Abstract
Members of the Hey family of transcriptional repressors are basic helix-loop-helix proteins that are thought to act downstream of Notch in diverse tissues. Although forced expression of Hey1, a target of Notch in myoblasts, is sufficient to recapitulate inhibitory effects of the pathway on differentiation, how Hey1 interferes with myogenic transcription has not been fully elucidated. We provide multiple lines of evidence that Hey1 does not target the intrinsic transcriptional activity of the skeletal muscle master regulator MyoD. Our results indicate instead that Hey1 is recruited to the promoter regions of myogenin and Mef2C, two genes whose induction is critical for myogenesis. Expression of Hey1 in C2C12 myoblasts correlates with reduced recruitment of MyoD to these promoters, arguing that Hey1 inhibits myogenesis by associating with and repressing expression of key myogenic targets.
Keywords: Development Differentiation/Muscle, Signal Transduction, Transcription, Transcription/Helix-loop-helix, Transcription/Target genes, Transcription/Tissue-specific Factors
Introduction
The Hey (Hesr/HERP/CHF) family of transcriptional repressors comprises three basic-helix-loop-helix proteins (Hey1, Hey2, HeyL) that function as downstream effectors of Notch signaling. As canonical direct targets of Notch (1–3), these factors have been implicated in the control of vascular and cardiac morphogenesis (4–9), neurogenesis (10), myogenesis (11), and inner ear and kidney development (12, 13).
Hey proteins have been studied extensively from a structure-function perspective. They dimerize with one another (and other basic helix-loop-helix proteins) via their helix-loop-helix motif and bind to E-box or N-box elements within DNA via their basic domain (14–16). In contrast to the closely related Hes family of inhibitors, which recruit corepressors such as TLE/Groucho via a C-terminal WRPW tetrapeptide, Hey factors lack this interaction domain and associate with the corepressors mSin3A, N-CoR, and histone deacetylase 1 by way of their basic domain (14). Hey family members also share a conserved Orange domain downstream of the basic helix-loop-helix motif, which is thought to mediate protein interactions and potentially serve as an extended dimerization interface (17). Despite the accumulation of this structural knowledge, the detailed molecular mechanisms by which Hey proteins function in specific biological contexts are still largely unknown.
Although Hey family members are capable of binding to DNA E-box or N-box elements in vitro, many reports have highlighted instead their apparent ability to physically associate with and antagonize the activity of other transcription factors. Hey1 has been shown to bind GATA1 and neutralize its ability to induce erythropoiesis (18), whereas all three Hey family members are capable of binding to GATA4/6 and inhibiting GATA-driven cardiac gene expression (19, 20). Inhibitory physical interactions between Hey proteins and Runx2 may prevent Runx-induced aortic calcification (21), whereas associations between Hey factors and Ptf1-p48 may account for Notch-directed inhibition of pancreatic exocrine differentiation (22). Other work has revealed Hey1 can act as a corepressor for androgen receptor and has linked nuclear exclusion of Hey1 to prostate cancer (23). By contrast, there have been only rare instances that implicate a role for DNA binding in Hey function. One report has presented evidence that Hey1 may target a specific E-box to shut off expression of p57 and maintain cell proliferation within the lens epithelium (24) and another has linked a single E-box within the interleukin-6 promoter to Hey1-mediated inhibition (25). More often, however, studies have demonstrated that binding to E-boxes within target promoters either does not occur or is not required for Hey-directed repression (19, 26–28).
Initial studies on Hey family members suggested that Hey1 might function in the regulation of skeletal muscle development, as it is expressed in the developing somites and limb buds (29, 30) and can inhibit the differentiation of C2C12 myoblasts in culture (11). More recent work has revealed important roles for Notch signaling in the control of both embryonic and post-natal skeletal myogenesis. Notch activity in the embryo is required to prevent the premature differentiation of myogenic precursors (31, 32), whereas in the adult it is induced upon injury and essential for the expansion of muscle stem cells or satellite cells (33, 34). The precise mechanisms by which Notch carries out these functions are not well understood. Our previous work has shown that ligand-mediated Notch signaling appears to act through multiple, redundant pathways to repress myogenesis (35). Of the two Hey proteins induced by Notch in myoblasts (Hey1 and HeyL), only Hey1 exhibited inhibitory activity. Interestingly, induction of Hey1 was not required for inhibition, consistent with the absence of overt muscle phenotypes in the Hey1 knock-out mouse (4). Rather than ruling out an important role for Hey1 in muscle, however, these findings likely reflect the fact that Hey1 functions together or in parallel with other inhibitors (e.g. MyoR or Id3) to mediate the effects of Notch. Indeed, genetic studies have provided a precedent for Hey1 working in collaboration with other transcription factors to control developmental processes. Specifically, the Hey1/Hey2 double knock-out mouse and the Hey1/HeyL double-knock-out exhibit vascular or cardiac phenotypes not observed in Hey1, Hey2, or HeyL single knock-out animals (4, 5). In systems where functional redundancy may be a defining feature, it is only by unraveling the modes of action of individual effectors that we can reach a complete understanding of the pathway as a whole.
With this rationale, we have investigated the question of how Hey1 inhibits skeletal muscle differentiation. On a transcriptional level, myogenesis is controlled by a group of four basic helix-loop-helix proteins known as muscle regulatory factors (MRFs); MyoD, Myf-5, myogenin, and MRF4. Upon dimerization with E-proteins (E12/E47, HEB, E2-2), muscle regulatory factors associate with E-box elements within target gene promoters and collaborate with a second family of transcription factors, Mef2, to activate muscle-specific gene expression (36). Previously, it was proposed that Hey1 functions by forming inactive heterodimers with the skeletal muscle master regulator MyoD (11). Contrary to this proposal, we present evidence that Hey1 does not repress the intrinsic transcriptional activity of MyoD but rather associates with chromatin in the vicinity of myogenic promoters to repress target gene expression.
EXPERIMENTAL PROCEDURES
Plasmids
G133-luciferase was provided by Vittorio Sartorelli (National Institutes of Health) and contains the 133-bp myogenin proximal promoter fused to luciferase (37). pcDNA3.1-Mef2C (α1β splice isoform) and 3x-Mef2-tk-luciferase were provided by Tod Gulick (Harvard Medical School). 3x-Mef2-tk-luciferase contains three copies of a Mef2 binding element fused to a minimal thymidine kinase (tk)2 promoter driving firefly luciferase (38). pEMSV-MyoD and 4RE-tk-luciferase were provided by Eric Olson (University of Texas). 4RE-tk-luciferase contains four copies of the muscle creatine kinase enhancer right E-box fused to a minimal tk promoter driving firefly luciferase (39). Mef2C-luciferase was generated by PCR amplification of the Mef2C proximal promoter (−158 to +106) from genomic DNA and insertion into the KpnI/BglII sites of pGL3-basic (Promega, Madison, WI). pcDNA3.1-TOPO- Hey1-V5 (C-terminal V5 epitope) was generated by inserting the full-length Hey1 cDNA (provided by Eric Olson) into the TOPO recognition site of pcDNA3.1D/V5-His-TOPO (Invitrogen). pcDNA3.1-Hey1-V5 (C-terminal V5 epitope) was generated by PCR subcloning the full-length Hey1 cDNA into the BamHI/EcoR1 sites of pcDNA3.1-V5/HisA (Invitrogen). pcDNA-Myc-Hey1 (N-terminal Myc epitope) has been described previously (28). G133-mutMef2-luciferase, G133-mutE1-luciferase, and G133-mutGATA-luciferase were generated by QuikChange-mediated mutagenesis of the G133-luciferase reporter construct. The Mef2 element was mutated from CTATATTTAT to CTATACTTTAT (40), the E1 element was mutated from CAGTTG to AATTCG, and the GATA element was mutated from ATTTATCT to ATTTATTG. CMV-E47 and pBABE-FLAG-Hey1 (N-terminal FLAG epitope) have been described (35, 41). DamID lentiviral vectors pLgw-RFC1-V5-EcoDam and pLgw-V5-EcoDam (42) were provided by Bas Van Steensel (Netherlands Cancer Institute). pLgw-MyoD-V5-EcoDam and pLgw-Hey1-V5-EcoDam were generated via the Gateway recombination system (Invitrogen). The MyoD and Hey1 cDNAs were first subcloned by PCR into the Gateway entry vector pENTR-3C. The resulting entry clones were then recombined using LR Clonase II with the lentiviral destination vector pLgw-RFC1-V5-EcoDam. pVSVG, pGag/Pol, and pRSV-REV were provided by Carl June (University of Pennsylvania). All plasmids generated by PCR were verified by sequencing.
Cell Culture
C2C12 myoblasts, C3H 10T1/2 fibroblasts, and 293T cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum supplemented with l-glutamine and penicillin-streptomycin (growth medium (GM)). For myoblast differentiation, cells were grown to near confluence and then shifted to Dulbecco's modified Eagle's medium containing 0.5% fetal bovine serum (differentiation medium (DM)).
Cell Transfections and Luciferase Assays
10T1/2 cells were transfected according to the FuGENE 6 protocol (Roche Diagnostics). For quantitative reverse transcription-PCR experiments, cells were seeded at a density of 5 × 104 cells per well in 6-well plates and transfected with a total of 1.5 μg of DNA per well (pcDNA3.1/V5-HisA empty vector was used to keep the total amount of DNA constant). Cultures were maintained in GM for 1 day post-transfection and then switched to DM for 24 h before isolation of RNA. For luciferase assays, cells were seeded at a density of 1 × 104 cells per well in 24-well plates and transfected with a total of 300 ng of DNA. Cultures were maintained for 1 day in GM post-transfection and then switched to DM for 24 h before harvesting lysates. Luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega). Transfections were normalized to Renilla luciferase (pRL-tk; Promega).
Quantitative Reverse Transcription-PCR
Total RNA was isolated from cultured cells using the RNeasy kit (Qiagen, Valencia, CA). 2 μg of RNA was used to generate cDNA with the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). TaqMan gene expression assays were employed for myogenin, cadherin-15, Mef2C, MyoD, and 18 S as an endogenous control (Applied Biosystems). 3% of a given cDNA reaction, 10 μl of 2× Taq Universal Mastermix, and 1 μl of the indicated 20× TaqMan assay were included in a 20-μl reaction volume per well. Reactions were performed in triplicate, and results were analyzed using SDS2.2 software (Applied Biosystems).
Co-immunoprecipitation Assays and Western Immunoblotting
For immunoprecipitations, 293T or 10T1/2 cells were harvested ∼48 h post-transfection by scraping into lysis buffer (20 mm Tris-HCl, pH 8.0, 100 mm KCl, 0.1% Nonidet P-40, 5 mm MgCl2, 10% glycerol) supplemented with freshly added protease inhibitor mixture (Roche Diagnostics), 10 mm sodium fluoride, and 400 μm sodium orthovanadate (14). Lysates were incubated on ice for 15 min and cleared by centrifugation. Protein concentrations were determined using the DC assay (Bio-Rad). Lysates were precleared in lysis buffer supplemented with 50 μl of protein A/G PLUS-agarose and 4 μg of normal rabbit IgG (Santa Cruz Biotechnology sc-2027, Santa Cruz, CA) for 2 h at 4 °C. 500–600 μg of precleared lysate was incubated with 2 μg of anti-MyoD (5.8A, Novocastra, Newcastle upon Tyne, UK), 2 μg of anti-E47 (G127-32, Pharmingen), or 2 μg of anti-Myc (#9E10, Santa Cruz Biotechnology, Santa Cruz, CA) antibody and 15 μl of protein A/G PLUS-agarose at 4 °C overnight. Immune complexes were washed 4× with lysis buffer, eluted in 2× SDS sample buffer, and boiled for 5 min before resolution by SDS-PAGE. Proteins were transferred to nitrocellulose and blotted with the following antibodies at the indicated dilutions: 1:5000 anti-V5 (Invitrogen), 1:1000 anti-E47 (Pharmingen G127-32), 1:500 anti-MyoD (5.8A, Novocastra), and 1:500 anti-Myc (9E10, Santa Cruz Biotechnology). After incubation with a 1:2000 dilution of horseradish peroxidase-conjugated anti-mouse secondary antibody (Amersham Biosciences), bands were visualized via the LumiLight or LumiLight-plus detection system (Roche Diagnostics).
For direct Western immunoblotting (Figs. 3B, supplemental Fig. S3A), cells were scraped into radioimmune precipitation lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with freshly added protease and phosphatase inhibitors as above. 25–50 μg of lysate was loaded per well. Blots were incubated with the following dilutions of primary antibodies: 1:500 anti-myogenin (M-225, Santa Cruz), 1:5000 anti-Mre11 (100-142G1, Novus NB, Littleton, CO), or 1:500 anti-GFP (sc-8334, Santa Cruz).
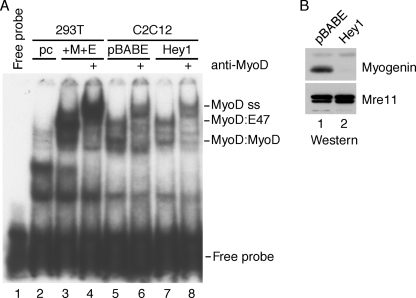
FIGURE 3.
Hey1 does not disrupt endogenous MyoD·E47 complexes in C2C12 cells. A, nuclear extracts were harvested from C2C12 cells stably transduced with pBABE-puro or pBABE-FLAG-Hey1 and maintained in DM for 24 h. Nuclear extracts were also isolated from 293T cells transiently transfected with either 6 μg of pcDNA3.1 (pc) empty vector or 2 μg of pEMSV-MyoD (M), 2 μg of CMV-E47 (E), and 2 μg of pcDNA3.1. Extracts were incubated with a 30-nucleotide 32P-labeled probe containing the high affinity MyoD E-box (CAGGTG) found within the Mef2C promoter and muscle creatine kinase enhancer. Protein-DNA complexes were incubated with or without MyoD-specific antibodies before resolution via non-denaturing SDS-PAGE. Supershifted MyoD-containing complexes are indicated MyoD ss. B, nuclear extracts from pBABE or pBABE-Hey1 C2C12 cells used in A were Western-blotted with antibodies specific for myogenin or Mre11 as a loading control.
Retroviral Infections
The infection of C2C12 myoblasts with pBABE-puro and pBABE-FLAG-Hey1 retroviruses has been described previously (35).
Electrophoretic Mobility Shift Assays (EMSAs)
Nuclear extracts for EMSAs were prepared from 293T or C2C12 cells using the NXTRACT CelLytic NuCLEAR extraction kit (Sigma). 293T cells were transiently transfected with either 2 μg of EMSV-MyoD and 2 μg of CMV-E47 or 4 μg of pcDNA3.1/Myc-HisC (Invitrogen) 48 h before harvesting extracts. C2C12 cells stably transduced with pBABE-puro or pBABE-FLAG-Hey1 were maintained in 0.5% serum for 24 h before extract isolation. Hey1-V5 was transcribed and translated in vitro using the TNT T7 Coupled Reticulocyte Lysate System (Promega). 32P-Labeled oligonucleotide probes containing the Mef2C E-box (43) or the Hey1 consensus target E-box (15, 16) were prepared by end-labeling annealed oligonucleotides with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). Labeled probes were purified through G-25 Quick Spin Sephadex Columns (Roche Diagnostics) as specified by the manufacturer. 8 μg of nuclear extract or 8 μl of transcribed and translated (TNT) lysate was incubated for 15 min at room temperature with 100,000 cpm of probe in a 15-μl binding reaction consisting of 0.2–1.0 μg of poly(dI·dC, 10 mm Tris, pH 7.5, 50 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, and 5.4% glycerol. Before the addition of probe, extract was preincubated in binding buffer at room temperature for 10 min. Where indicated, a 50× excess of cold competitor probe was added to the reaction. For supershifts, 1 μg of anti-MyoD (5.8A, Novocastra) or 1 μg of anti-V5 (Invitrogen) antibody was added to the sample and incubated for an additional 15 min. Binding reactions were run out on 5% non-denaturing polyacrylamide TBE Ready Gels (Bio-Rad) in 0.5× Tris borate-EDTA. Gels were dried and visualized by autoradiography. Oligonucleotide sequences used for the generation of labeled probes and cold competitors are as follows, with E-boxes or N-boxes underlined: MEF2C forward GAGTGACATGAACAGGTGCACCCTGGCCT; MEF2C reverse, AGGCCAGGGTGCACCTGTTCATGTCACTC; Hey1 consensus E-box (HCE) forward, TCCAATGGCACGTGCCACTGCC; HCE reverse, GGCAGTGGCACGTGCCATTGGA; ΔHCE forward, TCCAATGGGCCGTACCACTGCC; ΔHCE reverse, GGCAGTGGTACGGCCCATTGGA; E1 forward, CACCCAGCAGTTGGTGTGAG; E1 reverse, CTCACACCAACTGCTGGGTG; N1 forward, TGCCCTGTCCACCAGCTGCCTTG; N1 reverse, CAAGGCAGCTGGTGGACAGGGCA; E2 forward, GAAGGGGAATCACATGTAATCCACTG; E2 reverse, CAGTGGATTACATGTGATTCCCCTTC.
DamID
DamID assays were carried out essentially as described (42) with minor modifications. Briefly, lentiviral supernatants were harvested from 10-cm dishes of 293T cells on three consecutive days, 2 days after FuGENE 6-mediated transfection with 10 μg of the indicated pLgw lentiviral vector, 3.5 μg of pVSVG, 6.5 μg of pGag/Pol, and 2.5 μg of pRSV-REV. Supernatants were filtered (0.4 μm) to remove non-adherent 293T cells before storage at −80 °C. ∼18 h before infection, C2C12 cells were plated on 6-well plates at a density of 1 × 105 cells/well. Each well was incubated overnight with 1.5 ml of viral supernatant, diluted ∼2:1 in growth medium. After removal of virus, cultures were maintained for 2 days in GM and then switched to DM for an additional 24 h. Genomic DNA was isolated with the DNeasy Tissue kit (Qiagen). After ethanol precipitation of genomic DNA, DpnI digestion, ligation of adaptors, DpnII digestion, and ligation-mediated PCR (11 cycles of amplification in the final stage of PCR), samples were purified with Qiagen columns and diluted 1:60 in 10 mm Tris-Cl, pH 8.5, before quantitative PCR analysis (44). 8 μl of a diluted sample was mixed with 10 μl of 2× Power-SYBR Green mastermix (Applied Biosystems), 1 μl of 2 μm forward primer, and 1 μl of 2 μm reverse primer. PCR primers were first tested on genomic DNA via semiquantitative PCR to verify amplification of a single product of the expected size. Quantitative PCR reactions were also subjected to dissociation curve analysis. Primer sequences are as follows: Myog forward, GTGGACTGGCACAGGAGAAC; Myog reverse, GTGGACTTGGGACAAAGCAG; Mef2C forward, GAGAAGCAGAAAGGCACTGG; Mef2C reverse, CATTTCCAGCTCACTCATCATC; immunoglobulin heavy chain (IgH) forward, GTCATGTGGCAAGGCTATTTG; IgH reverse, TTTGCTCAGCCTGGACTTTC; GAPDH forward, CTCACGTCCCAACTCTCCAC; GAPDH reverse, GGCCTCCTATAGTATCCCTCCTC.
Primers for GAPDH and IgH are located directly within the proximal promoter and enhancer, respectively. Primers for Mef2C and myogenin are located ∼200 and ∼700 bp downstream of the transcriptional start sites, respectively, due to the unfavorable distribution of DpnI sites within the promoter regions; DpnI-generated fragments larger than 2 kb are not efficiently amplified in the ligation-mediated PCR step.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP was performed as previously described (45) with minor modifications. C2C12 cells stably transduced with pBABE-puro or pBABE-FLAG-Hey1 were seeded on 15-cm dishes at a density of 1.25–1.4 × 106 cells per plate, maintained in GM for 2 days, and then switched to DM for 40 h. Cultures were fixed in 1% formaldehyde for 10 min at room temperature, incubated for 5 min in 0.125 m glycine, washed twice in cold phosphate-buffered saline (PBS), and scraped into 3 ml of PBS. After a brief centrifugation, cell pellets were resuspended in 500 μl of cell lysis buffer (5 mm Pipes, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40) supplemented with 1 mm phenylmethylsulfonyl fluoride and protease inhibitor mixture (Roche Diagnostics), incubated on ice for 10 min, and Dounce-homogenized 15× to facilitate nuclei release. After a 5-min centrifugation at 5000 rpm, nuclei were resuspended in 300 μl of nuclei lysis buffer (50 mm Tris-Cl, pH 8.1, 10 mm EDTA, 1% SDS, 1 mm phenylmethylsulfonyl fluoride, protease inhibitor mixture) and incubated on ice for 10 min. Samples were sonicated in ice water using a Misonix 3000 sonicator for three 10-s intervals interrupted by 1-min rests followed by a 10-min centrifugation at 14,000 rpm at 4 °C. Supernatants were transferred to clean tubes, diluted 1:10 with dilution buffer (0.5% Triton X-100, 2 mm EDTA, 20 mm Tris-Cl, pH 8.1, 150 mm NaCl), and precleared with protein A/G-agarose single-stranded DNA (Upstate Biotechnology, Billerica, MA) for 2 h. 250 μg of precleared chromatin was incubated with 4 μg of normal rabbit IgG (Santa Cruz sc-2027, Santa Cruz, CA), 4 μg of anti-MyoD (Santa Cruz M-318X), or 4 μg of anti-RNA-Pol II (Santa Cruz Biotechnology H-224X) antibody overnight with rotation at 4 °C. Immune complexes were collected with bovine serum albumin-blocked protein A/G-agarose single-stranded DNA for 2 h. Beads were washed 8 times as follows: 2× with buffer 1 (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, pH 8.1, 150 mm NaCl), 2× with buffer 2 (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris, pH 8.1, 500 mm NaCl), 2× with buffer 1, 1× with buffer 3 (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris, pH 8.1), and 1× with Tris-EDTA. Washed beads were incubated twice in 150 μl of elution buffer (1% SDS, 100 mm NaHC03) for 15 min at 65 °C. Pooled eluates were treated with DNase-free RNase (Roche Diagnostics) and incubated at 65 °C overnight to reverse cross-links. After proteinase K treatment, phenol:chloroform extraction, and ethanol precipitation, samples were analyzed by quantitative PCR. 2 μl of a 50-μl sample was mixed with 10 μl of 2× Power-SYBR Green mastermix (Applied Biosystems), 6 μl of water, 1 μl of 2 μm forward primer, and 1 μl of 2 μm reverse primer. PCR primer sequences for the myogenin promoter and IgH enhancer have been published (46, 47). Primers for the Mef2C promoter are as follows: Mef2C forward2, GAGCAGTTCTGTGTTCTTTTGC; Mef2C reverse2, ATCCCTCTGCACAAGTGTCTG.
RESULTS
Hey1 Does Not Repress Intrinsic MyoD Transcriptional Activity
Past studies have demonstrated that Hey1 inhibits MyoD-mediated myogenic conversion of cultured fibroblasts and the differentiation of C2C12 myoblasts (11, 35). An important question relates to whether this inhibition reflects a generalized block to MyoD-driven transcription or, rather, a selective targeting of individual myogenic promoters. To address this, we performed transfection assays in 10T1/2 fibroblasts. When these cells are transfected with a vector expressing MyoD, they convert to a muscle phenotype and express a wide array of myogenic transcripts. We reasoned that if Hey1 were repressing the inherent ability of MyoD to activate gene expression, the induction of all targets downstream of MyoD should be compromised in the presence of Hey1. Two of the earliest markers activated in this system are the cell adhesion molecule cadherin-15 and the muscle regulatory factor myogenin. Although transfection of 10T1/2 cells with a MyoD expression plasmid robustly induced both of these genes as expected, co-expression of Hey1 strikingly inhibited the induction only of myogenin and not cadherin-15 (Fig. 1A). This apparent specificity in Hey1-mediated repression was also observed in luciferase reporter assays. Two reporter constructs were employed; one consisting of the proximal 133-bp myogenin promoter (G133-luciferase) and the other composed simply of four high affinity MyoD consensus E-box elements linked to a minimal promoter (4RE-tk-luciferase). 4RE-tk-luciferase has been used previously as a readout of “pure” MyoD activity, as this construct does not contain binding sites for any MyoD cofactors, such as Mef2 (39). Although MyoD robustly induced both reporters, Hey1 repressed the induction only of G133-luciferase and not 4RE-tk-luciferase (Fig. 1B). Together, these results strongly suggest that the Hey1 inhibitory effects on myogenesis reflect promoter-specific repression of select myogenic targets rather than generalized inhibition of MyoD activity.
FIGURE 1.
Hey1 exerts promoter-specific repression of myogenesis. A, 10T1/2 cells were transfected with 30 ng of pEMSV-MyoD alone or in combination with increasing amounts of pcDNA3.1-Hey1-V5 (60–180 ng). Myogenin (left) and cadherin-15 (right) RNA levels were determined by quantitative reverse transcription-PCR and plotted as the average of two replicate samples ±S.D. B, 10T1/2 cells were transfected with 25 ng of pRL-tk-Renilla and 25 ng of G133-luciferase (left) or 25 ng of 4RE-tk-luciferase (right), 25 ng of pEMSV-MyoD, and 25–100 ng of pcDNA3.1-Hey1-V5. Firefly luciferase values were normalized to Renilla luciferase and plotted as averages of three replicate samples ±S.D.
Hey1 Does Not Form Dimers with MyoD or E47 or Disrupt Endogenous MyoD·E47 Complexes
Although the above functional data revealed that not all MyoD-responsive promoters are subject to repression by Hey1, a past report had proposed that Hey1 targets MyoD itself by sequestering it into inactive heterodimers (11). To revisit this proposal, we evaluated whether Hey1 associates with MyoD or its binding partner E47 to repress myogenic transcription. We first performed co-immunoprecipitation assays in which we overexpressed either MyoD in combination with E47 or Hey1 (Fig. 2A) or E47 in combination with MyoD or Hey1 (Fig. 2B). 293T cells were used for these experiments, as they allow for very high transfection efficiencies and levels of expressed proteins, raising the likelihood that even weak interactions can be detected. These studies confirmed the expected binding of MyoD to E47 (Fig. 2, A, lane 5, B, lane 11) but failed to reveal any evidence of association between either of these factors and Hey1 (Fig. 2, A, lane 6, and B, lane 12). The ability of Hey1 to dimerize with itself was verified independently using the Hey1 construct employed in these assays in combination with a Myc-tagged Hey1 (supplemental Fig. S1). Similar results were obtained in 10T1/2 fibroblasts (supplemental Fig. S2).
FIGURE 2.
Hey1 does not form heterodimers with MyoD or E47. A, 293T cells were transfected as indicated with 2 μg of pEMSV-MyoD alone or in combination with 2 μg of CMV-E47 or 2 μg of pcDNA-3.1-TOPO-Hey1-V5. Lysates were harvested after 48 h and subjected to immunoprecipitation (IP) with MyoD-specific antibodies. Immunoprecipitates and input samples were immunoblotted with anti-V5, anti-E47, or anti-MyoD antibodies. B, 293T cells were transfected as indicated with 2 μg of CMV-E47 alone or in combination with 2 μg of pEMSV-MyoD or 2 μg of pcDNA-3.1-TOPO-Hey1-V5. Lysates were harvested after 48 h and immunoprecipitated with E47-specific antibodies followed by immunoblotting as in A. HC, Ig heavy chain.
In parallel, we used EMSAs to determine whether forced expression of Hey1 in C2C12 myoblasts would disrupt the formation of endogenous MyoD·E47 complexes. Nuclear extracts were prepared from C2C12 cells stably transduced with either the pBABE-puro or pBABE-FLAG-Hey1 retrovirus. As a control, we examined the extracts of control 293T cells or 293T cells co-transfected with MyoD and E47 expression plasmids. When these 293T extracts were incubated with a radiolabeled probe containing a high affinity E-box, complexes corresponding to MyoD·E47 heterodimers were readily observed (Fig. 3A, compare lanes 2 and 3). A MyoD antibody shifted the mobility of the complexes, confirming that they contain MyoD (lane 4). Similar complexes were observed in the C2C12 cell extracts (Fig. 3A, lanes 5–8). Importantly, these complexes were not appreciably affected by the presence of Hey1 (compare lanes 5 and 6 to lanes 7 and 8). The complex migrating just below what we tentatively identify as MyoD homodimers in extracts from the pBABE-transduced cells was not shifted by a MyoD antibody but was shifted with a myogenin antibody (data not shown). As expected, this complex was not observed in extracts from Hey1-expressing cells, consistent with the lack of myogenin protein (Fig. 3B). Taken together, our results indicate that Hey1 does not disrupt MyoD·E47 heterodimers, a finding fully consistent with our initial functional data demonstrating promoter-specific repression by Hey1.
Role for Mef2C Inhibition in Hey1-mediated Repression of Myogenesis
Our previous results (Fig. 1) showed that Hey1 inhibits MyoD-mediated induction of the endogenous myogenin gene and a transfected myogenin promoter but not the endogenous cadherin-15 gene or a transfected E-box-driven reporter. We sought to investigate the basis for this specificity. Past work in cultured fibroblasts by Tapscott and co-workers (46) demonstrated that cadherin-15, but not myogenin, is transcriptionally induced by MyoD in the absence of new protein synthesis. This finding suggested that MyoD must collaborate with a secondary mediator or coactivator to activate myogenin transcription. Indeed, other studies revealed that myogenin induction, which is absolutely required for myogenesis in vivo, requires both MyoD and Mef2 proteins (40, 48, 49). Mef2C is itself a target of MyoD (43, 50) and has been shown to be the only Mef2 family member transcriptionally up-regulated upon differentiation in C2C12 cells (35). We considered the possibility that Hey1 represses myogenesis primarily by repressing Mef2C activity and/or Mef2C gene transcription, not the myogenin promoter per se. To explore this possibility, we first confirmed that Mef2 is critical for myogenin promoter activity. Indeed, mutation of the single Mef2 element within the myogenin proximal promoter dramatically reduced MyoD-stimulated reporter activity (Fig. 4A). We next asked if Hey1 affects Mef2C activity. We transfected 10T1/2 cells with a Mef2C expression vector along with a reporter consisting of three Mef2 DNA binding elements upstream of a minimal promoter driving luciferase. Induction of this reporter by Mef2C was unaffected by increasing amounts of Hey1, indicating that Hey1 does not inhibit Mef2C transcriptional activity per se (Fig. 4B). By contrast, transcriptional induction of the endogenous Mef2C gene by MyoD was significantly repressed by Hey1 (Fig. 4C), a result consistent with those obtained with Hey1-transduced C1C12 cells (35). We further demonstrated that Hey1 repressed MyoD-mediated induction of a minimal 158-bp Mef2C promoter in luciferase reporter assays (Fig. 4D), suggesting that Hey1 likely targets the Me2C proximal promoter to inhibit gene expression.
FIGURE 4.
A role for inhibition of Mef2C expression in repression of myogenin by Hey1. A, 10T1/2 cells were transfected with 25 ng of pRL-tk-Renilla, 25 ng of G133-luciferase or G133-mutMef2-luciferase, and 25 ng of pEMSV-MyoD. Firefly luciferase values were normalized to Renilla luciferase and plotted as the averages of three replicate samples ±S.D. B, 10T1/2 cells were transfected with 25 ng of pRL-tk-Renilla, 25 ng of 3×-Mef2-tk-luciferase, 25 ng of pcDNA3.1-Mef2C (α1β), and 25–100 ng of pcDNA3.1-Hey1-V5. Firefly luciferase values were normalized to Renilla luciferase and plotted as the averages of three replicate samples ±S.D. C, 10T1/2 cells were transfected with 30 ng of pEMSV-MyoD alone or in combination with 180 ng of pcDNA3.1-Hey1-V5. Mef2C RNA levels were determined by quantitative reverse transcription-PCR and plotted as the average of two replicate samples ±S.D. D, 10T1/2 cells were transfected with 25 ng of pRL-tk-Renilla, 25 ng of Mef2C-luciferase, 25 ng of pEMSV-MyoD, and 25–100 ng of pcDNA3.1-Hey1-V5. Firefly luciferase values were normalized to Renilla luciferase and plotted as averages of three replicate samples ±S.D. E, 10T1/2 cells were transfected with 25 ng of pRL-tk-Renilla, 25 ng of G133-luciferase, 25 ng of pEMSV-MyoD, 25–100 ng of pcDNA3.1-Hey1-V5, and 100 ng of pcDNA3.1-Mef2C (α1β) as indicated. Firefly luciferase values were normalized to Renilla luciferase and plotted as the averages of three replicate samples ±S.D.
If Hey1 functions primarily by repressing Mef2C transcription, then one would expect exogenous Mef2C to rescue Hey1-mediated repression of myogenin. Reporter assays revealed that repression of the myogenin promoter was reduced but not eliminated by a Mef2C expression plasmid (Fig. 4E). These data suggest that inhibition of Mef2C expression likely contributes to Hey1-mediated repression of myogenesis but that Hey1 may also function through additional inhibitory mechanisms.
Evaluation of in Vitro DNA Binding by Hey1 to Myogenic Promoter Elements
We next asked if Hey1 inhibits transcription by binding DNA within target gene promoters. As a first step, we determined the ability of Hey1 to bind elements within the myogenin and Mef2C proximal promoters in vitro. Prior studies employing SELEX approaches derived an optimum binding site for Hey1, which is the E-box CACGTG. Closely related variants of this sequence bind Hey1 less well (14–16). The 133-bp myogenin promoter contains a single E-box (E1, CAGTTG), with an additional E-box (E2, CACATG) and N-box (N1, CACCAG) located within the 400 bp proximal to the start site. The Mef2C minimum promoter contains a single E-box (2C, CAGGTG). We performed EMSAs using in vitro TNT Hey1-V5 and a labeled probe containing the HCE. The Hey1 complex ran with the same mobility as a complex present in TNT lysates, so we evaluated Hey1 binding after shifting the complex to a slower mobility with an anti-V5 antibody (Fig. 5A, lanes 1–3). As expected, the addition of cold HCE competitor, but not mutant HCE, completely eliminated the binding of Hey1 to labeled probe (lanes 4 and 5). The addition of competitor DNA containing the various E-boxes and N-box found in the myogenin and Mef2C promoters only marginally reduced formation of the Hey1 complex despite a 50× molar excess of cold DNA (Fig. 5A, lanes 6–9). Other reports have also failed to show the association of Hey1 with the high affinity MyoD E-box (CAGGTG) (15, 16). These data argue against robust DNA binding by Hey1 to known control elements within these myogenic promoters. Consistent with this conclusion, when we mutated the E1 site within the myogenin promoter and carried out reporter assays in 10T1/2 cells, Hey1 still repressed MyoD-induced activity, indicating that inhibition is independent of this particular E-box (Fig. 5B). Efforts to map Hey1-responsive elements in the myogenin promoter distinct from those necessary for induced activity were unsuccessful (data not shown).
FIGURE 5.
Evaluation of in vitro DNA binding of Hey1 to myogenic promoter elements. A, TNT lysates programmed with either pcDNA3.1-V5/HisA empty vector or pcDNA3.1-TOPO-Hey1-V5 were incubated with a 22-nucleotide 32P-labeled probe containing the Hey1 consensus target E-box (CACGTG). Anti-V5 antibodies and cold competitor probes (50× excess relative to labeled probe) were added as indicated before resolution of complexes by non-denaturing SDS-PAGE.; ΔHCE, mutant Hey1-consensus E-box; E1, E2, E-boxes within the myogenin proximal promoter; N1, N-box ∼400 bp upstream of myogenin start site; 2C, high affinity MyoD E-box within the Mef2C proximal promoter. B, 10T1/2 cells were transfected with 25 ng of pRL-tk-Renilla, 25 ng of G133-luciferase or G133-mutE1-luciferase, 25 ng of pEMSV-MyoD, and 25–100 ng of pcDNA3.1-Hey1-V5. Firefly luciferase values were normalized to Renilla luciferase and plotted as the averages of three replicate samples ±S.D.
Evidence for Association of Hey1 with the Myogenin and Mef2C Promoters in Vivo
Despite the above findings in vitro, we sought to determine whether Hey1 associates with the myogenin and Mef2C promoters in vivo. Multiple rounds of ChIP assays failed to provide evidence of Hey1 recruitment; accordingly, we turned to DamID, an assay that is better suited for detecting potentially weak or indirect interactions with DNA in vivo (51, 52). In this assay a protein of interest is fused with the bacterial DNA adenine methyltransferase (Dam) and then expressed at low levels in mammalian cells. If the chimeric protein associates with particular regions of DNA, then only the targeted DNA becomes methylated as mammalian cells lack Dam. Methylated regions are revealed on the basis of cutting by the methylation-specific restriction enzyme DpnI and subsequent amplification of digested fragments (42, 44). This assay is capable of detecting even transient and weak interactions between the chimeric proteins and their target DNA.
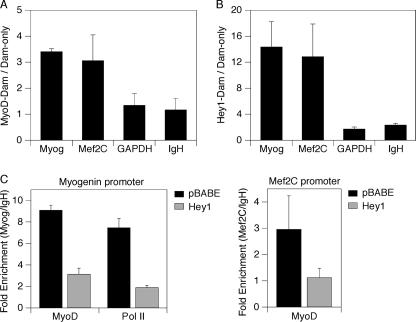
We generated lentiviruses that express MyoD-Dam and Hey1-Dam. Expression and activity of the fusion proteins were confirmed with Western immunoblots and reporter assays, respectively (supplemental Fig. S3). When transiently transfected, the pLgw-based lentiviral vectors employ a strong CMV promoter to drive high level expression. This promoter is deleted after infection and integration, and subsequent low-level expression from the virus DNA is controlled by an un-induced heat shock promoter. We infected C2C12 cells with individual lentiviruses, and genomic DNA was isolated and processed (see “Experimental Procedures”). Amplified (i.e. methylated) DNA was interrogated for genes of interest using quantitative PCR. Cells infected with a Dam-only virus were used to control for background methylation. As expected, cells infected with the MyoD-Dam virus gave rise to a ∼3-fold relative increase in methylation in the vicinity of the myogenin and Mef2C promoters but not in the vicinity of the GAPDH promoter or IgH enhancer (Fig. 6A). Importantly, cells infected with the Hey1-Dam virus also generated significantly increased relative methylation (∼12-fold) in the vicinity of the myogenin and Mef2C promoters (Fig. 6B). These data indicate that Hey1 associates specifically with the myogenin and Mef2C genes, and this likely leads to their transcriptional repression.
FIGURE 6.
Hey1 association with myogenic promoter regions in vivo correlates with reduced recruitment of MyoD. C2C12 cells were infected with pLgw-V5-EcoDam, pLgw-MyoD-V5-EcoDam, or pLgw-Hey1-V5-EcoDam lentiviruses, maintained for 2 days in GM, and then switched to DM for 24 h. Genomic DNA was harvested, subjected to the DamID protocol, and analyzed by quantitative PCR using primers in proximity to the promoter regions of myogenin, Mef2C, and GAPDH or within the IgH. PCR values are presented as ratios of the MyoD-Dam (A) or Hey1-Dam (B) signal to the Dam-only signal and represent averages of two independent experiments ± S.D. C, C2C12 cells stably transduced with either pBABE-puro or pBABE-FLAG-Hey1 retrovirus were switched to DM for 40 h before fixation and harvesting for ChIP assays. Chromatin was immunoprecipitated with IgG or antibodies specific for MyoD or RNA Pol II. Samples were analyzed by quantitative PCR using primers specific for the myogenin promoter (left) or Mef2C promoter (right) and IgH enhancer. Anti-MyoD to IgG or anti-Pol II to IgG ratios at the myogenin and Mef2C promoters were normalized to those at the IgH enhancer and presented as averages of two independent experiments ± S.D.
We next asked if targeting of Hey1 has consequences for the recruitment of MyoD. We transduced C2C12 cells with either a pBABE-puro or a pBABE-FLAG-Hey1 retrovirus and switched the cultures from growth medium to differentiation medium for 40 h. We then used ChIP to evaluate the recruitment of MyoD in each group of cells. We observed a reduction in MyoD recruitment in cells transduced with FLAG-Hey1 relative to those harboring the parental virus (Fig. 6C). As expected, Pol II recruitment was also reduced at the myogenin promoter (Pol II was not detected at the Mef2C promoter for unknown reasons). Only negligible recruitment of either factor was observed at the silent immunoglobulin heavy chain enhancer, which was used to normalize the data. Reduced MyoD recruitment in vivo is not likely to result from the down-regulation of MyoD expression by Hey1, as MyoD RNA levels were only marginally reduced (supplemental Fig. S4), and MyoD·E47 heterodimers were still observed in Hey1-expressing C2C12 cells (Fig. 3A). We conclude that forced expression of Hey1 results in compromised recruitment of the master regulator MyoD to its target gene promoters.
DISCUSSION
Notch signaling plays a critical role in embryonic and post-natal myogenesis (31–34); however, the molecular mechanisms by which the pathway represses myogenic transcription are not well defined. Our past work demonstrated that ligand-mediated Notch activity induces the expression of dozens of target genes in cultured myoblasts (35). The emerging picture is one of redundancy in which multiple Notch targets (e.g. the transcriptional repressors Hey1 and MyoR) are each capable of phenocopying the pathway effects. Although no single target may be essential, only by elucidating how these individual effectors impact myogenesis can we reach a complete functional understanding of the pathway as a whole. In this report we focused our analysis on Hey1, a canonical target of Notch and a known inhibitor of myogenic differentiation. We propose that Hey1 functions primarily by binding in the vicinity of the myogenin and Mef2C promoters to shut off target gene expression.
We present three independent and complementary lines of evidence that support this model and argue against a proposal that Hey1 sequesters MyoD into inactive heterodimers (11). First, MyoD-mediated induction of a direct MyoD target gene (cadherin-15) or of an E-box-driven reporter (4RE-tk-luc) was resistant to repression by Hey1. Second, MyoD did not form heterodimers with Hey1 under the conditions of our co-immunoprecipitation assays, yet readily formed heterodimers with E47 as expected. Importantly, Hey1 did form homodimers. Third, MyoD·E47 heterodimers were unaffected in cells expressing Hey1 despite the inability of these cells to differentiate. Although the basis for the discrepancy with a previous study (11) is currently unknown, it should be noted that Chin and co-workers (11, 26) employed an N-terminal Myc-tagged MyoD construct in their co-immunoprecipitation assays, whereas our own studies in two independent cell lines used an untagged version of MyoD. It remains possible that the tagged MyoD protein exhibits altered dimerization properties. Our functional and biochemical data in combination strongly argue that Hey1 does not repress the intrinsic ability of MyoD to activate transcription.
Because Hey1 repressed the induction of myogenin but not cadherin-15, we sought to determine the basis for this promoter specificity. Past work had implicated the Mef2 family of transcription factors as critical mediators of myogenin induction, in particular Mef2C (40, 43, 50). Although Hey1 did not inhibit Mef2C activity, it did repress Mef2C expression, suggesting that repression of myogenin might be due to the lack of Mef2C protein. However, forced expression of Mef2C only partially restored myogenin promoter activity in the presence of Hey1, suggesting additional mechanisms. Indeed, DamID assays showed that Hey1 associates with both the Mef2C and myogenin promoter regions, indicating that its ability to inhibit myogenesis may be due to the repression of multiple myogenic loci.
The resolution of DamID has been reported to be ∼1 kb at best (53) and, thus, cannot be used to identify specific binding sites occupied by Hey1 within these promoters. Our luciferase reporter assays suggest that Hey1 targets the myogenin and Mef2C proximal promoters, as minimal promoter fragments were sensitive to repression. Our EMSA data argue that Hey1 is not likely to bind directly to E-boxes or N-boxes at these loci, leaving open the possibility that Hey1 may associate instead through distinct DNA-bound regulatory proteins. Interestingly, the myogenin and Mef2C promoters contain conserved GATA sites, and a precedent exists for interactions between Hey1 and GATA proteins (18–20). We considered the possibility that Hey1 functions in conjunction with GATA3, which is induced by Notch in myoblasts (35)3 as well as other cell types (58, 59). However, our functional studies argue against this model, as mutation of the GATA binding site within the myogenin proximal promoter did not alleviate repression by Hey1 (data not shown). Aside from MyoD and Mef2, additional transcription factors such as Pbx1b/Meis1, MSY-3, Six1/4, and Ski also participate in the complex transcriptional control of the myogenin locus (54–57). We did not detect evidence of physical or functional interaction between Hey1 and Six1 (data not shown), yet Hey1 may interact with any of these other proteins present at the two promoters.
The presence of Hey1 correlates with reduced MyoD recruitment, yet it does not significantly affect MyoD expression. How this occurs is a matter of speculation. Hey proteins have been shown to bind histone deacetylases in vitro (14), but many studies have indicated Hey-mediated repression is insensitive to trichostatin A (18, 19, 21, 28), calling into question the relevance of such associations. We found that Hey1 did not repress the thymidine kinase promoter when linked with an upstream myogenin promoter (data not shown). This suggests that Hey1 is not a classic repressor and that the its repressive effects may be highly context-dependent and intimately linked to the process of promoter complex assembly and activation.
Although these questions require additional investigation, our results provide strong evidence that Hey1 functions by physically associating with the promoter regions of two critical myogenic genes, myogenin and Mef2C. It would appear that repressing one gene or the other would be sufficient for myogenic inhibition, thus raising the question, Why both? One possibility is that Hey1-mediated repression is inherently weak and that multiple targets are, therefore, necessary. A second possibility relates to the mechanism of Hey1 action and its relationship to activation as opposed to repression of the myogenic program. During the processes of embryogenesis and muscle regeneration, the Notch signal is initially on, allowing progenitor pools to expand, but then turned off to allow terminal myogenesis (34, 60). Repression of multiple genes by Hey1 (indeed, myogenin and Mef2C may represent only a fraction of the total) may be more relevant to the manner in which the myogenic program is resumed once the Notch signal is removed. It is possible that Hey1 target genes are poised to be rapidly induced and that the process of myogenesis is more efficient or robust if multiple genes are involved.
Supplementary Material
Acknowledgments
We thank Dr. Vittorio Sartorelli for providing a ChIP protocol and Drs. Sartorelli, Tod Gulick, Eric Olson, Bas van Steensel, and Carl June for generously providing reagents. We acknowledge Daan Peric Hupkes for expert assistance with DamID and Jamie Planck, Matt Culyba, and Yao-juan Liu for technical support.
This work was supported, in whole or in part, by National Institutes of Health Training Grants 5-T32-GM-008216-20 (to M. B.) and F32GM068394-01A1 (NRSA; to S. K.). This work was also supported by funds from the Muscular Dystrophy Association (MDA 3888).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
M. F. Buas, S. Kabak, and T. Kadesch, unpublished observations.
- tk
- thymidine kinase
- GM
- growth medium
- DM
- differentiation medium
- EMSA
- electrophoretic mobility shift assays
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TNT
- transcribed and translated
- HCE
- Hey1-consensus E-box
- Dam
- DNA adenine methyltransferase
- CMV
- cytomegalovirus
- Pol
- polymerase
- ChIP
- chromatin immunoprecipitation
- Pipes
- 1,4-piperazinediethanesulfonic acid
- IgH
- immunoglobulin heavy chain.
REFERENCES
- 1.Iso T., Chung G., Hamamori Y., Kedes L. (2002) J. Biol. Chem. 277, 6598–6607 [DOI] [PubMed] [Google Scholar]
- 2.Iso T., Sartorelli V., Chung G., Shichinohe T., Kedes L., Hamamori Y. (2001) Mol. Cell. Biol. 21, 6071–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maier M. M., Gessler M. (2000) Biochem. Biophys. Res. Commun. 275, 652–660 [DOI] [PubMed] [Google Scholar]
- 4.Fischer A., Schumacher N., Maier M., Sendtner M., Gessler M. (2004) Genes Dev. 18, 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer A., Steidl C., Wagner T. U., Lang E., Jakob P. M., Friedl P., Knobeloch K. P., Gessler M. (2007) Circ. Res. 100, 856–863 [DOI] [PubMed] [Google Scholar]
- 6.Kokubo H., Miyagawa-Tomita S., Nakazawa M., Saga Y., Johnson R. L. (2005) Dev. Biol. 278, 301–309 [DOI] [PubMed] [Google Scholar]
- 7.Kokubo H., Tomita-Miyagawa S., Hamada Y., Saga Y. (2007) Development 134, 747–755 [DOI] [PubMed] [Google Scholar]
- 8.Rutenberg J. B., Fischer A., Jia H., Gessler M., Zhong T. P., Mercola M. (2006) Development 133, 4381–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gessler M., Knobeloch K. P., Helisch A., Amann K., Schumacher N., Rohde E., Fischer A., Leimeister C. (2002) Curr. Biol. 12, 1601–1604 [DOI] [PubMed] [Google Scholar]
- 10.Sakamoto M., Hirata H., Ohtsuka T., Bessho Y., Kageyama R. (2003) J. Biol. Chem. 278, 44808–44815 [DOI] [PubMed] [Google Scholar]
- 11.Sun J., Kamei C. N., Layne M. D., Jain M. K., Liao J. K., Lee M. E., Chin M. T. (2001) J. Biol. Chem. 276, 18591–18596 [DOI] [PubMed] [Google Scholar]
- 12.Hayashi T., Kokubo H., Hartman B. H., Ray C. A., Reh T. A., Bermingham-McDonogh O. (2008) Dev. Biol. 316, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taelman V., Van Campenhout C., Sölter M., Pieler T., Bellefroid E. J. (2006) Development 133, 2961–2971 [DOI] [PubMed] [Google Scholar]
- 14.Iso T., Sartorelli V., Poizat C., Iezzi S., Wu H. Y., Chung G., Kedes L., Hamamori Y. (2001) Mol. Cell. Biol. 21, 6080–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer A., Leimeister C., Winkler C., Schumacher N., Klamt B., Elmasri H., Steidl C., Maier M., Knobeloch K. P., Amann K., Helisch A., Sendtner M., Gessler M. (2002) Cold Spring Harb. Symp. Quant. Biol. 67, 63–70 [DOI] [PubMed] [Google Scholar]
- 16.Pichon B., Taelman V., Bellefroid E. J., Christophe D. (2004) Biochim. Biophys. Acta 1680, 46–52 [DOI] [PubMed] [Google Scholar]
- 17.Taelman V., Van Wayenbergh R., Sölter M., Pichon B., Pieler T., Christophe D., Bellefroid E. J. (2004) Dev. Biol. 276, 47–63 [DOI] [PubMed] [Google Scholar]
- 18.Elagib K. E., Xiao M., Hussaini I. M., Delehanty L. L., Palmer L. A., Racke F. K., Birrer M. J., Shanmugasundaram G., McDevitt M. A., Goldfarb A. N. (2004) Mol. Cell. Biol. 24, 7779–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer A., Klattig J., Kneitz B., Diez H., Maier M., Holtmann B., Englert C., Gessler M. (2005) Mol. Cell. Biol. 25, 8960–8970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kathiriya I. S., King I. N., Murakami M., Nakagawa M., Astle J. M., Gardner K. A., Gerard R. D., Olson E. N., Srivastava D., Nakagawa O. (2004) J. Biol. Chem. 279, 54937–54943 [DOI] [PubMed] [Google Scholar]
- 21.Garg V., Muth A. N., Ransom J. F., Schluterman M. K., Barnes R., King I. N., Grossfeld P. D., Srivastava D. (2005) Nature 437, 270–274 [DOI] [PubMed] [Google Scholar]
- 22.Ghosh B., Leach S. D. (2006) Biochem. J. 393, 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belandia B., Powell S. M., García-Pedrero J. M., Walker M. M., Bevan C. L., Parker M. G. (2005) Mol. Cell. Biol. 25, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia J., Lin M., Zhang L., York J. P., Zhang P. (2007) Mol. Cell. Biol. 27, 7236–7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu X., Chung A. Y., Wu I., Foldi J., Chen J., Ji J. D., Tateya T., Kang Y. J., Han J., Gessler M., Kageyama R., Ivashkiv L. B. (2008) Immunity 29, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holderfield M. T., Henderson Anderson A. M., Kokubo H., Chin M. T., Johnson R. L., Hughes C. C. (2006) Biochem. Biophys. Res. Commun. 346, 637–648 [DOI] [PubMed] [Google Scholar]
- 27.Huang Q., Raya A., DeJesus P., Chao S. H., Quon K. C., Caldwell J. S., Chanda S. K., Izpisua-Belmonte J. C., Schultz P. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3456–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa O., McFadden D. G., Nakagawa M., Yanagisawa H., Hu T., Srivastava D., Olson E. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13655–13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leimeister C., Externbrink A., Klamt B., Gessler M. (1999) Mech. Dev. 85, 173–177 [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa O., Nakagawa M., Richardson J. A., Olson E. N., Srivastava D. (1999) Dev. Biol. 216, 72–84 [DOI] [PubMed] [Google Scholar]
- 31.Schuster-Gossler K., Cordes R., Gossler A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasyutina E., Lenhard D. C., Wende H., Erdmann B., Epstein J. A., Birchmeier C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4443–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conboy I. M., Conboy M. J., Smythe G. M., Rando T. A. (2003) Science 302, 1575–1577 [DOI] [PubMed] [Google Scholar]
- 34.Conboy I. M., Rando T. A. (2002) Dev. Cell 3, 397–409 [DOI] [PubMed] [Google Scholar]
- 35.Buas M. F., Kabak S., Kadesch T. (2009) J. Cell. Physiol. 218, 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molkentin J. D., Black B. L., Martin J. F., Olson E. N. (1995) Cell 83, 1125–1136 [DOI] [PubMed] [Google Scholar]
- 37.Xu Q., Wu Z. (2000) J. Biol. Chem. 275, 36750–36757 [DOI] [PubMed] [Google Scholar]
- 38.Zhu B., Gulick T. (2004) Mol. Cell. Biol. 24, 8264–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu J., McKinsey T. A., Zhang C. L., Olson E. N. (2000) Mol. Cell 6, 233–244 [DOI] [PubMed] [Google Scholar]
- 40.Edmondson D. G., Cheng T. C., Cserjesi P., Chakraborty T., Olson E. N. (1992) Mol. Cell. Biol. 12, 3665–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen C. P., Kadesch T. (1995) Mol. Cell. Biol. 15, 4518–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogel M. J., Peric-Hupkes D., van Steensel B. (2007) Nat. Protoc. 2, 1467–1478 [DOI] [PubMed] [Google Scholar]
- 43.Wang D. Z., Valdez M. R., McAnally J., Richardson J., Olson E. N. (2001) Development 128, 4623–4633 [DOI] [PubMed] [Google Scholar]
- 44.Reddy K. L., Zullo J. M., Bertolino E., Singh H. (2008) Nature 452, 243–247 [DOI] [PubMed] [Google Scholar]
- 45.Caretti G., Di Padova M., Micales B., Lyons G. E., Sartorelli V. (2004) Genes Dev. 18, 2627–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergstrom D. A., Penn B. H., Strand A., Perry R. L., Rudnicki M. A., Tapscott S. J. (2002) Mol. Cell 9, 587–600 [DOI] [PubMed] [Google Scholar]
- 47.Mal A., Harter M. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasty P., Bradley A., Morris J. H., Edmondson D. G., Venuti J. M., Olson E. N., Klein W. H. (1993) Nature 364, 501–506 [DOI] [PubMed] [Google Scholar]
- 49.Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., Nabeshima Y. (1993) Nature 364, 532–535 [DOI] [PubMed] [Google Scholar]
- 50.Dodou E., Xu S. M., Black B. L. (2003) Mech. Dev. 120, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 51.van Steensel B., Henikoff S. (2000) Nat. Biotechnol. 18, 424–428 [DOI] [PubMed] [Google Scholar]
- 52.van Steensel B., Henikoff S. (2003) Biotechniques 35, 346–350, 352,–354, 356–357 [DOI] [PubMed] [Google Scholar]
- 53.Greil F., Moorman C., van Steensel B. (2006) Methods Enzymol. 410, 342–359 [DOI] [PubMed] [Google Scholar]
- 54.Berkes C. A., Bergstrom D. A., Penn B. H., Seaver K. J., Knoepfler P. S., Tapscott S. J. (2004) Mol. Cell 14, 465–477 [DOI] [PubMed] [Google Scholar]
- 55.Spitz F., Demignon J., Porteu A., Kahn A., Concordet J. P., Daegelen D., Maire P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14220–14225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H., Stavnezer E. (2009) J. Biol. Chem. 284, 2867–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berghella L., De Angelis L., De Buysscher T., Mortazavi A., Biressi S., Forcales S. V., Sirabella D., Cossu G., Wold B. J. (2008) Genes Dev. 22, 2125–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amsen D., Antov A., Jankovic D., Sher A., Radtke F., Souabni A., Busslinger M., McCright B., Gridley T., Flavell R. A. (2007) Immunity 27, 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang T. C., Yashiro-Ohtani Y., Del Bianco C., Knoblock D. M., Blacklow S. C., Pear W. S. (2007) Immunity 27, 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brack A. S., Conboy I. M., Conboy M. J., Shen J., Rando T. A. (2008) Cell Stem Cell 2, 50–59 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.