF, MAECs from Asef−/− mice exhibited significantly lower migratory activity compared with those from wild-type mice (Fig. 2C). We also performed an in vitro tube formation assay on Matrigel (22). We found that Asef−/− MAECs generated fewer and shorter protrusions and fewer branches than wild-type cells (Fig. 2, D–G).
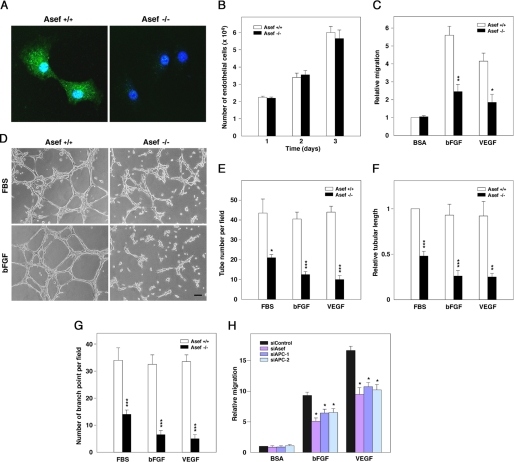
FIGURE 2.
Involvement of Asef in endothelial cell migration and tube formation. A, MAECs isolated from Asef+/+ and Asef−/− mice were stained with antibody against Asef (green). Nuclei were counterstained with TO-PRO3 (blue). B, a proliferation of Asef+/+ and Asef−/− endothelial cells is shown. MAECs were trypsinized and plated at densities of 2.0 × 104 cells into 6-well plates (day 0), and cell numbers were counted every 24 h. Results are expressed as the mean ± S.E. of four independent experiments. C, shown is the impaired cell migration in Asef−/− endothelial cells. Migration of MAECs derived from Asef+/+ and Asef−/−mice in response to bFGF or VEGF was quantified by transwell assays. The number of endothelial cells that had migrated to the lower surface of the transwell in 4.5 h was counted. Results are expressed as the mean ± S.E. of four independent experiments. *, p < 0.05; **, p < 0.01. D, MAECs isolated from Asef+/+ and Asef−/− mice were plated on Matrigel in the presence or absence of FBS or bFGF, and tube network formation was monitored. Scale bar, 50 μm. E–G, shown is the quantification of tube number (E), tubular length (F), and branch points (G) per field at ×100 magnification. Open bars, Asef+/+; filled bars, Asef−/−. Results are expressed as the mean ± S.E. of four independent experiments. *, p < 0.05; **, p < 0.01; and ***, p < 0.001. H, effects of Asef and APC knockdown on the migration of HAECs are shown. Cells were transfected with Asef- or APC-specific or control siRNA, respectively, and subjected to migration assays using Transwell migration chambers. Cells were allowed to migrate for 3.5 h in the presence or absence of bFGF or VEGF, respectively. Results are expressed as the means ± S.E. of four independent experiments. *, p < 0.05. BSA, bovine serum albumin.
We also examined the role of APC in bFGF- and VEGF-induced endothelial cell migration. We found that knockdown of either APC or Asef by siRNA resulted in a decrease in either bFGF- or VEGF-induced migration of HAECs (supplemental Fig. S1, A and B and Fig. 2H). HAECs treated with these siRNAs, however, barely showed morphological abnormalities. Similar results were obtained when we used Asef-ΔDH, a mutant that lacks the DH domain and acts as a dominant-negative mutant that competes with Asef for binding to APC (10) (supplemental Fig. S2). Because knockdown of APC is known to result in the stabilization of β-catenin, we examined whether overexpression of a stabilized mutant form of β-catenin, β-catenin S33Y, affects endothelial cell migration. We found that HAECs infected with adenovirus encoding β-catenin S33Y showed slightly increased motility compared with AdLacZ-infected cells in the presence of bFGF or VEGF (supplemental Fig. S3, A and B). Thus, β-catenin/TCF-dependent transcription does not appear to be important for the decrease in migration of HAECs that had been transfected with siRNA-targeting APC. Taken together, these results suggest that APC and Asef are required for bFGF- and VEGF-induced endothelial cell migration and tube formation.
Asef and APC Function Downstream of bFGF and VEGF
We next examined bFGF- and VEGF-induced changes of subcellular localization of Asef and APC. When HAECs were treated with bFGF and VEGF, Asef and APC were induced to accumulate and co-localize in membrane ruffles and lamellipodia (Fig. 3A). In addition, when Asef and APC were exogenously expressed in bFGF-stimulated HAECs, both proteins were found to accumulate in membrane ruffles and lamellipodia (Fig. 3B). Consistent with these results, pull-down assays revealed that the amounts of APC associated with Asef increased in response to bFGF stimulation (Fig. 3C). We also examined whether Asef is involved in bFGF- and VEGF-induced activation of Rac1 and Cdc42 in endothelial cells. When wild-type MAECs were treated with bFGF and VEGF, the amounts of the GTP-bound forms of both Rac1 and Cdc42 were increased (Fig. 3D). By contrast, treatment of Asef−/− MAECs with bFGF and VEGF barely induced Rac1 and Cdc42 activation. Furthermore, we found that knockdown of APC by siRNA resulted in a decrease in either bFGF- or VEGF-induced activation of Rac1 and Cdc42 in HAECs (Fig. 3E). Thus, Asef and APC are essential for bFGF- and VEGF-induced activation of both Rac1 and Cdc42. These results suggest that in response to either bFGF or VEGF treatment, the Asef-APC complex accumulates in membrane ruffles and lamellipodia, activates both Rac1 and Cdc42 and thereby stimulates endothelial cell migration and tube formation.
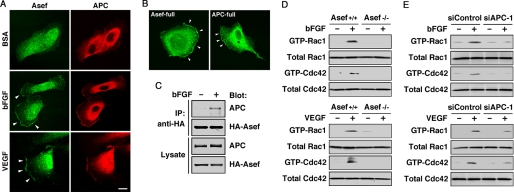
FIGURE 3.
The Asef-APC complex functions downstream of bFGF and VEGF. A, shown is the colocalization of endogeneous Asef and APC in HAECs treated with bFGF and VEGF, respectively. Cells were stimulated with bFGF (25 ng/ml for 30 min) or VEGF (10 ng/ml for 30 min) after starvation for 24 h and double-stained with anti-Asef and anti-APC antibodies. Arrowheads indicate the areas of colocalization of Asef and APC in membrane ruffles. Scale bar, 20 μm. B, shown is the subcellular localization of exogenously expressed Asef and APC in bFGF-stimulated HAECs. HAECs were transfected with expression plasmids encoding HA-tagged Asef or Myc-tagged APC and were treated with bFGF. Cells were stained with antibodies against HA or Myc. Arrowheads indicate ruffling membranes. C, a bFGF-stimulated interaction of Asef with APC is shown. Lysates from bFGF-stimulated or unstimulated HAECs infected with adenovirus encoding HA-tagged full-length Asef were immunoprecipitated (IP) with antibody against HA and then analyzed by immunoblot with antibodies against APC and HA. HA-Asef and APC expression levels were determined by immunoblotting of total cell lysates (bottom). D–E, Asef and APC are required for bFGF- and VEGF-induced activation of Rac1 and Cdc42 in endothelial cells. Wild-type and Asef−/− MAECs (for Asef; D), and siRNA-treated (si) HAECs (for APC; E) were stimulated with bFGF or VEGF and the amounts of active GTP-bound Rac1 and Cdc42 were measured using GST-PAK CRIB coupled to glutathione-Sepharose beads. The GTP-bound forms of GTPases were detected by immunoblotting with the indicated antibodies. The total amounts of GTPases in the lysates prior to pulldown assays were also analyzed by immunoblotting. BSA, bovine serum albumin.
Asef Is Required for Microvessel Formation
To evaluate the angiogenic properties of endothelial cells from Asef−/− mice, we next performed aortic ring assays. Aortic rings from wild-type and Asef−/− mice were embedded in Matrigel containing bFGF, EGF, VEGF, IGF-1, and serum, and the number of vascular sprouts per aortic ring was assessed. We found that the aortic rings from Asef−/− mice develop a smaller number of endothelial protrusions compared with those from wild-type mice (Fig. 4). These results raise the possibility that Asef may play an important role in angiogenesis.
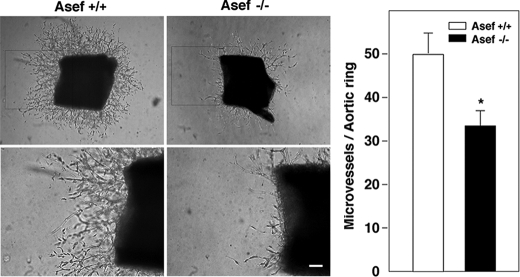
FIGURE 4.
Involvement of Asef in endothelial cell sprouting. Aortas from Asef+/+ and Asef−/− mice were embedded in Matrigel and cultured for 8 days in the presence of bFGF, EGF, VEGF, IGF-1, and 2% serum. The regions in boxes are shown magnified. Magnification, ×40 (upper panels), × 100 (lower panels). The graph shows the number of endothelial cell sprouts per aortic ring at day 8. Open bars, Asef+/+; filled bars, Asef−/−. Results are expressed as the mean ± S.E. of three independent experiments. *, p < 0.05. Scale bar, 100 μm.
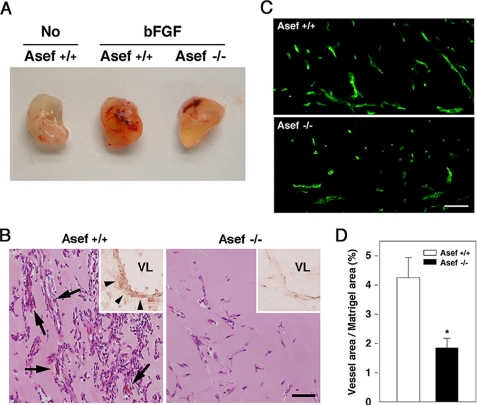
To prove this possibility, we performed in vivo Matrigel assays. We implanted Matrigel plugs supplemented with bFGF subcutaneously into mice and then removed these after 5 days. Matrigel plugs containing bFGF implanted in wild-type mice were reddish compared with control Matrigel plugs without bFGF. In contrast, Matrigel plugs containing bFGF implanted in Asef−/− mice were less reddish than those implanted in wild-type mice (Fig. 5A). Histological studies revealed that formation of blood vessel-like structures in bFGF-containing Matrigel plugs was significantly impaired in Asef−/− mice compared with wild-type mice (Fig. 5B). In addition, immunostaining with anti-Asef antibody showed that all vessels in Matrigel plugs from wild-type mice, but not Asef-null mice, were positive for Asef (Fig. 5B). Immunofluorescent labeling of microvessels with anti-CD31 antibody also revealed that microvessel formation in bFGF-containing Matrigel plugs is impaired in Asef−/− mice (Fig. 5C). Furthermore, quantification of endothelial cells in Matrigels confirmed that bFGF-induced microvessel formation is impaired in Asef−/− mice (Fig. 5D). These results suggest that Asef plays a critical role in capillary network formation and morphogenesis of endothelial cells.
FIGURE 5.
In vivo angiogenesis within Matrigel plugs. A, representative photographs of Matrigel plugs containing no growth factors (No) or bFGF. Plugs were removed from wild-type (Asef+/+) or Asef-deficient (Asef−/−) mice at 5 days post-implantation. B, immunohistochemistry of Matrigel plugs. Hematoxylin and eosin staining of sections from Matrigel plugs containing bFGF implanted in Asef+/+ and Asef−/− mice are shown. Insets, microvessels (arrowheads) in Matrigel plugs immunostained with anti-Asef antibody (brown reaction; nuclear fast red staining for morphology). Arrows point to microvessels containing red blood cells. VL, vessel lumen. Scale bar, 50 μm. C, immunofluorescent labeling of microvessels with anti-CD31 antibody. Scale bar, 50 μm. D, quantitative analysis of vessel density within bFGF-supplemented Matrigel plugs from Asef+/+ and Asef−/−mice (n = 4–5/genotype). Microvessel density was defined as the percentage of vessel area (CD31)/Matrigel plug area in 200× fields. Open bars, Asef+/+; filled bars, Asef−/−. Results are expressed as the mean ± S.E. *, p < 0.05.
Asef Is Not Required for Endothelial Cell Proliferation or Survival
We examined the proliferation and survival of endothelial cells in Matrigel plugs containing bFGF implanted in wild-type and Asef−/− mice. Immunostaining with anti-Ki67 antibody revealed that the number of endothelial cells in the active phases of the cell cycle per CD31-positive vessel was similar in Matrigel plugs from wild-type and Asef-null mice (supplemental Fig. S4A). Also, TUNEL staining revealed no endothelial cell apoptosis in Matrigel plugs from wild-type or Asef-null mice (supplemental Fig. S4B). Thus, Asef is not required for endothelial cell proliferation and survival.
Asef Is Required for Tumor Angiogenesis
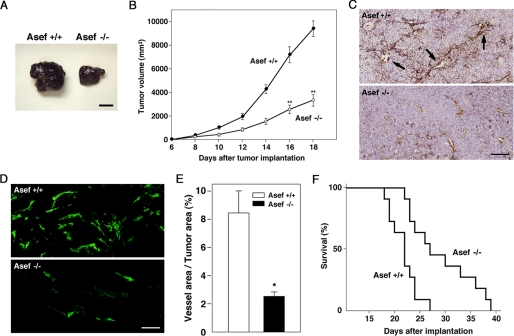
Angiogenesis is a critical event in the development of various tumors (20, 24). We therefore examined the significance of Asef in tumor angiogenesis using a mouse B16 melanoma model (25, 26). We implanted B16 melanoma cells subcutaneously in wild-type and Asef−/− mice and monitored tumor development and survival. B16 melanoma cells rapidly formed tumors in wild-type mice, whereas tumor growth was markedly retarded in the Asef-deficient mice (Fig. 6, A and B). Immunostaining with anti-CD31 antibody and histochemical analysis revealed that the density of microvessels in tumors of Asef−/− mice was markedly lower than those in tumors of wild-type mice (Fig. 6, C and D). Furthermore, quantification of endothelial cells stained with anti-CD31 antibody confirmed that microvessel formation was impaired in tumors implanted in Asef−/− mice (Fig. 6E). Asef−/− mice implanted with B16 melanoma cells survived significantly longer than wild-type controls (Fig. 6F). These results suggest that Asef plays a critical role in tumor angiogenesis and that the growth of B16 melanoma cells may be retarded due to the impairment of angiogenesis caused by Asef deficiency.
FIGURE 6.
Reduced tumor size and angiogenesis in Asef-deficient mice. A, representative photographs of subcutaneous melanoma tumors at 14 days post-implantation in Asef+/+ and Asef−/− mice. Scale bar, 10 mm. B, volumes of tumors developed in Asef+/+ and Asef−/− mice (n = 11/genotype). Filled circles, Asef+/+; open circles, Asef−/−. Results are expressed as the mean ± S.E. **, p < 0.01. C, tumor cross-sections were stained with anti-CD31 (brown) and counterstained with hematoxylin. Arrows indicate microvessels containing red blood cells. Scale bar, 50 μm. D, immunofluorescent labeling of microvessels with anti-CD31 antibody. Scale bar, 50 μm. E, quantitative analysis of vessel density within tumors from Asef+/+ and Asef−/− mice (n = 6–7/genotype) based on CD31 staining. Open bars, Asef+/+; filled bars, Asef−/−. Results are expressed as the mean ± S.E. *, p < 0.05. F, Kaplan-Meier survival curves of Asef+/+ and Asef−/− mice with subcutaneous melanoma (n = 11/genotype).
Macrophages Are Normally Recruited to Tumors of Asef−/− Mice
It has been shown that tumor-associated macrophages facilitate angiogenesis, extracellular matrix breakdown and remodeling, and promote tumor cell motility, thereby accelerating tumor progression and metastasis (27). We therefore examined whether macrophages are normally recruited to tumors of Asef−/− mice. Immunostaining with anti-F4/80 antibody revealed that there was no statistical difference in the number of macrophages accumulated in tumors between Asef−/− and wild-type mice (supplemental Fig. S5, A and B). Reverse transcription-PCR analysis revealed that Asef is expressed in macrophages from wild-type mice, but not Asef−/− mice (supplemental Fig. S5C). Because macrophages are known to release angiogenic factors, including VEGF, we performed immunostaining with anti-VEGF antibody. The results clearly showed that the levels of VEGF in tumors of Asef−/− mice are similar to those in tumors of wild-type mice (supplemental Fig. S5D). Thus, Asef may not be critical for migration of macrophages to tumors or for release of angiogenic factors by macrophages.
DISCUSSION
Our results showed that Asef is required for bFGF- and VEGF-induced endothelial cell migration and tube formation. Moreover, our Matrigel plug assays revealed that bFGF-induced microvessel formation is impaired in Asef−/− mice compared with wild-type mice. Aortic ring assays also showed that Asef is required for angiogenesis. These results suggest that Asef-mediated endothelial cell migration may be important for angiogenesis. Consistent with this notion, we found that retinal angiogenesis is modestly impaired in Asef−/− mice and that this is not caused by changes in VEGF or oxygen concentration or pericyte impairment.3 Asef−/− mice, however, do not show significant abnormalities, raising the possibility that other Dbl family genes might play redundant roles. In addition, we showed that APC is required for bFGF- and VEGF-induced endothelial cell migration. It remains to be determined whether APC is indeed involved in Asef-mediated angiogenesis.
Our findings that Asef is required for bFGF- and VEGF-mediated angiogenesis raise the possibility that Asef and APC function downstream of growth factors acting through receptor tyrosine kinases. In line with this notion, we have recently found that Asef functions downstream of EGF and hepatocyte growth factor in epithelial cells (19, 28). In this study, we showed that bFGF and VEGF induce the accumulation and colocalization of Asef and APC in membrane ruffles and lamellipodia and increase the amounts of the Asef-APC complex. We also demonstrated that Asef is required for bFGF- and VEGF-induced activation of Rac1 and Cdc42. These findings suggest that bFGF and VEGF activate the migratory activity of endothelial cells at least in part by inducing the accumulation of the Asef-APC complex in membrane ruffles and lamellipodia where Rac1 and Cdc42 are localized.
Consistent with our findings, it has been shown that several GEFs, including Vav2/3, P-REX and Tiam1, and the effector PAK2 function as downstream components of signal transduction pathways linking growth factors to Rac1/Cdc42-dependent endothelial cell spreading, migration, and angiogenesis (29–34). It is therefore intriguing to examine whether these GEFs and PAK2 function upstream or downstream of Asef in response to angiogenic stimulation. In addition, Cdc42 has been shown to regulate the localization of APC and Dlg1 at the leading edge of migrating astrocytes to control cell polarization and migration (35, 36). It would be interesting to assess the role of Asef in cell polarization and migration in which Cdc42, APC, and Dlg1 are known to be involved.
When we examined the significance of Asef in tumor angiogenesis using a mouse B16 melanoma model, we found that the density of microvessels in tumors of Asef−/− mice was markedly lower than those in tumors of wild-type mice. The growth of B16 melanoma cells was significantly retarded presumably due to the impairment of angiogenesis caused by Asef deficiency. Although tumor-associated macrophages are known to facilitate angiogenesis (27), we found that the number of macrophages accumulated in tumors of Asef−/− mice is similar to that in tumors of wild-type mice. Thus, Asef-mediated endothelial cell migration may be also important for tumor angiogenesis.
In conclusion, we have shown that Asef plays a critical role in angiogenesis. Our findings suggest that Asef may contribute to tumorigenesis not only by inducing aberrant migration of tumor cells (10) but also by promoting tumor angiogenesis. We speculate that compounds that target Asef could hold promise as novel anti-tumor reagents. Of particular interest is the fact that Asef−/− mice survive as long as wild-type mice and are fertile. Thus, reagents targeting Asef would be expected to have few serious side effects. In addition, it should also be examined whether Asef is involved in various other pathogenesis, including inflammatory, ischemic, infectious, and immune disorders.
Supplementary Material
This work was supported by grants-in-aid for Scientific Research on Priority Areas and the Organization for Pharmaceutical Safety and Research and in part by Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms), Ministry of Education, Culture, Sports, Science and Technology, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Y. Kawasaki, T. Jigami, S. Furukawa, M. Sagara, K. Echizen, Y. Shibata, R. Sato, and T. Akiyama, unpublished observation.
- APC
- adenomatous polyposis coli
- GEF
- guanine-nucleotide exchange factor
- SH3
- Src homology 3
- DH
- Dbl homology domain
- ES
- embryonic stem
- HAEC
- human aortic endothelial cell
- MAEC
- mouse aortic endothelial cell
- EGF
- epidermal growth factor
- HA
- hemagglutinin
- mAb
- monoclonal antibody
- siRNA
- small interfering RNA
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- bFGF
- basic fibroblast growth factor
- VEGF
- vascular endothelial growth factor
- EGM-2
- endothelial growth medium-2
- IGF-1
- insulin-like growth factor-1
- GST
- glutathione S-transferase
- PAK
- p21-activated kinase
- CRIB
- Cdc42/Rac interactive binding.
REFERENCES
- 1.Fodde R., Smits R., Clevers H. (2001) Nat. Rev. Cancer 1, 55–67 [DOI] [PubMed] [Google Scholar]
- 2.Kinzler K. W., Vogelstein B. (1996) Cell 87, 159–170 [DOI] [PubMed] [Google Scholar]
- 3.Bienz M., Clevers H. (2000) Cell 103, 311–320 [DOI] [PubMed] [Google Scholar]
- 4.Brembeck F. H., Rosário M., Birchmeier W. (2006) Curr. Opin. Genet. Dev. 16, 51–59 [DOI] [PubMed] [Google Scholar]
- 5.Cadigan K. M., Nusse R. (1997) Genes Dev. 11, 3286–3305 [DOI] [PubMed] [Google Scholar]
- 6.Peifer M., Polakis P. (2000) Science 287, 1606–1609 [DOI] [PubMed] [Google Scholar]
- 7.Akiyama T., Kawasaki Y. (2006) Oncogene 25, 7538–7544 [DOI] [PubMed] [Google Scholar]
- 8.Jimbo T., Kawasaki Y., Koyama R., Sato R., Takada S., Haraguchi K., Akiyama T. (2002) Nat. Cell Biol. 4, 323–327 [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki Y., Senda T., Ishidate T., Koyama R., Morishita T., Iwayama Y., Higuchi O., Akiyama T. (2000) Science 289, 1194–1197 [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki Y., Sato R., Akiyama T. (2003) Nat. Cell Biol. 5, 211–215 [DOI] [PubMed] [Google Scholar]
- 11.Kawasaki Y., Sagara M., Shibata Y., Shirouzu M., Yokoyama S., Akiyama T. (2007) Oncogene 26, 7620–7627 [DOI] [PubMed] [Google Scholar]
- 12.Mimori-Kiyosue Y., Shiina N., Tsukita S. (2000) J. Cell Biol. 148, 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Näthke I. S., Adams C. L., Polakis P., Sellin J. H., Nelson W. J. (1996) J. Cell Biol. 134, 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe T., Wang S., Noritake J., Sato K., Fukata M., Takefuji M., Nakagawa M., Izumi N., Akiyama T., Kaibuchi K. (2004) Dev. Cell 7, 871–883 [DOI] [PubMed] [Google Scholar]
- 15.Hamann M. J., Lubking C. M., Luchini D. N., Billadeau D. D. (2007) Mol. Cell. Biol. 27, 1380–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotthardt K., Ahmadian M. R. (2007) Biol. Chem. 388, 67–71 [DOI] [PubMed] [Google Scholar]
- 17.Murayama K., Shirouzu M., Kawasaki Y., Kato-Murayama M., Hanawa-Suetsugu K., Sakamoto A., Katsura Y., Suenaga A., Toyama M., Terada T., Taiji M., Akiyama T., Yokoyama S. (2007) J. Biol. Chem. 282, 4238–4242 [DOI] [PubMed] [Google Scholar]
- 18.Mitin N., Betts L., Yohe M. E., Der C. J., Sondek J., Rossman K. L. (2007) Nat. Struct. Mol. Biol. 14, 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawasaki Y., Tsuji S., Sagara M., Echizen K., Shibata Y., Akiyama T. (2009) J. Biol. Chem. 284, 22436–22443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrara N., Kerbel R. S. (2005) Nature 438, 967–974 [DOI] [PubMed] [Google Scholar]
- 21.Shi W., Wang N. J., Shih D. M., Sun V. Z., Wang X., Lusis A. J. (2000) Circ. Res. 86, 1078–1084 [DOI] [PubMed] [Google Scholar]
- 22.Maffucci T., Piccolo E., Cumashi A., Iezzi M., Riley A. M., Saiardi A., Godage H. Y., Rossi C., Broggini M., Iacobelli S., Potter B. V., Innocenti P., Falasca M. (2005) Cancer Res. 65, 8339–8349 [DOI] [PubMed] [Google Scholar]
- 23.Bout D., Kühner A. L., Klimetzek V., Remold H. G., David J. R. (1981) Cell. Immunol. 63, 198–202 [DOI] [PubMed] [Google Scholar]
- 24.Folkman J. (1995) Nat. Med. 1, 27–31 [DOI] [PubMed] [Google Scholar]
- 25.Egami K., Murohara T., Shimada T., Sasaki K., Shintani S., Sugaya T., Ishii M., Akagi T., Ikeda H., Matsuishi T., Imaizumi T. (2003) J. Clin. Invest. 112, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao W. M., Seng W. L., Duquette M., Lawler P., Laus C., Lawler J. (2001) Cancer Res. 61, 7830–7839 [PubMed] [Google Scholar]
- 27.Lewis C. E., Pollard J. W. (2006) Cancer Res. 66, 605–612 [DOI] [PubMed] [Google Scholar]
- 28.Itoh R. E., Kiyokawa E., Aoki K., Nishioka T., Akiyama T., Matsuda M. (2008) J. Cell Sci. 121, 2635–2642 [DOI] [PubMed] [Google Scholar]
- 29.Birukova A. A., Alekseeva E., Mikaelyan A., Birukov K. G. (2007) FASEB J. 21, 2776–2786 [DOI] [PubMed] [Google Scholar]
- 30.Garrett T. A., Van Buul J. D., Burridge K. (2007) Exp. Cell Res. 313, 3285–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavard J., Gutkind J. S. (2006) Nat. Cell Biol. 8, 1223–1234 [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez E., Kou R., Michel T. (2006) J. Biol. Chem. 281, 3210–3216 [DOI] [PubMed] [Google Scholar]
- 33.Lee S. H., Kunz J., Lin S. H., Yu-Lee L. Y. (2007) Cancer Res. 67, 11045–11053 [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Paik J. H., Wang Z., Hla T., Wu D. (2005) Prostaglandins Other Lipid Mediat. 76, 95–104 [DOI] [PubMed] [Google Scholar]
- 35.Etienne-Manneville S., Hall A. (2003) Nature 421, 753–756 [DOI] [PubMed] [Google Scholar]
- 36.Etienne-Manneville S., Manneville J. B., Nicholls S., Ferenczi M. A., Hall A. (2005) J. Cell Biol. 170, 895–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.