Abstract
Sepsis is encoded by a sequel of transcription activation and repression events that initiate, sustain, and resolve severe systemic inflammation. The repression/silencing phase occurs in blood leukocytes of animals and humans following the initiation of systemic inflammation due to developing endotoxin tolerance. We previously reported that NF-κB transcription factor RelB and histone H3 lysine methyltransferase G9a directly interact to induce facultative heterochromatin assembly and regulate epigenetic silencing during endotoxin tolerance, which is a major feature of sepsis. The general objective of this study was to assess whether dynamic temporal, structural, and positional changes of nucleosomes influence the sepsis phenotype. We used the THP-1 sepsis cell model to isolate mononucleosomes by rapid cell permeabilization and digestion of chromatin with micrococcal nuclease and then compared tumor necrosis factor α (TNFα) proximal promoter nucleosome alignment in endotoxin-responsive and -tolerant phenotypes. We found differential and dynamic repositioning of nucleosomes from permissive to repressive locations during the activation and silencing phases of transcription reprogramming and identified the following mechanisms that may participate in the process. 1) Two proximal nucleosomes repositioned to expose the primary NF-κB DNA binding site in endotoxin-responsive cells, and this “promoter opening” required the ATP-independent chaperone NAP1 to replace the core histone H2A with the H2A.Z variant. 2) During RelB-dependent endotoxin tolerance, the two nucleosomes repositioned and masked the primary NF-κB DNA binding site. 3) Small interfering RNA-mediated inhibition of RelB expression prevented repressive nucleosome repositioning and tolerance induction, but the “open” promoter required endotoxin-induced NF-κB p65 promoter binding to initiate transcription, supporting the known requirement of p65 posttranslational modifications for transactivation. 4) Sustaining the permissive promoter state after RelB knockdown required ATP-dependent nucleosome remodeler BAF complex. Moreover, we found that forced expression of RelB in responsive cells induced repressive nucleosome positioning and silenced TNFα transcription, demonstrating the plasticity of nucleosome remodeling and its dependence on RelB. Our data suggest that nucleosome repositioning controls both the induction and epigenetic silencing phases of TNFα transcription associated with sepsis.
Keywords: Chromatin Remodeling, Epigenetics, Inflammation, Innate Immunity, Sepsis, Transcription
Introduction
Nucleosomes are dynamically and constantly remodeled to allow or prevent access to regulatory factors and cofactors. Each nucleosome, the fundamental building unit of chromatin, is composed of ∼147 bp of genomic DNA wrapped 1.65 times around an octamer of the core histone proteins H2A, H2B, H3, and H4 (1–4). Because the majority of genomic DNA is wrapped in nucleosomes (2), the presence of nucleosomes on genomic DNA inhibits the binding of sequence-specific regulatory factors and basal cofactors. For example, access to DNA wrapped in nucleosome is occluded for RNA polymerase and regulatory complexes (5–7), although nucleosomes may recruit other protein complexes through interactions with their histone tails (8). Because transcription factor binding sites often cluster in linker DNA between nucleosomes, the precise locations of nucleosomes and the accessible linker DNA may play a role in transcriptional control.
Recent studies showed that promoter nucleosomes frequently adopt selective positions to functionally regulate transcription factor binding (3). In addition, nucleosomes can be displaced from promoter DNA by promoter-binding transcription factors or in combination with chromatin-remodeling complexes and histone chaperones (4, 9). Previous studies showed that the selective nucleosomal organization observed in many systems underlies the differential accessibility and transcriptional potential of chromatin structure in active versus inactive promoters (10–13). Because nucleosomal positioning is often associated with discrete changes within regulatory regions (14), remodeling of promoter nucleosomes is a key mechanism of gene activation or silencing (15, 16).
Posttranslational modifications on histone components of nucleosomes play a role in changing chromatin structure by altering histone-DNA interactions and help in the recruitment of chromatin-remodeling complexes (17, 18). These complexes alter chromatin configuration by nucleosomal sliding or eviction, thereby promoting access to transcription factors (3, 4, 16). The mammalian ATP-dependent SWI/SNF chromatin-remodeling complexes are tumor suppressors and function as transcriptional coactivators or corepressors (19–22). Because nucleosome assembly by histone chaperones tends to place nucleosomes over low energy nucleosome-positioning sequences, these remodeling complexes use ATP energy to move nucleosome away from complex-preferred positions, depending on the DNA regulatory sequence and histone modifications (3, 4, 20, 23). Thus, recruitment of a remodeling complex could invert the normal accessibility pattern at a promoter and therefore act as an on-off switch for transcription (3). Other chromatin remodelers like the ATP-independent nucleosome assembly protein NAP1 bind and sequester histone complexes through exchanging histone dimers, resulting in nucleosomal sliding (24, 25).
We discovered that transcription silencing of proinflammatory TNFα2 and IL-1β genes in endotoxin-tolerant THP-1 monocytes, a phenotype present in blood leukocytes after the initiation of severe systemic inflammation (26–28), is mediated by selective changes in transcription factor binding and chromatin structure. This gene reprogramming event can be remodeled in vitro by generating a state of lipopolysaccharide (LPS) tolerance in cultured cell lines by the prolonged stimulation with LPS (28–30). The transcription silencing phase is initiated and maintained through a combinatorial silencing mechanism that involves interactions between the transcription repressor RelB and chromatin-associated proteins (26, 31, 32). The silencing mechanism requires dimethylation on histone H3 lysine 9 (H3K9me2) by G9a, increased binding of heterochromatin protein HP1, and formation of silent facultative heterochromatin structure (26, 31, 32). This process correlates with diminished binding of the active NF-κB factor p65 and increased binding of feedback repressor transcription factor RelB to the proximal promoters. RelB is essential initiator of silencing by directly interacting with and recruiting G9a to promote and maintain a silent heterochromatin structure (27, 32). Our finding that inhibiting RelB expression in tolerant cells reactivated TNFα and IL-1β transcription (26, 27, 31) raised the possibility that nucleosome remodeling physically contributes to transcriptional silencing of proinflammatory genes in SSI.
In this study, we examined nucleosomal organization around the TNFα promoter and analyzed its contribution to the induction and silencing of the TNFα promoter, using micrococcal nuclease digestion combined with chromatin immunoprecipitation. The results revealed two positioned nucleosomes encompassing the promoter element required for TNFα induction as well as silencing. We report that nucleosome occupancy and positioning around the promoter displays a strikingly different profile that correlates with the level of expression. Upon activation of THP-1-responsive cells by LPS, two proximal nucleosomes are rapidly remodeled to open the promoter, suggesting that selective remodeling is essential for TNFα induction. In tolerant cells, one nucleosome is repositioned to a repressive location, covering the κB site. This differential nucleosome positioning was dependent on the presence of RelB at the promoter sequence in tolerant cells. We also show that nucleosome positioning in responsive cells is mediated by the ATP-independent chromatin remodeler NAP1 (nucleosome assembly protein 1), whereas the ATP-dependent remodeler BAF complex is required to reposition nucleosomes and open the promoter DNA in tolerant cells. The results suggest that nucleosomal occupancy and positioning at the TNFα promoter correlates with the level of expression, and that chromatin remodeling is an intrinsic and dynamic mechanism that controls proinflammatory gene expression during the course of development and maintenance of severe systemic inflammation.
MATERIALS AND METHODS
SSI Cell Model
The human monocytic cell line, THP-1, obtained from the American Type Culture Collection, was maintained in RPMI medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine at a density of 0.5–1.0 × 106 cells/ml at 37 °C and 5% CO2. For induction of LPS tolerance (29), cells were incubated overnight with 1 μg/ml Gram-negative bacterial LPS from Escherichia coli 0111:B4 (Sigma). LPS-tolerant and LPS-responsive (normal) cells were washed with incomplete medium and cultured at 0.5–1.0 × 106 cells/ml and stimulated with 1 μg/ml LPS for the indicated times.
RNA Interference
Cells were plated at 0.5 × 106 cells/ml 1 day before transfection. Transfection with pools of control or RelB-, NAP1-, or BAF47-specific small interfering RNAs (siRNAs) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was performed by electroporation using 5 μl (0.5 μm final concentration) of siRNA in 100 μl of transfection medium (Nucleofector; Amaxa, Gaithersburg, MD). Immediately after transfection, cells were transferred to culture medium at 0.5 × 106 cell/ml and left unstimulated or stimulated with 1 μg/ml LPS to induce tolerance. After 36 h, cells were harvested, washed with incomplete medium, and then stimulated with 1 μg/ml LPS for the indicated times.
Plasmids
Wild-type and mutant RelB constructs (truncated RelB with amino acids 264–379 deleted) were generated by GenScript Corp. (Piscataway, NJ). DNA was transfected by electroporation as described above. This truncation disrupts the Rel homology domain of RelB and therefore affects its dimerization and interaction with G9a (26).
RNA Analysis
Expression of TNFα was evaluated by quantitative real-time PCR. Total RNA was isolated using the STAT-60 extraction kit, according to the manufacturer's protocol (Tel-Test, Friendswood, TX). Two micrograms of RNA were reverse-transcribed to cDNA in a 25-μl volume containing 0.2 μm dNTPs, 2.5 μm oligo(dT), 5 mm MgCl2, and 0.25 units/μl murine leukemia reverse transcriptase (Applied Biosystems, Foster City, CA). The reverse transcription reaction was incubated for 1 h at 42 °C and 5 min at 99 °C. The PCR was performed using 5 μl of cDNA product and TNFα predesigned TaqMan primer/probe sets (Applied Biosystems). The PCR conditions were as follows: 2 min at 50 °C, 10 min at 95 °C, followed by 40 cycles with 15 s at 95 °C and 1 min at 60 °C (combined annealing and extension), using ABI Prism 7000 Sequence Detection System (Applied Biosystems). Sample data were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA and are presented as -fold change.
Preparation of Mononucleosome and Input DNA and Immunoprecipitation
Cells grown to 70–80% confluence were harvested and treated with 1% formaldehyde for 10 min at room temperature to cross-link chromatin. Cross-linking was stopped by adding glycine at 125 mm. Cells were washed once with cold phosphate-buffered saline and suspended in lysis buffer (30 mm Hepes, pH 7.5, 60 mm KCl, 300 mm sucrose, 5 mm K2HPO4, 5 mm MgCl2, 2 mm EDTA, and 0.5 mm DTT), followed by Dounce homogenization. The nuclear suspension was then centrifuged for 15 min at 3,000 rpm. Nuclei were collected, and a portion (1:20) were used to isolate input DNA using the DNeasy kit (Qiagen, Valencia, CA). The remainder of the nuclei was resuspended in micrococcal nuclease (MNase) digestion buffer (10 mm Tris (pH 7.4), 15 mm NaCl, 1 mm CaCl2, 60 mm KCl), to which 0.2 mm phenylmethylsulfonyl fluoride and 0.5 mm spermidine were added immediately prior to use.
For preparation of mononucleosomes, we used an optimized MNase digestion protocol (34) with some modifications. Five hundred microliters of nuclei suspension (at 2 × 106 nuclei/ml) were incubated with 5–25 units of MNase (stock solution, 20 units/μl; EMD Calbiochem), gently mixed, and then incubated at room temperature for 5 min. The digestion was stopped by adding 1 ml of MNase stop buffer containing 100 mm EDTA, 1% SDS, 50 mm Tris, pH 8.0, 0.1 mg/ml proteinase K, and 100 μl of 10% SDS. The lysate was incubated overnight at 37 °C. DNA was then extracted by phenol/chloroform extraction and ethanol precipitation (with glycogen).
For chromatin immunoprecipitation, immediately after digestion with MNase, nuclei lysis buffer (Active Motif, Carlsbad, CA) and protease inhibitor mixture were added for 30 min on ice. After centrifugation for 5 min at 10,000 rpm, chromatin solution was precleared by incubation with protein G-agarose beads and then subjected to immunoprecipitation with antibody against H3K9me2, H3K4me3 (Millipore, Temecula, CA), H2A.Z (Active Motif), H2A, NAP1 (Santa Cruz Biotechnology, Inc.), or BAF47 (BD Biosciences). Chromatin was decross-linked, and DNA was extracted as described previously (27).
PCR
Semiquantitative PCR was performed in a 50-μl volume containing 5 μl of mononucleosomal, input, or immunoprecipitated (chromatin-immunoprecipitated) DNA, 1 μm each primer, 2 mm MgCl2, 0.2 μm dNTPs, and 0.04 unit/μl AmpliTag Gold DNA polymerase (Applied Biosystems). The PCR conditions were as follows: 1 cycle at 94 °C for 5 min, 30 cycles at 94 °C, 58 °C, and 72 °C for 30 s each, and a final cycle at 72 °C for 5 min. Equal amounts of PCR products were run on 1.2% ethidium bromide-stained agarose gel, and images were captured using a Quantity One imager (Bio-Rad). The primers used in PCR were designed to amplify sequences in the human TNFα regulatory region from −800 to +200 bp relative to the transcription start site (35, 36).
Western Blot
Nuclear proteins were extracted by incubating cells on ice for 15 min in a buffer containing 10 mm HEPES (pH 7.9), 1.5 mm MgCl2, 10 mm KCl, 0.2 mm EDTA, 20 mm NaF, 1 mm Na4P2O7, 1 mm Na3VO4, 0.5 mm dithiothreitol, 0.1% Triton X-100, and 1× protease inhibitor mixture. Nuclei were pelleted by centrifugation at 5,000 rpm for 10 min at 4 °C and then resuspended in lysis buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) and incubated on ice for 30 min. Extracts were cleared by centrifugation, and protein concentration was determined. Whole cell extracts were prepared using the same nuclei lysis buffer. Equal amounts (50 μg) of protein were separated on SDS-PAGE and transferred to polyvinylidene difluoride membranes (Pierce). Membranes were blocked and probed overnight at 4 °C with appropriate dilutions of primary antibodies against RelB, NAP1, or BAF47. After washing, blots were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc.). Proteins were visualized using an ECL detection system (Pierce). Blots were stripped and reprobed with control antibody.
Statistical Analysis
Data were analyzed by Microsoft Excel 2003 and are presented as the mean ± S.E. of three independent experiments. Student's t test was used to determine significant differences, with p values of ≤0.05 being significant.
RESULTS
Pattern of Nucleosome Positioning at the Human TNFα Promoter
TNFα transcription is significantly induced after LPS activation of THP-1 cells but is silenced once cells become LPS-tolerant despite a second dose of LPS (27, 28). Therefore, we sought to determine whether the chromatin structure contributes to TNFα induction or silencing by comparing the nucleosomal organization across a 1-kb promoter fragment in the different cellular phenotypes. LPS-responsive (normal) THP-1 cells were made tolerant (T) by pretreatment with LPS overnight and then washed and restimulated, together with the responsive (R) cells, for 1 h. We chose the 1 h time point because our previous work demonstrated that after 1 h, TNFα mRNA is significantly increased in responsive cells but was not expressed in tolerant cells (27). We consider resting, responsive cells (R0) as the base-line response; therefore, nucleosome positions in these cells will be identified as “in native/default positions.” Because TNFα mRNA is restored in tolerant cells in which RelB expression is inhibited by siRNA, we included this phenotype in our analysis.
MNase preferentially digests protein-free DNA, including linker regions between nucleosomes. We first digested nucleosomal DNA with varying concentrations of MNase and then choose the conditions that produced mononucleosome particles (∼180 bp). DNA was purified and separated on 3% agarose gel. The optimal digestion conditions were determined by detecting the purified DNA as one main band of ∼180 bp (not shown). Conditions that produced several bands were indicative of partial digestion and were excluded. Optimal digestion conditions were not changed throughout our analyses. The extent of nucleosome occupancy was analyzed by comparing the abundance of the promoter DNA in total mononucleosomal DNA relative to genomic DNA by PCR. We elected not to digest genomic DNA extracted before MNase digestion because MNase may cut randomly and generate several bands, which makes it difficult to quantitate the level of DNA abundance. These measurements provide an assessment of the extent of DNA protection via packaging into nucleosomes.
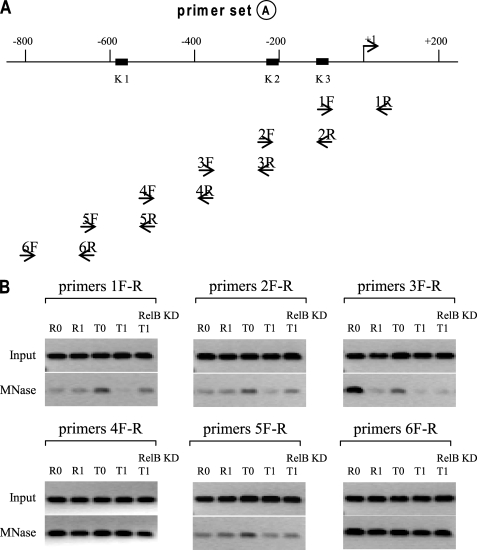
We employed a reductive approach by designing a series of PCR primers, namely primer set A–C (see Fig. 1A). Each primer pair encompass ∼200-bp sequence. Using primer set A, we observed one nucleosome between −200 and −400 that was detected with primer pair 3F-R in unstimulated responsive (R0) cells only (Fig. 1B). The experiment also revealed the presence of two nucleosomes, one close to −400 (using primer pair 4F-R) and one between −600 and −800 (using primer pair 6F-R). These two nucleosomes were uniformly positioned in all of the cell phenotypes examined. The faint bands shown with T0 cells using primer pairs 1F-R, 2F-R, and 3F-R may result from the incomplete digestion with MNase, meaning that the corresponding DNA is not occupied by nucleosomes.
FIGURE 1.
Mapping nucleosome positions across TNFα upstream regulatory region. A, diagram of the 800-bp upstream regulatory sequence involved in the transcriptional regulation of TNFα gene. The locations of the three NF-κB binding sites (K2 at −98; K2 at −216; K1 at −598) are shown. The primer sets used for PCR experiments presented in Figs. 1–3 are all shown here for illustration, but only those used in Fig. 1 are highlighted in boldface type. B, to map nucleosome positions, THP-1 cells were made tolerant by pretreatment with 1 μg/ml LPS overnight. Responsive (R) and tolerant (T) cells were washed and left unstimulated (0) or stimulated with 1 μg/ml LPS for 1 h (1). For RelB KD, cells were transfected with control or RelB-specific siRNA and then made tolerant, as described above (T1-RelB KD). Chromatin was cross-linked, and nuclei were isolated and then digested with MNase. A portion of the nuclei (1:20) was reserved before MNase digestion to extract genomic DNA and served as control (Input). MNase reaction conditions were adjusted to have average DNA sizes of ∼180 bp, as determined by several MNase dilutions and a PCR assay (not shown). After digestion, mononucleosomal DNA was extracted, and the level of DNA enrichment was determined by PCR. Results are representative of 2–3 experiments.
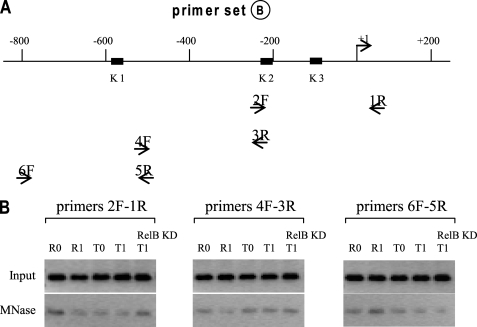
We next used primer pairs (primer set B) that encompass longer sequences (∼300 bp), which is longer than the mononucleosomal DNA (∼180 bp). As shown in Fig. 2, we did not detect any nucleosome in all of the cell phenotypes. Although this result may suggest that these sequences are nucleosome-free, it also suggests that they contain nucleosomes, but because MNase cuts DNA at the nucleosomal edges, they may not be amplified by PCR if the cut falls in the middle of the sequence the primers cover.
FIGURE 2.
Mapping nucleosome positions (continued). THP-1 cells were treated, and mononucleosomal and input DNAs were prepared as described in the legend to Fig. 1. The primers used to amplify the DNA are highlighted in boldface type (A), and the PCR results are shown in B. Note that no bands were detected with this primer set, indicating that MNase cut within the sequences encompassing these primers. Results are representative of 2–3 experiments.
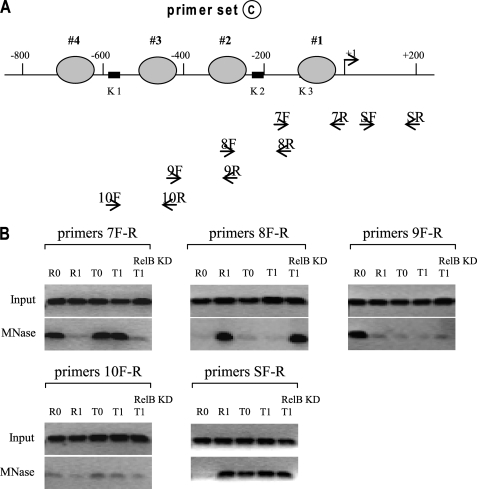
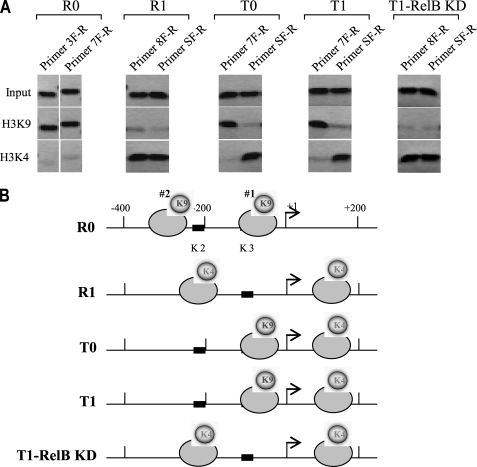
In order to narrow down the locations of the three nucleosomes identified in Fig. 1, we designed a primer set C that overlaps primer set A (Fig. 3A). Using these primers, we located one nucleosome between +1 and −200 in R0, T0, and T1 cells with primer pair 7F-R (Fig. 3B). Primer 8F-R detected one nucleosome in R1 cells and also in T1 cells after RelB knockdown, whereas primer 9F-R detected one nucleosome in R0 cells only. In addition, primer SF-R detected one nucleosome between +1 and +200 in all cell phenotypes except R0 cells.
FIGURE 3.
Mapping nucleosome positions (continued). THP-1 cells were treated, and mononucleosomal and input DNAs were prepared as described in the legend to Fig. 1. The primers used are highlighted in boldface type (A), and the PCR results are shown in B. The results are representative of 2–3 experiments.
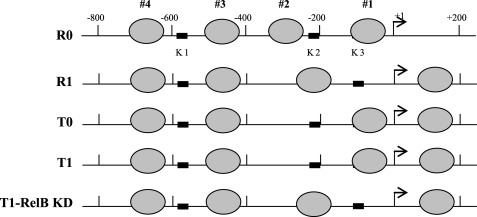
Nucleosome Occupancy around TNFα Promoter Correlates with Its Transcription State
Based on the results presented in Figs. 1–3, we tentatively mapped four nucleosomes (namely nucleosomes 1–4) within the 1-kb TNFα regulatory region (Fig. 4). Nucleosomes 3 and 4 were regularly spaced, and their positions were not changed upon transcription induction or silencing, whereas nucleosomes 1 and 2 were repositioned. After LPS activation of responsive cells (R1), nucleosome 2 was repositioned to cover the κB binding site (K2), whereas nucleosome 1 was repositioned downstream of the start site, leaving the proximal κB binding site (K3) exposed. Once cells entered the tolerant state (T0 cells), nucleosome 2 was repositioned again from the K3 to the K2 position, and this pattern did not change even in the presence of LPS (T1 cells). Interestingly, when RelB was knocked down in tolerant cells (T1-RelB knockdown (KD)), nucleosome 2 was repositioned to cover the K2 site, giving a pattern similar to that seen in R1 cells. In addition, we noticed that once nucleosome 1 displaced to upstream position after LPS stimulation of R0 cells, it was not repositioned again regardless of the transcription state. Together, these results show that nucleosome occupancy around the proximal promoter is selectively remodeled and that nucleosome positioning upstream of the transcription start site correlates with the transcription level of TNFα. Because the proximal κB-binding site (K3) is essential to the induction of TNFα by LPS, the results also suggest that nucleosome repositioning from permissive (in R1 cells) to repressive (in T cells) positions may play a key role in the transcription regulation of TNFα.
FIGURE 4.
Transcription activation and epigenetic silencing of TNFα expression involve differential repositioning of two proximal nucleosomes. Shown is a map of the nucleosome positions across the regulatory region. Based on the PCR results presented in Figs. 1–3, we tentatively localized four nucleosomes across the regulatory region. We identify them as nucleosomes 1–4. Note that after LPS stimulation, nucleosomes 1 and 2 were repositioned in responsive (R1) cells. In tolerant cells, however, LPS stimulation did not change the positioning of the two nucleosomes from the locations where tolerance was first induced (compare T0 and T1). After LPS stimulation of tolerant cells, in which RelB was knocked down (T1-RelB KD), one nucleosome was moved again to a position similar to that seen in R1 cells. Note that the positions of nucleosomes 3 and 4 were not changed in all cell phenotypes. Therefore, all following analyses will be focused on nucleosomes 1 and 2 only.
Differential Histone Modifications Correlate with Nucleosome Occupancy and Transcription State
Distinct histone modifications have been associated with transcriptional activation or silencing (37). In addition, we have previously shown that the histone methyltransferase G9a binds to the TNFα proximal promoter nucleosome and induces DNA and histone methylation in tolerant cells (32). To further characterize the remodeled nucleosome and examine whether histone modifications associate with nucleosome repositioning, we compared the extent of histone methylation of the repositioned nucleosomes, specifically H3K4me3 (which marks actively transcribed genes) and H3k9me2 (which correlates with silenced genes) by chromatin immunoprecipitation analysis. After MNase digestion, mononucleosomes were immunoprecipitated with the respective antibody, and mononucleosomal DNA was then extracted. Because the results described above showed that only nucleosomes 1 and 2 were repositioned, we will focus our next analyses on these two nucleosomes.
We observed that both nucleosomes were dimethylated on H3K9 in the native positions (R0 cells) (Fig. 5). After activation by LPS (R1 cells), this mark was replaced by H3K4me3 on the two repositioned nucleosomes. In tolerant cells, however, we observed a mixed methylation pattern. Nucleosome 1 contained the H3K4me3 mark, whereas nucleosome 2 contained the H3K9me2 mark. Interestingly, H3K4me3 was detected on both nucleosomes after RelB knockdown in tolerant cells (T1-RelB KD). Importantly, we noticed that the methylation mark (H3K4me3) of nucleosome 1 did not change once this nucleosome was repositioned downstream of the start site regardless of the transcription state. Because the nucleosomes acquired distinct histone marks after they were repositioned (i.e. transition from R0 to R1 or from R1 to T0 and T1), these results imply that the changes in histone modifications may occur subsequent to the nucleosome repositioning.
FIGURE 5.
Repositioned nucleosomes acquire new histone marks. To investigate interactions between nucleosome repositioning and histone modifications, we measured the levels of H3K9me2 and H3K4me3 on nucleosomes 1 and 2, considering their positions in R0 cells as native/default positions. Chromatin was isolated and subjected to MNase digestion. The suspension was then precleared with protein G-agarose beads, and mononucleosomes were immunoprecipitated with antibody against H3K9me2 or H3K4me3. Mononucleosomal DNA was then extracted and analyzed by PCR. A, PCR was performed using primer pairs that we knew from the previous experiment spanned the different locations of nucleosomes 1 and 2 in the different cell phenotypes. For example, the two primer pairs 7 and 3 were used for R0 cells, and primer pairs 8 and S were used for R1 cells (see Figs. 3 and 4 for alignment). B, mapping histone methylation on the two proximal nucleosomes. Note that the nucleosome downstream of the start site acquired the H3K4me3 mark after LPS stimulation, but this mark and the location of that nucleosome did not change even in tolerant cells. Results are representative of three experiments.
RelB Remodels Nucleosomes and Induces Tolerance in LPS-responsive Cells
We have shown that RelB binds to the TNFα and IL-1β promoter nucleosomes in tolerant cells only and that RelB knockdown reactivates their transcription (26, 27). In addition, our finding that RelB knockdown resulted in nucleosomal occupancy profile similar to that seen in R1 cells (see Fig. 4) suggested that nucleosome positioning in tolerant cells might be established (or at least maintained) by RelB. To test this possibility, we performed the reverse experiment in which RelB cDNA was introduced into the responsive cells (which do not normally express RelB). After 24 h, cells were washed and left unstimulated or stimulated with LPS for 1 h.
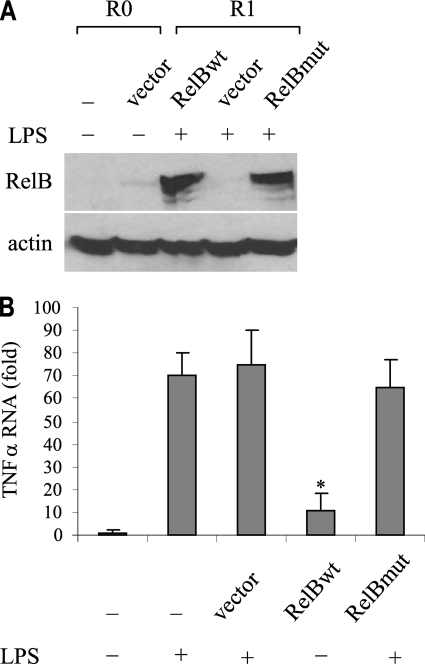
We first confirmed RelB expression by Western blot (Fig. 6A). We also measured TNFα mRNA in responsive cells expressing or lacking RelB. As shown in Fig. 6B, wild-type but not mutant expression significantly reduced TNFα mRNA. This result clearly shows that RelB induces tolerance and transcription silencing of TNFα in responsive cells.
FIGURE 6.
Forced expression of RelB in responsive cells induces tolerance and silences TNFα expression. THP-1-responsive (R) cells were transfected with pcDNA3 vector alone or with vector encoding full-length wild-type (wt) or mutant (mut) RelB. After 24 h, cells were washed and left unstimulated (R0) or stimulated with 1 μg/ml LPS for 1 h (R1). A, Western blot of RelB protein after RelB transfection. B, TNFα mRNA level. RNA was isolated and analyzed by real-time PCR. Values were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA level and are presented as -fold change relative to non-transfected, unstimulated cells (assigned 1-fold). Data represent the mean ± S.E. from three experiments. *, significant difference (p ≤ 0.05) compared with vector alone.
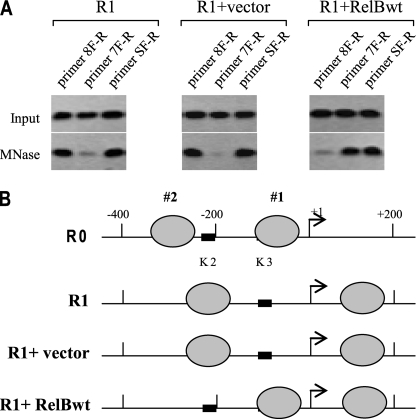
Next, we examined the effect of RelB forced expression in responsive cells on nucleosome positioning. We used the primer pairs that cover the three proximal nucleosomal positions. We show R0 cells here again as a base-line position for comparison. The results (Fig. 7) showed that in responsive cells without transfection or those transfected with empty vector, nucleosomes 1 and 2 were displaced after LPS stimulation to the same permissive positions seen earlier (compare Fig. 7B with Fig. 4). Interestingly, stimulation of responsive cells expressing RelB resulted in repositioning of nucleosome 2 to the same repressive position seen in tolerant cells (compare with Fig. 4). Despite the expression of RelB, unstimulated responsive cells (R0-RelBwt) did not exhibit any change in the occupancy pattern seen in native positions (i.e. in R0 cells) (not shown). Collectively, the results presented in Figs. 6 and 7 strongly suggest that RelB induction by LPS plays a key role in nucleosome remodeling and transcription silencing of TNFα in tolerant cells.
FIGURE 7.
Forced expression of RelB in responsive cells induces chromatin remodeling through repositioning promoter nucleosomes. THP-1 cells were transfected and treated as described in the legend to Fig. 6. Chromatin was isolated and digested with MNase, and the mononucleosomal DNA was isolated. A, PCR analysis of mononucleosomal DNA. The primers used were those known (from the previous experiments) to cover the two repositioned nucleosomes (see Fig. 4). The results represent three experiments. B, map of the two proximal nucleosomes. R0 cells are shown here again for comparison. Note the repositioning of nucleosomes 1 and 2 after the introduction of RelB DNA into responsive cells (compare with Fig. 4).
The Nucleosome Assembly Protein NAP1 Promotes Nucleosome Positioning in LPS-responsive Cells
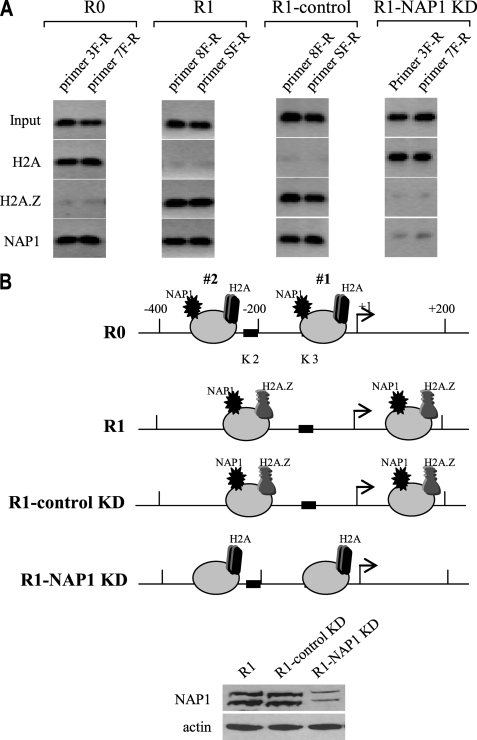
Chromatin-remodeling complexes promote nucleosome positioning through dynamic histone exchange, resulting in nucleosome sliding or eviction (24). NAP1 enhances chromatin fluidity through exchanging histone dimers containing an H2A variant with H2A.Z variant and also interacts with transcriptional activators (25, 38). Our results suggested that LPS stimulation in responsive cells promoted repositioning of nucleosomes 1 and 2 from the native/default position (R0) to an open/permissive position (R1). In addition, our previous work3 showed that the histone variants H2A and NAP1 were not bound to the transcriptionally active TNFα promoter (i.e. in R1 cells), suggesting that the nucleosomes might be remodeled through histone variant exchange. To test this possibility, we measured the H2A and H2A.Z content of, and the binding of NAP1 to, nucleosomes 1 and 2 before and after LPS activation of responsive cells with LPS. In these experiments, chromatin was digested with MNase and then immunoprecipitated with the respective antibody. Nucleosomal DNA was then extracted and analyzed by PCR. As shown in Fig. 8, the H2A variant was detected in both nucleosomes in the basal state (in R0 cells, at native positions) along with NAP1. Interestingly, H2A was replaced with H2A.Z variant (which is normally enriched at transcriptionally active promoters (39)) in the repositioned nucleosomes after LPS stimulation (R1), whereas NAP1 remained bound, suggesting that NAP1 may be involved in histone variant exchange.
FIGURE 8.
NAP1 is required for LPS-induced histone variant exchange and nucleosome repositioning in responsive cells. THP-1 cells were transfected with control or NAP1-specific siRNA. After 36 h, cells were left unstimulated (R0) or stimulated with 1 μg/ml LPS for 1 h (R1). Chromatin was digested with MNase, and mononucleosomes were immunoprecipitated with antibody against H2A, H2A.Z or NAP1. Mononucleosomal DNA was extracted and analyzed. A, PCR analysis of mononucleosomal DNA. The primers used were those known to cover the two repositioned nucleosomes (see Fig. 4). The results represent three experiments. B, map of the histone variant H2A and H2A.Z and NAP1 binding to the two repositioned nucleosomes. Western blot of NAP1 protein after NAP1 KD is shown (bottom).
Next we knocked down NAP1 by RNA interference and then measured the levels of H2A and H2A.Z. NAP1 depletion was confirmed by Western blot (not shown). As shown in Fig. 8, NAP1 inhibition prevented H2A exchange with H2A.Z and inhibited nucleosome repositioning after LPS stimulation (R1-NAP1 KD), because both nucleosomes remained constrained in the native positions despite the presence of LPS (compare with R0). These results clearly demonstrate that NAP1 promotes histone variant exchange and consequently nucleosome positioning around the TNFα promoter after LPS stimulation.
BAF Chromatin-remodeling Complex Is Required for Nucleosome Positioning in LPS-tolerant Cells
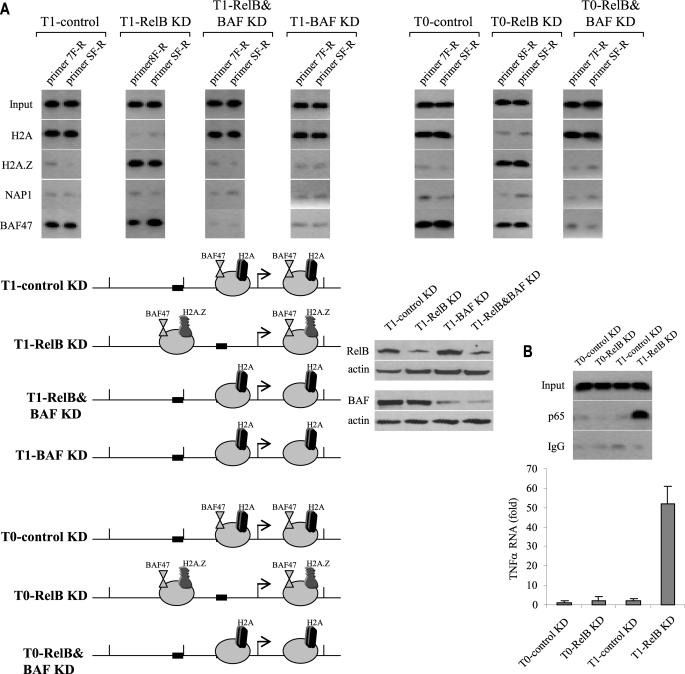
Our results suggested that nucleosome remodeling by histone variant exchange activity of NAP1 was required for nucleosome repositioning from native positions in R0 cells to open/permissive locations in R1 cells. In addition, our results (see Fig. 4) also showed that nucleosomes were also repositioned to permissive locations after RelB knockdown in tolerant cells. To determine whether the same remodeling mechanism, by NAP1, that was observed in responsive cells occurs in tolerant cells after RelB knockdown, we inhibited RelB expression and measured the histone variant content 1 h after LPS stimulation. As shown in Fig. 9A, RelB knockdown resulted in nucleosome repositioning to a permissive location. Interestingly, although H2A was replaced with H2A.Z, NAP1 was not detected either before or after RelB knockdown, suggesting that NAP1 is not involved in histone exchange and nucleosome repositioning in tolerant cells.
FIGURE 9.
The chromatin-remodeling complex BAF replaces the core histone H2A with H2A.Z variant and repositions nucleosomes in tolerant cells depleted of RelB. THP-1 cells were transfected with control or RelB- or RelB plus BAF47-specific siRNAs and stimulated with LPS to induce tolerance. After 36 h, cells were left unstimulated (T0) or stimulated with 1 μg/ml LPS for 1 h (T1). Chromatin was digested with MNase, and mononucleosomes were immunoprecipitated with antibody against H2A, H2A.Z, NAP1, or BAF47. Mononucleosomal DNA was extracted and analyzed. A, PCR analysis of mononucleosomal DNA. The primers used were those known to cover the two repositioned nucleosomes (see Fig. 4). A map of the histone variant H2A and H2A.Z and NAP1 and BAF47 binding to the two repositioned nucleosomes is shown (below). Note the inability of nucleosomes to reposition in the absence of BAF47 expression. Western blots of RelB and BAF proteins after KD are shown (right). B, LPS signal is required to induce p65 binding and transcription activation of TNFα in tolerant cells after RelB knockdown. THP-1 cells were transfected with control or RelB- or RelB-specific siRNAs and stimulated with LPS to induce tolerance. After 36 h, cells were left unstimulated (T0) or stimulated with 1 μg/ml LPS for 1 h (T1). Chromatin isolated and directly immunoprecipitated p65 antibody. DNA was extracted and analyzed by PCR (top). We used a primer pair covering an ∼200-bp proximal promoter sequence that includes the K3 site that binds p65 during transcription activation. The results represent three experiments. In the panel below, RNA was isolated and analyzed by real-time PCR. Values were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA level and are presented as -fold change relative to unstimulated cells (T0-control KD) (assigned 1-fold). Data represent the mean ± S.E. from three experiments.
One of the best characterized examples of mammalian chromatin remodelers is the BAF complexes, the mammalian orthologue of the yeast SWI/SNF (40), which are composed of 10 subunits that use Brg or Brm ATPase activity to move nucleosomes. Because BAF47 is a core subunit of BAF complexes (22, 40) and a previous study reported that siRNA-mediated inhibition of BAF47 prevented IFN-β induction in HeLa cells (41), we measured BAF47 binding to nucleosomes 1 and 2 in tolerant cells before and after RelB knockdown. As shown in Fig. 9A, BAF47 was detected before RelB knockdown along with H2A. After RelB knockdown, which we showed to reposition nucleosomes to permissive locations (see Fig. 4), H2A was replaced with H2A.Z, and BAF47 remained bound. In addition, when both RelB and BAF47 were knocked down, nucleosomes remained in the repressive locations, suggesting that BAF47 participates in nucleosome positioning in tolerant cells but only in the absence of RelB, because BAF47 knockdown alone failed to reposition nucleosomes (Fig. 9A).
To determine whether LPS signal is required for the nucleosome repositioning promoted by BAF complex, we simultaneously knocked down RelB and BAF47 in unstimulated tolerant cells (T0). As shown in Fig. 9A, we noticed the same pattern of nucleosome occupancy seen in stimulated cells (compare all T0 with all T1 phenotypes). This result suggested that, in contrast to responsive cells (see Fig. 8), LPS is not required for nucleosome repositioning in tolerant cells.
Our previous studies showed that RelB knockdown induced p65 binding to the proximal promoters of TNFα and IL-1β and, consequently, reactivated their transcription (27, 42). To assess whether nucleosome repositioning to the permissive locations seen in T0 and T1 cells after RelB knockdown was associated with the induction of p65 binding, we measured p65 binding to the proximal NF-κB (K3) site, which we previously showed is required for the transcription activation (27). As shown in Fig. 9B, p65 was bound in tolerant cells to the open promoter after RelB knockdown but only when cells were stimulated with LPS (compare T0-RelB KD with T1-RelB KD). This binding pattern correlated with the level of TNFα mRNA induction (Fig. 9B, bottom), suggesting that although RelB removal is necessary for nucleosomal remodeling and for opening the promoter in tolerant cells, LPS is still required to induce p65 binding and transcriptional activation. Taken together, the results presented in Fig. 9 suggest that the ATP-dependent chromatin-remodeling complex BAF promotes histone variant exchange and nucleosomal repositioning in epigenetically silenced LPS-tolerant cells. BAF promotes this process only in the absence of RelB. Our results also suggest that nucleosome repositioning from the repressive to permissive location in tolerant cells is not enough to reactivate TNFα transcription and that LPS signal is necessary for transcriptional activation, provided that the promoter nucleosome are repositioned to transcriptionally permissive locations.
DISCUSSION
The gene reprogramming paradigm associated with SSI in animals and humans is typified by transcription induction, followed by sustained silencing of proinflammatory TNFα and IL-1β genes (28, 43, 44). Here, we showed that chromatin remodeling through nucleosomal positioning plays an essential role in the transcription induction as well as silencing of proinflammatory genes. We found that two proximal nucleosomes at the TNFα promoter were repositioned from native/default locations in the basal state to permissive locations after LPS stimulation of responsive cells and then to repressive locations in endotoxin-tolerant cells, thus allowing and preventing access, respectively, to the transcriptional activator NF-κB p65. We observed that ectopic expression of RelB in LPS-responsive cells promoted nucleosome repositioning to repressive locations, whereas RelB inhibition in tolerant cells repositioned the same nucleosomes to permissive locations and reactivated TNFα transcription. We further showed that the ATP-independent chromatin remodeler NAP1 promoted nucleosome positioning in endotoxin-responsive cells, whereas repositioning in tolerant cells required the ATP-dependent BAF-remodeling complex. Together, these results suggest that the TNFα promoter nucleosomes are selectively and constantly remodeled to promote transcription induction and silencing during the course of SSI.
We further found that the TNFα regulatory region is occupied by four positioned and regularly spaced nucleosomes in the basal phenotype (R0 cells). Recent in vivo mapping studies have established that a large fraction of nucleosomes over promoter regions tend to be well positioned and that this positioning appears to be largely dependent on the presence of genetically encoded nucleosome-positioning sequences (reviewed in Ref. 3). Thus, repressed but inducible promoters may block access to some factors critical for transcriptional initiation depending on the sequence of the DNA in nucleosome (3). In other words, nucleosomes may have DNA sequence preferences. Although nucleosome positioning by intrinsically flexible DNA sequences (in cis) may play a role in locating a small subset of nucleosomes (45), recent studies have suggested that nucleosome positions might be regulated in trans by DNA-binding proteins (such as transcription factors) and nucleosome-remodeling complexes, which might override the sequence preferences of nucleosomes and move them to new locations whenever needed (7, 45).
The TNFα promoter contains three NF-κB (K1, K2, and K3) binding sites, with the K3 site indispensable for transcriptional induction, whereas K1 has no role (36). In addition, the K2 site contribution has been unclear. Some studies showed that K2 contributes slightly to the promoter activity, but others suggest that it is dispensable for the promoter activation (35, 36). Interestingly, this K2 site was open only in the basal state (i.e. before transcription induction by LPS) and also in transcriptionally silenced cells, suggesting that it might play a negative role in transcription. Alternatively, it might bind a transcription cofactor that might recruit a remodeling complex and promoter sequence-specific redistribution of nucleosomes. In this study, LPS stimulation led to repositioning of the two proximal nucleosomes. Nucleosome 1 was repositioned upstream of the start site, whereas nucleosome 2 was repositioned to cover the K2 site, leaving the K3 site open and nucleosome-free. In support of our results, recent genome-wide studies indicate that human promoters display reduced nucleosome occupancy (46, 47), and expressed genes often have nucleosome-free regions at their transcription start sites (48, 49).
That transcription factors and nucleosomes may compete for binding to the same DNA sequence has implications for regulation of gene activity (45). The presence of a nucleosome over the transcription factor binding site results in competition between the two types of proteins. Transient dissociation of nucleosomes at the promoter would allow binding of transcription factor and transcription initiation (45). The nucleosome-positioning sequence-directed arrangement of nucleosomes supports the tendency of chromatin assembly factors to assemble histone octamers in low energy binding sites (3). This could explain the basal nucleosome occupancy (i.e. in R0 cells) we observed at the TNFα promoter. The two proximal nucleosomes repositioned after LPS stimulation to different positions, especially nucleosome 2, may depend on transcription factors or chromatin cofactors rather than nucleosome-positioning sequences.
Importantly, we found that forced expression of RelB into responsive cells without LPS stimulation displaced nucleosomes to the same repressive positions observed in tolerant cells, whereas RelB knockdown in tolerant cells moved nucleosomes back to their permissive positions. We discovered that development of endotoxin tolerance and transcription silencing depends on RelB de novo induction and promoter binding (26, 27, 32). The current results clearly demonstrate that nucleosome positioning may be one mechanism by which RelB promotes transcription silencing and tolerance. Although it is well established that the overall stability of nucleosome depends on its constituent DNA sequence and histone modifications (18, 50), how such stability takes place is not clearly understood (51). Our striking finding that H3K9me2 modification always associated with stable nucleosomes, either in a basal/silent position (R0) or in a repressive position (T0 and T1), whereas H3K4me3 associated with permissive/open positions, suggests that histone methylation may play a role in nucleosome positioning. We previously reported that H3K9 dimethylation is catalyzed by G9a, which interacts with and is recruited to the promoter by RelB (32). Since responsive cells do not express RelB, it is unclear how H3K9me2 is generated in responsive cells. It is likely that this chromatin mark is induced by the histone methyltransferase SUV39h, because we found that it is not required to maintain histone methylation in tolerant cells.4 In contrast to the H3K9 silencing mark, the H3K4me3 mark associated with actively transcribed genes (37) existed on nucleosome 1 upstream of the start site in both the activated and silenced phenotype. Together with the finding that nucleosome 1 was never repositioned after LPS stimulation, this suggests that silencing does not require this mark's removal. This does not exclude the possibility that H3K4me3 may contribute to transcription activation, provided that nucleosome 2 is displaced to a permissive position (i.e. after the RelB knockdown). This also supports conclusions (3, 52) that nucleosome positioning on the promoter region are more important than nucleosomes downstream of the start site.
Other reports suggest that one or more positioned promoter nucleosome are repositioned during transcriptional activation (e.g. during activation of IL-12 and cyclin A promoters) (53, 54). These effects are reversible, because nucleosomes were rapidly replaced when the genes were repressed. The emerging picture is that histone replacement or removal requires chromatin-remodeling complexes and histone chaperones (3). The remodeling complex may remove the normal H2A/H2B and replace it with dimers containing the anti-silencing histone variant H2A.Z (21). Since different remodeling complexes may have distinct sequence preferences (55), each complex might promote a unique arrangement of promoter nucleosomes. In addition, remodeling complexes may antagonize each other by promoting an opposing arrangement of nucleosomes (3). Thus, promoters may adopt permissive or repressive nucleosome positions, depending on the type of remodeling complex recruited and whether specific DNA-binding proteins influence remodeling (3).
Our results suggest that NAP1 remodels the TNFα promoter nucleosomes from native/default positions to permissive positions after LPS stimulation of responsive cells. In contrast, NAP1 was not detected on the promoter in tolerant cells before or after LPS stimulation. This suggests that NAP1 promoted nucleosome repositioning in responsive cells only. NAP1 is capable of transiently removing the histone H2A variant from folded nucleosomes and replacing it with the H2A.Z variant (24, 25). The presence of H2A.Z alters nucleosome stability and is highly enriched at gene promoters both upstream and downstream of start sites, where its binding levels correlate positively with transcription activity (39, 56–58). H2A.Z increases in the nucleosome repositioned at the GAL1 promoter, contributing to its transcription activation (57). H2A.Z/H2B dimers are less stable than H2A/H2B dimers (59). H2A.Z-containing promoters have repositioned nucleosomes, whereas promoters lacking H2A.Z do not (57). Although this may not be the case with the TNFα promoter (because nucleosomes in native/default positions (R0 cells) were well positioned, yet they did not have H2A.Z), these previous studies and the current results clearly demonstrate that transcriptionally active chromatin is depleted in the H2A variant and that NAP1 plays an important role in histone exchange and nucleosomal sliding (24, 60, 61). Our finding that H2A.Z associated with H3K4me3 mark suggests that H2A.Z may influence the epigenetic nature of the target chromatin loci, as previously demonstrated (56). In addition, our results indicated that LPS was required for histone variant exchange and nucleosome repositioning by NAP1, because unstimulated responsive cells had no histone variant exchange or nucleosome repositioning despite NAP1 binding (not shown).
RelB knockdown in tolerant cells invoked a distinct mechanism for nucleosome repositioning. Nucleosome repositioning in tolerant cells required BAF complex, because BAF displaced nucleosomes to permissive positions in tolerant cells only after RelB knockdown, and this did not require an LPS signal. Another novel observation of our work is that transcription of the open promoter required an LPS signal that induced NF-κB activation and p65 binding to the K3 site (see Fig. 9).
BAF complex is the mammalian SWI/SNF-related chromatin-remodeling complex (40) where BAF47 is a core subunit. The BAF complex uses ATPase activity of Brg1 or Brm subunit to remodel chromatin (40, 41). Our study suggests that BAF complex promoted nucleosome repositioning in endotoxin-tolerant cells, because the nucleosomes remained in repressive positions when BAF47 was knocked down, despite the inhibition of RelB.
BAF complex regulation of nucleosome remodeling and transcription activation of other cytokines occur. The human IL-12(p35) gene activation during dendritic cell maturation involves selective remodeling of a single positioned promoter nucleosome with a critical SP1-binding site (62). SP1 recruits CREB-binding protein/p300 acetyltransferase activity and interacts with the BAF complex (63). BAF complex control of IFN-β transcription is reported (41), which is induced by viral infection (64). A precisely positioned nucleosome at the IFN-β promoter must be repositioned for such induction. The BAF complex contacts CREB-binding protein (which assembles at the promoter as a part of a large nuclear complex, including NF-κB and HMGB1) to induce sliding of this nucleosome, leading to the induction of IFN-β. In our study, BAF bound the two proximal nucleosomes in tolerant cells (where RelB was bound) and remained bound after RelB knockdown (Fig. 9), suggesting that the BAF complex may be recruited independently of RelB, probably through a chromatin cofactor.
Recently, Smale and co-workers (52) performed a genome-wide nucleosome remodeling study using LPS-stimulated bone marrow-derived macrophages. Their knockdown experiments of Brg1 and Brm (the catalytic subunits of the mammalian nucleosome-remodeling complex) suggest that promoter nucleosomes of many primary response genes are differentially remodeled, depending on the underlying DNA sequence. Remodeling of CpG-rich promoters, including TNFα, from the basal to the activated state, was ATP-independent, whereas remodeling at promoters lacking the CpG island were ATP-dependent and required the ATPase activity of Brg1 and Brm subunits of the remodeling complex. CpG-rich promoters may be unstable due to the nucleotide content and be ATP-independent during transcription activation. In our model, TNFα promoter nucleosomes were remodeled in responsive cells independently of ATP activity, in agreement with the study described above. In tolerant cells, however, nucleosome remodeling was mediated by ATP-dependent BAF complex.
From the results of this study, we propose a new model of nucleosome repositioning in gene expression coupled to euchromatin versus heterochromatin (33). We discovered that endotoxin tolerance generates stably but reversibly silenced heterochromatin from “poised” euchromatin to generate epigenetic silencing of SSI, a process controlled by the RelB transcription factor acting as a repressor (26, 32, 42). When heterochromatin opens after RelB removal by knockdown or perhaps through resolution of SSI,3 the TNFα promoter is again poised for rapid transcription after an LPS signal recruits p65. Unlike the basal state euchromatin and its NAP1 dependence for nucleosome repositioning, ATP-dependent nucleosome repositioning from compacted but reversible heterochromatin requires BAF ATPase activity to reposition the nucleosome and reopen the primary NF-κB binding site. This hypothetical model may have implications for therapeutic interventions and reversing the immunosuppressive epigenetically silenced state associated with SSI.
Acknowledgments
We thank Jean Hu and Sue Cousart for assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI-065791 and R01AI-079144.
M. El Gazzar, T. Liu, B. K. Yoza, and C. E. McCall, unpublished observations.
M. El Gazzar, T. Liu, B. K. Yoza, and C. E. McCall, unpublished data.
- TNFα
- tumor necrosis factor α
- siRNA
- small interfering RNA
- MNase
- micrococcal nuclease
- H3K9me2
- dimethylation on histone H3 lysine 9
- H3K4me3
- trimethylation on histone H3 lysine 4
- KD
- kinase-dead
- R
- responsive
- R0
- unstimulated responsive
- R1
- responsive, stimulated for 1 h
- T
- tolerant
- T0
- unstimulated tolerant
- T1
- tolerant, stimulated for 1 h
- IL
- interleukin
- LPS
- lipopolysaccharide
- CREB
- cAMP-response element-binding protein
- SSI
- severe systemic inflammation.
REFERENCES
- 1.Gottesfeld J. M., Luger K. (2001) Biochemistry 40, 10927–10933 [DOI] [PubMed] [Google Scholar]
- 2.Richmond T. J., Davey C. A. (2003) Nature 423, 145–150 [DOI] [PubMed] [Google Scholar]
- 3.Schnitzler G. R. (2008) Cell Biochem. Biophys. 51, 67–80 [DOI] [PubMed] [Google Scholar]
- 4.Workman J. L. (2006) Genes Dev. 20, 2009–2017 [DOI] [PubMed] [Google Scholar]
- 5.Bondarenko V. A., Steele L. M., Ujvári A., Gaykalova D. A., Kulaeva O. I., Polikanov Y. S., Luse D. S., Studitsky V. M. (2006) Mol. Cell 24, 469–479 [DOI] [PubMed] [Google Scholar]
- 6.Kireeva M. L., Hancock B., Cremona G. H., Walter W., Studitsky V. M., Kashlev M. (2005) Mol. Cell 18, 97–108 [DOI] [PubMed] [Google Scholar]
- 7.Segal E., Fondufe-Mittendorf Y., Chen L., Thåström A., Field Y., Moore I. K., Wang J. P., Widom J. (2006) Nature 442, 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 9.Steger D. J., Workman J. L. (1996) BioEssays 18, 875–884 [DOI] [PubMed] [Google Scholar]
- 10.Benezra R., Cantor C. R., Axel R. (1986) Cell 44, 697–704 [DOI] [PubMed] [Google Scholar]
- 11.Buckle R., Balmer M., Yenidunya A., Allan J. (1991) Nucleic Acids Res. 19, 1219–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G., Chandler S. P., Wolffe A. P., Hall T. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4772–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sewack G. F., Hansen U. (1997) J. Biol. Chem. 272, 31118–31129 [DOI] [PubMed] [Google Scholar]
- 14.Pazin M. J., Bhargava P., Geiduschek E. P., Kadonaga J. T. (1997) Science 276, 809–812 [DOI] [PubMed] [Google Scholar]
- 15.Kadonaga J. T. (1998) Cell 92, 307–313 [DOI] [PubMed] [Google Scholar]
- 16.Krebs J. E., Peterson C. L. (2000) Crit Rev. Eukaryot. Gene Expr. 10, 1–12 [PubMed] [Google Scholar]
- 17.Berger S. L. (2007) Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 18.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 19.Chen J., Kinyamu H. K., Archer T. K. (2006) Mol. Endocrinol. 20, 1–13 [DOI] [PubMed] [Google Scholar]
- 20.de la Serna I. L., Ohkawa Y., Imbalzano A. N. (2006) Nat. Rev. Genet. 7, 461–473 [DOI] [PubMed] [Google Scholar]
- 21.Saha A., Wittmeyer J., Cairns B. R. (2006) Nat. Rev. Mol. Cell Biol. 7, 437–447 [DOI] [PubMed] [Google Scholar]
- 22.Simone C. (2006) J. Cell. Physiol. 207, 309–314 [DOI] [PubMed] [Google Scholar]
- 23.Sims H. I., Baughman C. B., Schnitzler G. R. (2008) Nucleic Acids Res. 36, 6118–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park Y. J., Chodaparambil J. V., Bao Y., McBryant S. J., Luger K. (2005) J. Biol. Chem. 280, 1817–1825 [DOI] [PubMed] [Google Scholar]
- 25.Park Y. J., Luger K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X., El Gazzar M., Yoza B. K., McCall C. E. (2009) J. Biol. Chem. 284, 27857–27865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Gazzar M., Yoza B. K., Hu J. Y., Cousart S. L., McCall C. E. (2007) J. Biol. Chem. 282, 26857–26864 [DOI] [PubMed] [Google Scholar]
- 28.McCall C. E., Yoza B. K. (2007) Am. J. Respir. Crit. Care Med. 175, 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaRue K. E., McCall C. E. (1994) J. Exp. Med. 180, 2269–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virca G. D., Kim S. Y., Glaser K. B., Ulevitch R. J. (1989) J. Biol. Chem. 264, 21951–21956 [PubMed] [Google Scholar]
- 31.El Gazzar M. (2007) Inflamm. Res. 56, 162–167 [DOI] [PubMed] [Google Scholar]
- 32.El Gazzar M., Yoza B. K., Chen X., Hu J., Hawkins G. A., McCall C. E. (2008) J. Biol. Chem. 283, 32198–32208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trojer P., Reinberg D. (2007) Mol. Cell 28, 1–13 [DOI] [PubMed] [Google Scholar]
- 34.Chen C., Yang T. P. (2001) Mol. Cell. Biol. 21, 7682–7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trede N. S., Tsytsykova A. V., Chatila T., Goldfeld A. E., Geha R. S. (1995) J. Immunol. 155, 902–908 [PubMed] [Google Scholar]
- 36.Yao J., Mackman N., Edgington T. S., Fan S. T. (1997) J. Biol. Chem. 272, 17795–17801 [DOI] [PubMed] [Google Scholar]
- 37.Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 38.Asahara H., Tartare-Deckert S., Nakagawa T., Ikehara T., Hirose F., Hunter T., Ito T., Montminy M. (2002) Mol. Cell. Biol. 22, 2974–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 40.Chi T. (2004) Nat. Rev. Immunol. 4, 965–977 [DOI] [PubMed] [Google Scholar]
- 41.Cui K., Tailor P., Liu H., Chen X., Ozato K., Zhao K. (2004) Mol. Cell. Biol. 24, 4476–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoza B. K., Hu J. Y., Cousart S. L., Forrest L. M., McCall C. E. (2006) J. Immunol. 177, 4080–4085 [DOI] [PubMed] [Google Scholar]
- 43.Cavaillon J. M., Adib-Conquy M., Fitting C., Adrie C., Payen D. (2003) Scand. J. Infect. Dis. 35, 535–544 [DOI] [PubMed] [Google Scholar]
- 44.Mathison J. C., Virca G. D., Wolfson E., Tobias P. S., Glaser K., Ulevitch R. J. (1990) J. Clin. Invest. 85, 1108–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radman-Livaja M., Rando O. J. (2009) Dev. Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 47.Ozsolak F., Song J. S., Liu X. S., Fisher D. E. (2007) Nat. Biotechnol. 25, 244–248 [DOI] [PubMed] [Google Scholar]
- 48.Anderson J. D., Widom J. (2000) J. Mol. Biol. 296, 979–987 [DOI] [PubMed] [Google Scholar]
- 49.Mito Y., Henikoff J. G., Henikoff S. (2005) Nat. Genet. 37, 1090–1097 [DOI] [PubMed] [Google Scholar]
- 50.Cosgrove M. S., Boeke J. D., Wolberger C. (2004) Nat. Struct. Mol. Biol. 11, 1037–1043 [DOI] [PubMed] [Google Scholar]
- 51.Hall M. A., Shundrovsky A., Bai L., Fulbright R. M., Lis J. T., Wang M. D. (2009) Nat. Struct. Mol. Biol. 16, 124–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramirez-Carrozzi V. R., Braas D., Bhatt D. M., Cheng C. S., Hong C., Doty K. R., Black J. C., Hoffmann A., Carey M., Smale S. T. (2009) Cell 138, 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X., Wang J., Woltring D., Gerondakis S., Shannon M. F. (2005) Mol. Cell. Biol. 25, 3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coisy M., Roure V., Ribot M., Philips A., Muchardt C., Blanchard J. M., Dantonel J. C. (2004) Mol. Cell 15, 43–56 [DOI] [PubMed] [Google Scholar]
- 55.Rippe K., Schrader A., Riede P., Strohner R., Lehmann E., Längst G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15635–15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gervais A. L., Gaudreau L. (2009) BMC Mol. Biol. 10, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guillemette B., Bataille A. R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L. (2005) PLoS Biol. 3, e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zlatanova J., Thakar A. (2008) Structure 16, 166–179 [DOI] [PubMed] [Google Scholar]
- 59.Placek B. J., Harrison L. N., Villers B. M., Gloss L. M. (2005) Protein Sci. 14, 514–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonaldi T., Längst G., Strohner R., Becker P. B., Bianchi M. E. (2002) EMBO J. 21, 6865–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krude T. (1999) Eur. J. Biochem. 263, 1–5 [DOI] [PubMed] [Google Scholar]
- 62.Goriely S., Demonté D., Nizet S., De Wit D., Willems F., Goldman M., Van Lint C. (2003) Blood 101, 4894–4902 [DOI] [PubMed] [Google Scholar]
- 63.Liu H., Kang H., Liu R., Chen X., Zhao K. (2002) Mol. Cell. Biol. 22, 6471–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goodbourn S., Didcock L., Randall R. E. (2000) J. Gen. Virol. 81, 2341–2364 [DOI] [PubMed] [Google Scholar]