Abstract
Regulation of gene expression in kinetoplastid mitochondria is largely post-transcriptional and involves the orchestration of polycistronic RNA processing, 3′-terminal maturation, RNA editing, turnover, and translation; however, these processes remain poorly studied. Core editing complexes and their U-insertion/deletion activities are relatively well characterized, and a battery of ancillary factors has recently emerged. This study characterized a novel DExH-box RNA helicase, termed here REH2 (RNA editing associated helicase 2), in unique ribonucleoprotein complexes that exhibit unwinding and guide RNA binding activities, both of which required a double-stranded RNA-binding domain (dsRBD) and a functional helicase motif I of REH2. REH2 complexes and recently identified related particles share a multiprotein core but are distinguished by several differential polypeptides. Finally, REH2 associates transiently, via RNA, with editing complexes, mitochondrial ribosomes, and several ancillary factors that control editing and RNA stability. We propose that these putative higher order structures coordinate mitochondrial gene expression.
Introduction
Unique gene expression mechanisms in kinetoplastid flagellates include U-insertion/deletion RNA editing by concerted cycles of cleavage, U-addition/removal, and ligation that can create hundreds of amino acid codons in most mitochondrial mRNAs (1, 2). The RNA editing core complex (RECC)4 contains 18–20 subunits (3–6), although a few subunits seem to exchange in substrate-specific variants of this complex (7). The RECC acronym was recently introduced by Simpson et al. (55). Editing complexes recognize partial helices between pre-mRNA and complementary guide RNAs (gRNAs) initially stabilized by a short “anchor” duplex (6, 8, 9). Substrate determinants for duplex binding and nuclease specificity (6, 10, 11) and substrate structure in solution (12–14) have been characterized.
Several accessory factors, mostly in multisubunit arrays, have been proposed to modulate RNA editing during catalysis, substrate production, or RNA turnover. The MRP complex has RNA annealing activity in vitro and may promote mRNA and gRNA pairing (15, 16). Post-transcriptional mRNA terminal 3′-poly(A)/(U) and gRNA 3′-poly(U) maturation are mediated by KPAP1 and RET1 complexes (17, 18). MRB1, TbRGG1, and GRBC complexes proposed to contain between 14 and 24 proteins (termed here MRB-related complexes) share several components, but their functional relationship remains unclear. Repression of a few common subunits inhibited RNA editing and in some cases also decreased the level of total gRNA. GRBC1 and GRBC2 co-purified with RECC subunits (18–24). MERS1, MRP, and RBP16 proteins were associated with mRNA stability (23, 25). RBP16 also stimulated RNA insertion in vitro (26, 27). DEAD-box mHel61 (also termed REH1) is the only predicted helicase known to impact RNA editing (28). Most of these proteins are likely to have additional roles outside editing. RNA helicases are common across species and typically multifunctional; however, only a few examples have been studied in mitochondria. This work characterized the protein and RNA interactions of a factor REH2 (Tb927.4.1500) that we initially found in native editing complexes of Trypanosoma brucei. REH2 has a conserved dsRNA-binding (dsRBD) and DExH-helicase domains and forms novel ribonucleoprotein complexes (RNPs) containing helicase activity, gRNA, and a protein array that overlaps with MRB-related complexes. The integrity of REH2 RNPs as well as their helicase and gRNA binding activities require the dsRBD. REH2 associates, via RNA, with RECC, a battery of accessory editing factors, and mitochondrial ribosomes; thereby, we propose that REH2 RNPs are integral components of RNA-linked supramolecular networks that orchestrate the expression of the mitochondrial genome.
EXPERIMENTAL PROCEDURES
TAP and RNAi Constructs and Site-directed Mutagenesis of REH2
A TAP-REH2 construct was made by PCR amplification of the entire open reading frame from procyclic genomic DNA using a proofreading thermostable polymerase mix (AccuTaq, Sigma) and cloning into the XhoI and BamHI sites of pLew79-ada-TAP. PCR-based site-directed mutagenesis was performed directly on this plasmid to alter the helicase motif I with oligonucleotides F-REH2-mI and R-REH2-mI and to delete the dsRBD with oligonucleotides F-dsRBD-Δ and R-dsRBD-Δ. An RNAi construct was obtained by cloning an REH2 fragment of 1665 bp into p2T7-177 (29). All constructs were confirmed by DNA sequencing, linearized with NotI, and transfected in procyclic 29-13 trypanosomes (30). REH2-TAP expression and RNAi were induced with tetracycline at 1 μg/ml.
Protein Purification and Analysis
Native editing complexes were purified by ion-exchange chromatography from mitochondrial extracts (3, 6), and TAP purifications were performed essentially as reported (31) with some modifications. Sedimentation fractions were obtained from freshly made mitochondria or whole-cell extracts in 10–30% glycerol gradients. Although our protocols to prepare the extracts include DNase I, a sample indicated in the text was subjected to an extra DNase treatment (DNA-free kit, Ambion) prior to sedimentation. Catalase and thyroglobulin were used as ∼10 S and ∼20 S markers, and Western blots of Tbmp45 (formerly termed REAP1) were used to determine the ∼40 S region (32). Affinity-purified REH2 antibodies were produced against the peptide CSHTPTTSAEAGGDS (Bethyl Laboratories, Inc). Immunoprecipitations of endogenous and ectopic REH2 used antibody-conjugated protein A-Dynabeads (Invitrogen). Ectopic REH2 was specifically immunopurified using anti-rabbit IgG Dynabeads (Invitrogen). All washes were performed at 150 mm KCl. For mass spectrometry analyses, the antibodies were cross-linked to the beads with 25 mm dimethyl pimelimidate in 0.2 m triethanolamine, pH 8.2.
Enzymatic Assays and Photocross-linking
The conditions and substrates to assay for full-round (33) and precleaved (1) editing were as described. Photoreactive substrates containing a single thio-U and 32P at the editing site were prepared (33, 34), and gRNA labeling was performed as reported (35). RNA helicase assays used a dsRNA substrate consisting of the pre-mRNA fragment A6-tag annealed to the cognate gRNA gA6[14] (9). The dephosphorylated mRNA was 5′-end-labeled with [γ-32P]ATP and annealed with a 10-fold excess of gRNA in RNA folding buffer (25 mm Tris-HCl, pH 8.0, 250 mm KCl, 10 mm Mg(OAc)2, 0.5 mm EDTA) by incubation at 95 °C for 10 min followed by a gradual return to room temperature over the course of 2 h. The annealed form was purified by native gel electrophoresis in an 8% polyacrylamide gel in 1× TBE supplemented with 10 mm Mg(OAc)2. The standard RNA helicase assay consisted of 50 cps (∼10 fmol) of dsRNA in 25 mm Tris-HCl, 22 mm KCl, 10 mm Mg(OAc)2, 0.5 mm EDTA, 3 mm dithiothreitol, 1 unit/μl RNase inhibitor, 1 mm ATP, 50 ng/μl bovine serum albumin, a 20-fold excess of an unlabeled trap single strand RNA that complements ∼33 bp of gRNA, and 10 μl of beads in a final volume of 20 μl. Reactions were incubated for 30–60 min at 26 °C with constant flicking to mix the beads. This was followed by addition of 4 μl of 6× stop solution as follows: 0.12% xylene cyanol, 0.12% bromphenol blue, 3% SDS, 125 ng/μl proteinase K, 17% glycerol, and incubation was at room temperature for 10 min. Samples were then loaded onto an 8% polyacrylamide gel, 1× TBE supplemented with 10 mm Mg(OAc)2. Protein RNA photocross-linking was performed as described (34) except that it was scaled up 10-fold. Denaturation of complexes was accomplished by the addition of SDS to 1% final and sequential incubations at 95 °C for 10 min and at 70 °C for 30–60 min. After allowing the sample to reach room temperature, Triton-X-100 was added to 5% and incubated for 10 min at room temperature. Samples were then passed through a gel filtration spin column (Bio-spin 6, Bio-Rad 732 6221) according to the manufacturer's instructions and immunoprecipitated as above.
RNase Treatment of TAP Purifications and Immunoprecipitations
Samples coupled to Dynabeads were treated with the following nucleases as indicated in the text at the given final concentration: RNase A (0.1 unit/μl), T1 (0.125 unit/μl), V1 (0.001/μl), and micrococcal nuclease (0.03 unit/μl) for 60 min in ice.
Mass Spectrometry
Trichloroacetic acid precipitates were resuspended in digestion buffer (100 mm Tris-HCl, pH 8.5, 8 m urea) and digested by the sequential addition of Lys-C and trypsin proteases as described previously (36). Digested samples were fractionated using a five-step on-line separation method during which peptides were eluted directly into an LTQ-Orbitrap mass spectrometer (Thermo Fisher) in which tandem mass spectra were collected (37–39). SEQUEST and DTASelect algorithms were used to identify peptide sequences from tandem mass spectra (40, 41). Proteins were considered present in a sample if at least two peptides were identified per protein using a peptide level false-positive rate of 5% as determined using a decoy data base strategy (42).
Note
Detailed protocols and oligonucleotide sequences used during this work are available upon request.
RESULTS
DExH-box Helicase Associates with RNA Editing Complexes
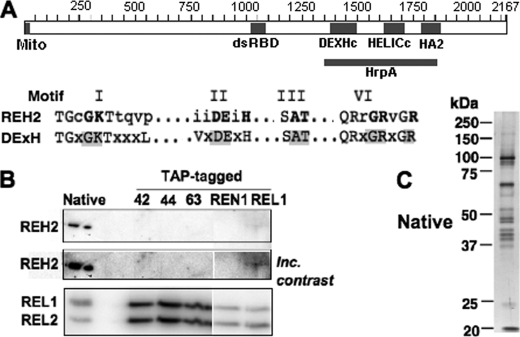
In a mass spectrometric analysis of native editing complexes purified from T. brucei mitochondria, we detected multiple unique peptides of most RECC subunits and the accessory MRP factors (6). However, we also found a single peptide for a 241-kDa protein, termed REH2, with highly conserved DExH-helicase domains, a dsRBD, and an N-terminal mitochondrial import sequence (Fig. 1A). Western blot analyses clearly detected an ∼250-kDa protein in native but not in TAP-isolated editing complexes (Fig. 1B), although a weak signal was apparent in TAP-REL1 complexes (see the middle panel). This suggested a transient interaction of REH2 with RECC that is disrupted during high stringency affinity purifications. Silver staining of native editing complexes did not evidently detect components near 250 kDa suggesting that REH2 may be substoichiometric relative to RECC subunits (Fig. 1C).
FIGURE 1.
REH2 gene organization and co-purification with native editing complexes. A, T. brucei REH2 has 2167 amino acids, including a conserved mitochondrial import signal (Mito), dsRBD, and domains typically associated with DExH-box helicases (drawn at scale): DExHc, HELICc, HA2, and HrpA. The first domain, which defines this protein family, contains six motifs, including the motif II signature amino acids DExH, where “X” is any residue. Four such motifs are evident in REH2 (I, II, III, and VI) and shown with the most conserved residues in gray (56). B, REH2 Western blot of native and TAP-purified editing complexes tagged at various subunits. The middle panel is shown at increased (Inc) contrast. Adenylylated ligase subunits REL1 and REL2 are shown as loading control. C, silver-staining of purified native editing complexes.
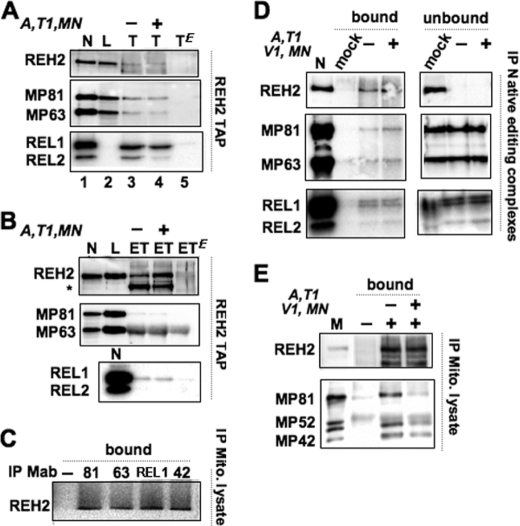
To confirm the physical interaction of REH2 with editing subunits, we expressed in procyclic trypanosomes the complete 6504-nucleotide REH2 gene with a C-terminal TAP tag (31) under the control of a tetracycline-inducible promoter (Fig. 2A). REH2 mRNA and protein increased 5- and 2–3-fold, respectively, at day 3 of induction (Fig. 2, B and C), and a slight reduction in cell growth was seen at day 5 (Fig. 2D). Consistent with its predicted mitochondrial localization (supplemental Fig. S1), REH2 was enriched in a mitochondrial lysate (Fig. 2E) (43). REH2 antibodies reacted well with purified native editing complexes and mitochondrial lysates but weakly with TAP pulldowns due to a partial occlusion of the tag and consequent low purification efficiency from mitochondrial lysates. A small amount of RECC was detected in both TEV (Fig. 3A) and concentrated EGTA eluates (Fig. 3B) but not in mock pulldowns from empty vector control cells (last lane in each panel). Interestingly, this low level of RECC in the eluates was resistant to a pretreatment with RNases A and T1 and micrococcal nuclease (MN). In a converse approach, REH2 was detected in mitochondrial extract pulldowns of several RECC subunits (Fig. 3C). Also, REH2 antibodies immunoprecipitated a small fraction of preisolated native editing complexes after an extensive RNase-MN treatment including the dsRNA-specific RNase V1. However, most RECC remained in the unbound fraction (Fig. 3D), suggesting that relatively few purified complexes were stably bound to REH2.
FIGURE 2.
REH2 expression in procyclic T. brucei. A, scheme of the C-terminal TAP tag and anti-(CBP) Western blot of total cell lysates with or without 3 days of induction with increasing tetracycline (Tc) concentrations. B, quantitative reverse transcription-PCR of REH2 normalized to 18 S rRNA and α-tubulin ± tetracycline. C, REH2 Western blot of induced empty-TAP vector (−) and TAP-REH2 cells for 1 and 3 days (d). Cytosolic α-enolase (α-enol) was used as control. Overexp., overexpression. D, growth curves of procyclic trypanosome cell lines expressing TAP constructs of wild-type REH2, dsRBD-Δ, and mot 1 mutants. E, Western blot showing REH2 enrichment in a mitochondrial (mito) lysate. Mitochondrial MP81 and cytosolic (cyto) α-enolase markers are shown.
FIGURE 3.
REH2 co-purification with RECC is largely sensitive to extensive nuclease treatments. REH2 TAP purification and detection of editing subunits before or after (+/−) treatments with RNases A, T1 (and V1 as indicated), and micrococcal nuclease (MN). A, TEV (T) eluates from IgG-Sepharose. B, EGTA (ET) eluates from calmodulin-agarose. Control lanes include native editing complex (N), diluted whole-cell lysate (L) and empty-vector controls (TE or ETE; last lane). EGTA eluates were concentrated by acetone precipitation. Immunoblots of REH2 and editing subunits and auto-adenylylation of REL ligases are shown. ATPases in whole-cell lysates often inhibit the latter activity (e.g. A, lane 2). MP63 may co-migrate with antibody cross-reacting bovine serum albumin added as precipitation carrier (see the ETE control lane). REH2 fragmentation (*) is observed in the eluates. C, mitochondrial lysate immunoprecipitated (IP) pulldowns with antibodies to RECC subunits (MP81, MP63, REL1, and MP42) or a mock reaction without antibodies. D, REH2 pulldowns from purified native editing complexes. E, REH2 pulldowns from mitochondrial (M) lysate (Mito lysate). Mock reactions used pre-immune serum. An unaccounted band near MP52 is visible in the lysate lane.
Besides the above examination of affinity-purified eluates and isolated native editing complexes, we further analyzed the REH2/RECC association in mitochondrial lysates. Importantly, although RECC subunits were present in nuclease-treated REH2 pulldowns, most RECC was released by the treatment (Fig. 3E). This suggests that the transient association observed is largely mediated by RNA. Also, in line with transient contacts, REH2 purifications exhibited some precleaved but not full-round editing activity, which is less sensitive due to a limiting cleavage step (supplemental Fig. S2).
Heterodisperse REH2-associated Complexes Are Stabilized by RNA and dsRBD
Sedimentation analyses of mitochondrial lysates showed significant heterodispersion of REH2 with a broad peak at ∼20–30 S. Western blots of these fractions with REH2 antibodies and the peroxidase anti-peroxidase reagent (to score ectopic REH2 only) showed that the tag does not affect the sedimentation of REH2 (Fig. 4A, top and middle panels). The RECC subunit MP63 was also dispersed, but in contrast to REH2 it was not detected in light fractions (Fig. 4A, bottom panel). Interestingly, REH2 at ∼20 S and >40 S co-immunopurified with RECC subunits (see Fig. 4D). A pretreatment of the mitochondrial lysate with RNases/MN disrupted most high S value REH2 complexes generating a discrete peak at ∼15 S, whereas a significantly sharpened peak of editing complexes remained at ∼20 S (Fig. 4B). As described above, RNase treatment eliminated most REH2 association with RECC (Fig. 3E). To examine the relevance of the conserved dsRBD, we expressed a construct with a deletion of the entire motif (dsRBD-Δ) (see supplemental Fig. S3). Notably, this construct severely compromised cell growth (Fig. 2D) and reduced the sedimentation of both endogenous and ectopic REH2 to an extent comparable with the RNase treatment (Fig. 4C). Finally, we established that DNA does not largely contribute to the observed broad sedimentation of REH2 in a sample treated with DNase (“Experimental Procedures”; data not shown). Thus, REH2 forms heterodisperse particles that include editing complexes and are stabilized by RNA and the dsRBD, as well as relatively low density particles that resist RNase.
FIGURE 4.
Heterodispersion of REH2 complexes requires RNA and the dsRBD. Glycerol gradients of mitochondrial lysates in Western blots of endogenous plus ectopic REH2 (REH2) or ectopic REH2 (peroxidase anti-peroxidase (PAP) reaction) and MP63 as a RECC marker. A–C, mitochondrial lysates untreated (A) or treated with RNases A, T1, and MN (B), or whole lysates of REH2 dsRBD-Δ cells (C). D, REH2-IPs of gradient fractions 3 (∼10 S), 7–8 (∼20 S), and 12–13 (>40 S) from panel A. IP, immunoprecipitation.
REH2 Co-purifies with gRNA in a Manner Dependent on the dsRBD and Helicase Motif I
Weng et al. (23) recently reported that GRBC complexes, which co-purified with REH2, bind gRNA. We determined whether REH2 immunopurified complexes associate with gRNA, and we further examined the importance of conserved domains of this protein. To this end, we analyzed IgG-Dynabead pulldowns of ectopically expressed REH2 wild type and mutant dsRBD-Δ or motif I (GK-to-AQ) (Fig. 1A). The motif I mutated residues have been associated with ATP binding and hydrolysis in other DExH proteins (44). Although a significant amount of total gRNA co-purified with wild-type REH2, little if any was associated with either mutant (Fig. 5, A and B). It is of interest, however, that relatively large RNA species accumulated in both mutants. Furthermore, as shown by Hashimi et al. (22), RNAi down-regulation of REH2 decreased the steady-state levels of gRNA (Fig. 5, C and D). Importantly, REH2 pulldowns from wild-type cells contained gRNA, demonstrating that endogenous REH2 RNPs bind gRNA and that this association is not an artifact of overexpression (Fig. 5, E and F). Thus, gRNA binding by REH2 RNPs requires the dsRBD and wild-type motif I of REH2.
FIGURE 5.
REH2 co-purification with gRNA requires the dsRBD and wild-type helicase motif I. A, IgG-Dynabead pulldowns (IP, immunoprecipitation) of whole-cell lysates expressing TAP-REH2: wild-type (w.t.) or mutants dsRBD-Δ (ds) and motif I (m-I). Western blots with REH2 antibodies or the peroxidase anti-peroxidase (PAP) reagent of pulldowns normalized for the amount of REH2. B, gRNA labeling assays with guanylyltransferase of the pulldowns in A. Whole-cell lysate (L) from wild-type cells or an REH2 pulldown from “empty vector” (ev) cells were used as controls. A few RNA species accumulated in the REH2 mutants (arrowhead). A typical artifact in guanylyltransferase assays (*) serves as loading control. C, Western blots of REH2 RNAi cells before or after the indicated days of tetracycline (Tc) induction. Mitochondrial MP81 and cytosolic α-enolase (α-eno) controls. D, gRNA labeling assays in REH2 RNAi cells before or after 6 days of tetracycline induction. E, Western blot of a REH2 pulldown from 29.13 cells lacking the ectopic REH2 construct. A mock reaction with preimmune serum and whole lysate (L) are controls. F, gRNA labeling of the pulldowns in E.
REH2 Is Associated with RNA Helicase Activity and Appears to Photocross-link with RNA
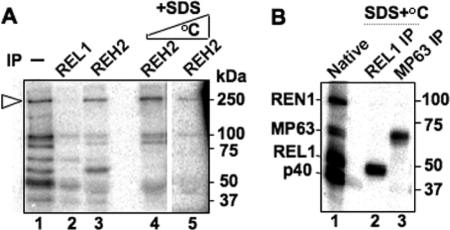
We examined the REH2 immunoprecipitated pulldown from mitochondrial extracts for possible RNA helicase activity and RNA photocross-linking. Notably, a model A6 pre-edited mRNA, preannealed with cognate gRNA, was efficiently unwound by the REH2 pulldown in a reaction requiring ATP hydrolysis at its β-γ phosphodiester linkage (Fig. 6A). Although the dsRNA substrate in these assays was gel-isolated, some dissociation is visible in input and mock lanes in the absence of REH2 complexes. A continuous duplex with 3′-overhangs was also unwound (Fig. 6B) but not an identical helix with 5′- or no overhangs (data not shown), consistent with the substrate specificity of the vast majority of SF2 RNA helicases with the exception of DEAD-box proteins (45). Importantly, REH2 pulldowns from the dsRBD-Δ and motif I cell lines had no detectable unwinding activity (Fig. 6C). We analyzed the sedimentation distribution of this helicase activity relative to REH2 and the DEAD-box helicase REH1. Interestingly, most helicase activity sedimented in fractions containing REH2 but away from REH1, which localizes at the top of the gradient in fractions 1–3 (Fig. 6, D–F) (28). An REH2 pulldown of fraction 8 exhibited unwinding activity (data not shown). Together with our above studies of REH2 mutants, these data suggest that REH2 is linked with the observed RNA helicase activity. To examine the possibility that REH2 may directly bind RNA, we cross-linked a protein fraction enriched in RECC (3) with the pre-mRNA/gRNA substrate described above but substituted with a photoreactive thio-U and 32P at the editing site (34). This protein fraction produced a cross-link at ∼250 kDa (Fig. 7A, lane 1) that was enriched in a pulldown with REH2 but not REL1 antibodies (lanes 2 and 3). Importantly, REH2 pulldowns of cross-linked reactions that were treated with SDS and increasing temperature to dissociate the RNPs further enriched the ∼250-kDa cross-link (Fig. 7A, lanes 4 and 5), suggesting that the reacting protein is REH2. As a proof of concept for the above denaturation protocol (Fig. 7B), we isolated the RNA photocross-linked RECC subunits REL1 (lane 2) and reported MP63 (lane 3) (6, 34) using specific antibodies against these proteins. Additional studies are needed to confirm that the ∼250-kDa cross-link represents direct binding by REH2, but these data suggest that REH2 contacts the RNA duplex near the photoreactive moiety in the model editing site (≤4 Å) (46).
FIGURE 6.
REH2 is associated with helicase activity. RNA helicase assays of REH2 pulldowns from mitochondrial lysate were supplemented with gel-isolated dsRNA (ds) substrates. A, A6 pre-mRNA and cognate gRNA with 1 mm ATP, ADP-CP, or no nucleotide (no nuc). IP, immunoprecipitation. The pre-mRNA was 5′-end-radiolabeled. The gRNA 3′-oligo(U) hybrid is presumably unstable (arrowheads). B, short duplex with symmetrical 3′-overhangs and 1 mm ATP. C, IgG-Dynabead pulldowns of TAP-REH2 wild-type (w.t.) and mutants (see Fig. 5 legend). RNA input before (−) and after boiling, and mock preimmune serum pulldowns from empty vector (e.v.) cell lysates are controls. D–F, sedimentation fractions of whole-cell lysate examined for helicase activity and Western blots of REH2 and REH1. nt, nucleotides.
FIGURE 7.
A ∼250-kDa protein in REH2 pulldowns photocross-links with RNA. A, cross-linking of an A6 pre-mRNA/gRNA pair bearing a single photoreactive thio-U at the editing site. Mitochondrial Q-Sepharose fraction enriched with RECC (lane 1) or immunoprecipitation (IP) pulldowns of this material by antibodies to REL1 or REH2 (lanes 2 and 3, respectively). Lanes 4 and 5 are repeats of lane 3 but after a treatment with 0.1% SDS at 70 and 90 °C, respectively, that enriches a cross-link at ∼250 kDa (arrowhead). The REL1 pulldown shows at least four reported cross-linking RECC subunits (6, 34) (see B). B, proof of concept of the denaturation protocol used in A, showing immunopurification of cross-linked RECC subunits. Cross-linked subunits of purified RECC (lane 1) and specific RECC subunits (lanes 2 and 3) after SDS dissociation at 90 °C and immunopurification with the indicated antibodies. The efficiency of this procedure depends on the antibodies affinity, polyclonal for REH2 and monoclonal for RECC subunits.
At Least Seven RNase/MN-resistant REH2-associated Proteins Are Also Present in MRB1, TbRGG1, and GRBC Complexes
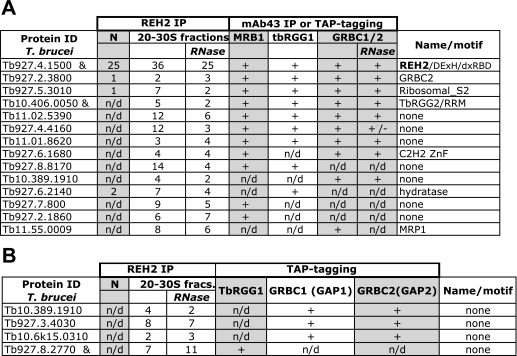
We performed mass spectrometric studies of REH2 immunoprecipitated pulldowns of both purified native editing complexes and 20–30 S sedimentation fractions (Fig. 4A, fractions 7–10), the latter before or after RNase/MN treatment. A large number of REH2 unique peptides were found in all pulldowns reflecting the relatively large size of this protein. The 20–30 S fractions contained 22 proteins (besides REH2) previously found in one or more of the reported MRB1, TbRGG1, and GRBC complexes (21, 23, 24). These complexes significantly overlap but also exhibit important compositional differences (Table 1 and supplemental Fig. S4 show RNase-resistant and RNase-sensitive REH2 interactions, respectively). Seven RNase-resistant proteins (out of 13) were common to the known MRB-related complexes, namely REH2, GRBC2, ribosomal S2 homolog, and TbRGG2 and the hypothetical proteins Tb11.02.5390, Tb927.4.4160, and Tb11.01.8620 (Table 1, A). Likely, GRBC1 is also in this group, but it was not detected with the RNase treatment implying that its proposed 1:1 ratio with GRBC2 in GRBC complexes (23) is not strictly conserved in the immunopurified REH2 complexes. Apparent differences with known MRB-related complexes include five proteins found in at least one of these complexes, but not in the REH2 pulldown (supplemental Fig. S4), and 15 RNase-resistant proteins exclusively found in the REH2 pulldown, including the putative helicase REH1 (mHel61; supplemental Fig. S5) (28) and hypothetical proteins with unrecognized motifs (supplemental Fig. S6). The TbRGG1-associated complex described by Hashimi et al. (21) consists of proteins detected in TAP purifications of all three TbRGG1, GAP1 (GRBC1), and GAP2 (GRBC2). Among the proteins from Hashimi et al. (21) that were absent in at least one of their purifications, we found four RNase-resistant REH2-associated proteins (Table 1, B). These four proteins were not found in MRB1 and GRBC complexes isolated by other laboratories (23, 24), except for Tb10.389.1910, which was present in the latter complex (see Table 1). Thus, some proteins may be preferential or even unique components of REH2 complexes, but this has to be confirmed by subsequent cross-tagging/purification studies. Thus, besides a common core of at least 7 to 8 polypeptides in REH2 and other MRB-related assemblies, there may be significant compositional differences among these novel multiprotein particles.
TABLE 1.
RNase-resistant REH2-associated proteins that were found in MRB1, TbRGG1, or GRBC1 complexes
A, mass spectrometric analyses of proteins were identified in REH2 antibody pulldowns from ∼20 to 30 S mitochondrial lysate fractions, and unique peptides (criteria, two peptides minimum) are shown before or after an extensive nuclease treatment with RNases A, T1, V1, and MN while proteins were bound to the beads. REH2 pulldowns of native editing complexes (N) and a low efficiency REH2 TAP purification (&) (criteria, one unique peptide minimum) are shown. Reported MRB1-immunopurified complex (using monoclonal 43 antibodies) and TAP-purified TbRGG1 and GRBC1/2 complexes (21, 23, 24) are shown. GRBC1/2 (+) indicates proteins found in both GRBC1 and GRBC2 purifications, and +/− indicates detection only in the GRBC1 purification. Some proteins listed as putative GRBC components by Weng et al. (23) are ribosomal proteins and were RNase-sensitive in the REH2 pulldowns (supplemental Fig. S8). B, RNase-resistant REH2-associated proteins that were not considered as subunits of MRB-related complexes but were found in independent TAP purifications are shown. Many proteins in a study by Hashimi et al. (21) were not present in at least one of the TAP purifications for TbRGG1, GBP1, and GBP2 (i.e. were not considered as TbRGG1 complex subunits). The identified REH2-associated components correspond to hypothetical proteins with unknown motifs (criteria, two unique peptides). Proteins in native editing complexes and an REH2 TAP purification (&) (criteria, one unique peptide) are shown. Some reported proteins are ribosomal components and were added to supplemental Fig. S8.
In contrast to the above examined sedimentation fractions, an REH2 pulldown of purified native editing complexes contained only six reported components of MRB-related complexes (including three RNase-resistant interactions). This could reflect differences in REH2 complex composition or relative abundance of REH2 in the samples examined, as the editing complexes had a somewhat smaller number of unique peptides (Table 1). Interestingly, the protein Tb927.5.2930 may represent an REH2-specific cofactor as it was detected in REH2 pulldowns of both native editing complexes and sedimentation fractions, but it was not found in other MRB-related complexes (supplemental Fig. S6). In fact, affinity purification of this protein contained REH2, helicase activity, and RECC subunits.5 Finally, REH2 co-purified with most known subunits of both RNA editing complexes and mitochondrial ribosomes (4, 47), largely via RNase-sensitive associations (supplemental Figs. S7 and S8). An association with mitochondrial ribosomes was not implied for other MRB-related complexes, although four ribosomal proteins were inadvertently included in a model of the GRBC complex core (23). Importantly, none of the protein components discussed above were found in mock immunopurifications or using an “empty”-TAP vector. Other RNase-resistant interactions in the pulldowns correspond to common metabolic proteins or proteins that were detected in our mock purifications (supplemental Fig. S9). Thus, the REH2 RNPs described here and previously reported MRB-related complexes are similar but not identical in composition and apparent RNA-linked interactions with other mitochondrial components.
DISCUSSION
RNA helicases may be the largest group of enzymes in RNA metabolism from bacteria to humans, and they are usually assembled in macromolecular RNPs such as spliceosomes and ribosomes. Helicases are often multifunctional within the same cell, and understanding the basis of their specificity, particularly the relevant cofactors and substrates, remains a major challenge in biology (44). REH2 is a novel ∼241-kDa factor in kinetoplastids that bears an unusual dual combination of DExH-box helicase and dsRBD motifs only previously observed in the eukaryotic RNA helicase A (48), and to our knowledge it represents the only reported example of a DExH-box helicase in mitochondria besides Suv3p in yeast and its orthologues (49). Recent RNAi studies by Hashimi et al. (22) and by our laboratory showed that repression of REH2 decreases RNA editing and total gRNA levels (Fig. 5, C and D, and data not shown). The REH2 complexes we described appear to be novel particles that overlap in composition with recently published “MRB-related complexes,” namely MRB1, TbRGG1, and GBRC complexes. These complexes exhibit compositional differences (Table 1), but they share a few subunits that resisted extensive RNase treatment in REH2 purifications, which we propose form a scaffold core for the assembly of more dynamic protein-protein and protein-RNA interactions. RECC binds REH2 and GRBC RNPs (23), but an association with MRB1 and TbRGG1 complexes was not detected, probably reflecting compositional or stoichiometric differences that control this interaction. TbRGG1 itself is bound via RNA (supplemental Fig. S4) (21).
Association of REH2 with RECC subunits was observed in REH2 antibody pulldowns of prepurified native editing complexes and in converse immunoprecipitations (Fig. 3 and Fig. 4E). Consistent with a transient interaction, (a) REH2 was readily detected in native but not in affinity-purified editing complexes; (b) TAP-REH2 purifications showed only a small amount of editing subunits after concentration of the final elutes, and (c) the majority of editing complexes appeared in the unbound fraction of REH2 pulldowns. A small fraction of REH2 consistently associated with editing proteins despite extensive treatments with MN and RNases A, T1, and V1, suggesting that at least some REH2 may directly bind RECC. The majority, however, was clearly RNA bridged (e.g. Fig. 3E and Fig. 4B). Alternatively, a high affinity RNA linker, not completely removed by protein purification and single-/double-stranded nucleases, could mediate the association with editing complexes. A TAP-GRBC co-purification with REL1 and RET1 editing subunits was partially sensitivity to RNase A and thus thought to involve transient contacts (23).
GRBC complexes reported by Weng et al. (23) and the REH2 complexes described here contain gRNA; however, we also found helicase activity in the REH2 pulldowns. Our data suggest that REH2 is directly responsible for gRNA binding and the helicase activity as both activities required the dsRBD and a wild-type motif I of REH2 (Fig. 6C). This helicase activity immunopurified with REH2 from sedimentation fractions that contained no visible REH1 (Fig. 6, D–F, and data not shown). Interestingly, Missel et al. (28) found a helicase activity of mitochondrial lysates, which largely sedimented away from REH1, localized at the top of the gradient. Consistent with the motif I mutation effect, the unwinding activity utilized ATP but not a nonhydrolyzable analog, and it dissociated a model pre- mRNA/gRNA hybrid and a continuous helix with 3′- but not 5′-overhangs. The molecular basis of this substrate selectivity remains to be studied; however, our data suggest that REH2 translocates in a 3′ to 5′ direction from a single-stranded loading region into a helical structure (50). We hypothesize that the dsRBD and DExH-helicase domains of REH2 act in concert to mediate selective binding of properly folded gRNAs. Helical elements of gRNA (51) may be targeted by the 70-amino acid dsRBD, although other REH2 sequences or its co-factors likely contribute to substrate specificity. Subsequent ATP-dependent winding and unwinding remodeling cycles may generate specific gRNA folds with increased affinity for REH2, thereby becoming protected from degradation. Thus, the conformation of gRNAs may be subject to regulation. Consistent with the idea that REH2 and other associated factors cooperate to provide gRNA-binding specificity, RNAi of REH2 and GRBC (GAP) factors severely decreased the gRNA steady-state levels (Fig. 5, C and D) (22, 23).
The precise compositional and functional relationship of REH2 complexes with known MRB-related complexes needs to be further studied. The number of RNase-resistant interactions that co-purified with REH2 contrasts with the significantly decreased S value of REH2 after RNase treatments or dsRBD deletion. To reconcile these seemingly discordant observations, we propose that REH2 may form multiple particles of variable composition or stoichiometry. In this line of thought, we indicated that related complexes, isolated via either REH2, GRBC, or TbRGG1 proteins, for example, share a common scaffold core but differ in multiple dynamic components. Such components could include specificity factors that link these particles with various aspects of mitochondrial gene expression. A putative collection of purified REH2 complexes may include most if not all proteins in Table 1 and potentially other RNase-resistant components we observed, but additional studies are underway to examine this further.
The broad heterodispersion of REH2 in sedimentation gradients likely reflects higher order assemblies linked by RNA that include REH2 RNPs, other MRB-related assemblies, RECC, and several factors, including KPAP1, MERS1, RET1, MRP, and PPR1. The pentatricopeptide protein PPR1 was associated with processing/stability of mitochondrial RNA, including editing and ribosomal transcripts, specifically with regulation of long poly(A) tails (52–54). Consistent with the above model: (a) these factors were found in REH2 pulldowns before but not after an extensive RNase treatment (supplemental Fig. S4); (b) either RNase or dsRBD deletion decreased the S value of REH2, and (c) REH2 co-immunoprecipitated with RECC subunits in ∼20 S and >40 S fractions (Fig. 4). Importantly, mitochondrial ribosomes were a major component of REH2 purifications via specific antibodies or TAP tagging (but were not found in mock preparations; supplemental Fig. S8), and their association was bridged by RNA. We found a few ribosomal proteins in reported TAP purifications of GRBP, KPAP1, MERS1, MRP, TbRGG1, and editing proteins (21, 23), although this had passed unnoticed as the composition of mitochondrial ribosomes was reported after these studies (47). Furthermore, Osato et al. (55) found rRNA in TAP isolations of editing complexes. Collectively, data by us and by others laboratories are consistent with a model whereby transient and dynamic RNA-interlinked networks in mitochondria functionally integrate and coordinate mitochondrial machineries in RNA maturation, stability, and translation (schematized in Fig. 8). In this context, REH2 may be multifunctional. Relevant to this model, and the integration of ribosomes in particular, is the presence of an S2 ribosomal homolog in the core of REH2 RNPs and other MRB-related complexes (Table 1, A; Fig. 8). Interestingly, S2 was not found in isolated mitochondrial ribosomes of T. brucei (47), and we speculate that this protein may help to functionally link MRB-related complexes with ribosomes. Finally, the multicomponent networks we propose may be similar to the L*b complexes recently detected in Leishmania using blue native gel electrophoresis by Osato et al. (55), and which the authors proposed represent the active holoenzyme or “editosome.”
FIGURE 8.

Proposed RNA-interlinked molecular networks in kinetoplastid mitochondria. Machineries and factors for RNA processing, stability, and translation that co-purify with REH2 RNPs via RNA, which we propose reflect transient supramolecular assemblies that coordinate mitochondrial gene expression. For simplicity, RNA bridges (double lines) irradiate from the center, but alternative or additional RNA and protein contacts could exist across components in the network (dotted lines). The arbitrary center of the model includes seven RNase-resistant proteins in the REH2 pulldown that are common to the known MRB-related complexes. They were abbreviated as follow: REH2 (H2), GRBC2 (GRBC), ribosomal. S2 homolog (S2), TbRGG2 (RGG2), and hypothetical proteins Tb11.01.8620 (8620), Tb11.02.5390 (5390), and Tb927.4.4160 (4160) (Table 1, A). At least GRBC1 may also be included in this protein array (see text).
Supplementary Material
Acknowledgments
We thank Paul Straight at Texas A&M University for the careful comments and discussion on the manuscript. We are also grateful to the following colleagues: Larry Simpson for T. brucei genomic DNA; Jason Carnes for advice on quantitative reverse transcription-PCR, and Ken Stuart for monoclonal antibodies against core subunits of editing complexes; H. Ulrich Goringer, Paul Michels, and Susan Madison-Antenucci for mHel61, α-enolase, and Tbmp45 antibodies, respectively; Kirk Gaston and Juan D. Alfonzo for their generous assistance in the subcellular localization studies; Achim Schanufer and Ruslan Aphasizhev for sharing their TAP purification protocols; and Neili Cooksey for assisting with the preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM067130 (to J. C.-R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
B. R. Madina and J. Cruz-Reyes, unpublished data.
- RECC
- RNA editing core complex
- dsRBD
- dsRNA-binding domain
- gRNA
- guide RNA
- RNP
- ribonucleoprotein
- RNAi
- RNA interference
- TAP
- tandem affinity-purified
- GRBC
- guide RNA binding complex
- MRB
- mitochondrial RNA binding
- MN
- micrococcal nuclease
- GAP
- guide RNA-associated protein
- TEV
- tobacco etch virus protease.
REFERENCES
- 1.Carnes J., Stuart K. D. (2007) Methods Enzymol. 424, 25–54 [DOI] [PubMed] [Google Scholar]
- 2.Cruz-Reyes J., Hernandez A. (2008) Protein-Protein and RNA-Protein Interactions in U-Insertion/Deletion RNA Editing Complexes in RNA and DNA Editing (Smith H. C. ed) pp. 71–98, John Wiley & Sons, Inc., New York [Google Scholar]
- 3.Rusché L. N., Cruz-Reyes J., Piller K. J., Sollner-Webb B. (1997) EMBO J. 16, 4069–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panigrahi A. K., Schnaufer A., Stuart K. D. (2007) Methods Enzymol. 424, 3–24 [DOI] [PubMed] [Google Scholar]
- 5.Aphasizhev R., Aphasizheva I., Nelson R. E., Gao G., Simpson A. M., Kang X., Falick A. M., Sbicego S., Simpson L. (2003) EMBO J. 22, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez A., Panigrahi A., Cifuentes-Rojas C., Sacharidou A., Stuart K., Cruz-Reyes J. (2008) J. Mol. Biol. 381, 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnes J., Trotter J. R., Peltan A., Fleck M., Stuart K. (2008) Mol. Cell. Biol. 28, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum B., Bakalara N., Simpson L. (1990) Cell 60, 189–198 [DOI] [PubMed] [Google Scholar]
- 9.Seiwert S. D., Stuart K. (1994) Science 266, 114–117 [DOI] [PubMed] [Google Scholar]
- 10.Cifuentes-Rojas C., Halbig K., Sacharidou A., De Nova-Ocampo M., Cruz-Reyes J. (2005) Nucleic Acids Res. 33, 6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cifuentes-Rojas C., Pavia P., Hernandez A., Osterwisch D., Puerta C., Cruz-Reyes J. (2007) J. Biol. Chem. 282, 4265–4276 [DOI] [PubMed] [Google Scholar]
- 12.Koslowsky D. J., Reifur L., Yu L. E., Chen W. (2004) RNA Biol. 1, 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reifur L., Koslowsky D. J. (2008) RNA 14, 2195–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu L. E., Koslowsky D. J. (2006) RNA 12, 1050–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller U. F., Lambert L., Göringer H. U. (2001) EMBO J. 20, 1394–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aphasizhev R., Aphasizheva I., Nelson R. E., Simpson L. (2003) RNA 9, 62–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aphasizhev R., Sbicego S., Peris M., Jang S. H., Aphasizheva I., Simpson A. M., Rivlin A., Simpson L. (2002) Cell 108, 637–648 [DOI] [PubMed] [Google Scholar]
- 18.Etheridge R. D., Aphasizheva I., Gershon P. D., Aphasizhev R. (2008) EMBO J. 27, 1596–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisk J. C., Ammerman M. L., Presnyak V., Read L. K. (2008) J. Biol. Chem. 283, 23016–23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acestor N., Panigrahi A. K., Carnes J., Zíková A., Stuart K. D. (2009) RNA 15, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimi H., Zíková A., Panigrahi A. K., Stuart K. D., Lukes J. (2008) RNA 14, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimi H., Cicová Z., Novotná L., Wen Y.Z., Lukes J. (2009) RNA 15, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng J., Aphasizheva I., Etheridge R. D., Huang L., Wang X., Falick A. M., Aphasizhev R. (2008) Mol. Cell 32, 198–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panigrahi A. K., Zíková A., Dalley R. A., Acestor N., Ogata Y., Anupama A., Myler P. J., Stuart K. D. (2008) Mol. Cell. Proteomics 7, 534–545 [DOI] [PubMed] [Google Scholar]
- 25.Vondrusková E., van den Burg J., Zíková A., Ernst N. L., Stuart K., Benne R., Lukes J. (2005) J. Biol. Chem. 280, 2429–2438 [DOI] [PubMed] [Google Scholar]
- 26.Pelletier M., Read L. K. (2003) RNA 9, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller M. M., Halbig K., Cruz-Reyes J., Read L. K. (2006) RNA 12, 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Missel A., Souza A. E., Nörskau G., Göringer H. U. (1997) Mol. Cell. Biol. 17, 4895–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickstead B., Ersfeld K., Gull K. (2002) Mol. Biochem. Parasitol. 125, 211–216 [DOI] [PubMed] [Google Scholar]
- 30.Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 31.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 32.Cruz-Reyes J., Sollner-Webb B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8901–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cruz-Reyes J. (2007) Methods Enzymol. 424, 107–125 [DOI] [PubMed] [Google Scholar]
- 34.Sacharidou A., Cifuentes-Rojas C., Halbig K., Hernandez A., Dangott L. J., De Nova-Ocampo M., Cruz-Reyes J. (2006) RNA 12, 1219–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blum B., Simpson L. (1990) Cell 62, 391–397 [DOI] [PubMed] [Google Scholar]
- 36.Florens L., Carozza M. J., Swanson S. K., Fournier M., Coleman M. K., Workman J. L., Washburn M. P. (2006) Methods 40, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 38.Wohlschlegel J. A. (2009) Methods Mol. Biol. 497, 33–49 [DOI] [PubMed] [Google Scholar]
- 39.Wolters D. A., Washburn M. P., Yates J. R., 3rd (2001) Anal. Chem. 73, 5683–5690 [DOI] [PubMed] [Google Scholar]
- 40.Eng J., McCormack A., Yates J. (1994) J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 41.Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elias J. E., Gygi S. P. (2007) Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 43.McManus M. T., Shimamura M., Grams J., Hajduk S. L. (2001) RNA 7, 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jankowsky E. (2007) Nature 449, 999–1000 [DOI] [PubMed] [Google Scholar]
- 45.Bleichert F., Baserga S. J. (2007) Mol. Cell 27, 339–352 [DOI] [PubMed] [Google Scholar]
- 46.Fabre A. (1990) in Photobiochemistry and Nucleic Acids (Morrison H. ed) pp. 379–425, John Wiley & Sons, Inc., New York [Google Scholar]
- 47.Zíková A., Panigrahi A. K., Dalley R. A., Acestor N., Anupama A., Ogata Y., Myler P. J., Stuart K. (2008) Mol. Cell. Proteomics 7, 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robb G. B., Rana T. M. (2007) Mol. Cell 26, 523–537 [DOI] [PubMed] [Google Scholar]
- 49.Margossian S. P., Li H., Zassenhaus H. P., Butow R. A. (1996) Cell 84, 199–209 [DOI] [PubMed] [Google Scholar]
- 50.Fairman M. E., Maroney P. A., Wang W., Bowers H. A., Gollnick P., Nilsen T. W., Jankowsky E. (2004) Science 304, 730–734 [DOI] [PubMed] [Google Scholar]
- 51.Hermann T., Schmid B., Heumann H., Göringer H. U. (1997) Nucleic Acids Res. 25, 2311–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mingler M. K., Hingst A. M., Clement S. L., Yu L. E., Reifur L., Koslowsky D. J. (2006) Mol. Biochem. Parasitol. 150, 37–45 [DOI] [PubMed] [Google Scholar]
- 53.Pusnik M., Small I., Read L. K., Fabbro T., Schneider A. (2007) Mol. Cell. Biol. 27, 6876–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koslowsky D. J. (2009) Trends Parasitol. 25, 252–255 [DOI] [PubMed] [Google Scholar]
- 55.Simpson L., Aphasizhev R., Lukes J., Cruz-Reyes J. (2009) Protist, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jankowsky E., Jankowsky A. (2000) Nucleic Acids Res. 28, 333–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.