Abstract
The biology of the α subunits of hypoxia-inducible factors (HIFα) has expanded from their role in angiogenesis to their current position in the self-renewal and differentiation of stem cells. The results reported in this article show the discovery of FM19G11, a novel chemical entity that inhibits HIFα proteins that repress target genes of the two α subunits, in various tumor cell lines as well as in adult and embryonic stem cell models from rodents and humans, respectively. FM19G11 inhibits at nanomolar range the transcriptional and protein expression of Oct4, Sox2, Nanog, and Tgf-α undifferentiating factors, in adult rat and human embryonic stem cells, FM19G11 activity occurs in ependymal progenitor stem cells from rats (epSPC), a cell model reported for spinal cord regeneration, which allows the progression of oligodendrocyte cell differentiation in a hypoxic environment, has created interest in its characterization for pharmacological research. Experiments using small interfering RNA showed a significant depletion in Sox2 protein only in the case of HIF2α silencing, but not in HIF1α-mediated ablation. Moreover, chromatin immunoprecipitation data, together with the significant presence of functional hypoxia response element consensus sequences in the promoter region of Sox2, strongly validated that this factor behaves as a target gene of HIF2α in epSPCs. FM19G11 causes a reduction of overall histone acetylation with significant repression of p300, a histone acetyltransferase required as a co-factor for HIF-transcription activation. Arrays carried out in the presence and absence of the inhibitor showed the predominant involvement of epigenetic-associated events mediated by the drug.
Keywords: Cell/Differentiation, Development Differentiation/Stem Cell, Diseases/Neurological, Histones/Acetylase, Oxygen/Hypoxia, Stem Cells, Stem Cells/Neural
Introduction
Hypoxia-inducible transcription factors (HIFs)3 have been the subject of numerous research studies, as they are the key regulators of cell reaction to the lack of cell oxygen. They are widely referred to, in the context of pathological processes of cancer, inflammation, cardiovascular, and neurodegenerative diseases and, in general, all the angiogenic pathologies (reviewed in Refs. 1–4). More recently, HIF biology has progressed due to its interactions with cell pathways that regulate stem cell self-renewal and differentiation, suggesting a new mechanism whereby HIF proteins may drive tumor growth through the generation of tumor-initiating cells or cancer stem cells (5, 6). HIFα proteins, a hallmark of different tumor types, were the focus of many drug discovery efforts, but most inhibitors did not comply with the pharmacological properties required for approval of the drug by the regulatory agencies. Thus, even after more than 20 years of research, there is still room for intervention with novel small molecules that modulate HIF. Strategies for HIF inhibitors include the wide area of angiogenic pathologies and, within the field of regenerative medicine, promising treatments for degenerative diseases and/or the pre-conditioning of the stem cells used for cell transplantation therapies.
HIF is a heterodimer consisting of an oxygen-regulated α subunit (1α, 2α, or 3α) and a constitutively expressed β subunit, or ARNT. HIF proteins are members of the basic helix loop helix-PAS family and bind to canonical DNA sequences (hypoxia-regulated elements or HREs) in the promoters or enhancers of target genes. Despite the existing similarities, α subunits trigger overlapping and specific genes and are therefore involved in different molecular pathways with different physiological consequences for the cells with non-redundant or compensatory function (7–9). Briefly, HIF1α, but not HIF2α, induces genes involved in the glycolysis process (10), whereas HIF2α regulates the angiogenic route, even in the absence of hypoxia (11). HIF2α is also seen as the physiological regulator of Epo production in adult mice (12). HIF3α, however, forms an abortive transcriptional complex with HIF-2α and prevents the engagement of HIF-2 with the HREs acting as negative feedback regulators (13). HIF activity is mainly regulated at the protein level, due to the hydroxylation of key proline residues present in the oxygen-dependent degradation domain of the α subunits by the prolyl-hydroxylases (PHDs) triggering polyubiquitination and rapid degradation of the HIFα proteins through an E3 ubiquitin ligase complex (14). Depletion of oxygen prevents destruction, leading to stabilized α proteins that interact with ARNT in the nucleus, recognize the HRE sequences in the DNA, and activate the transcription mediated by the p300·CBP complexes.
HIFα proteins affect self-renewal and differentiation processes of stem cells by specific regulation of relevant genes and the key transcription factors involved in these processes. It is now known that lowered oxygen concentration enhances neurogenesis and delays certain differentiation processes (15, 16). For instance, HIF1α interacts with Notch1 to maintain undifferentiated cell states (5), whereas HIF2α binds to the marker of the undifferentiated state Oct4 promoter, inducing its expression and transcriptional activity (8). Sox2 controls pluripotency by direct modulation of Oct4 levels in embryonic stem cells of mice (17, 18). Recent articles have shown how pluripotency can be acquired through only a few genetic modifications. Interestingly, the experiments of Takahashi et al. (19, 20) showed that somatic cells can be reprogrammed into pluripotent stem cells by transduction of four defined transcription factors, c-MYC, KLF4, SOX2, and OCT4, two of which (OCT4 and c-MYC) are directly activated by HIF2α (8, 21).
One of the most challenging objectives in cell therapy is to restore neurological function after spinal cord injury (SCI). After SCI, there is a significant cell proliferation of ependymal-derived stem/progenitor cells (epSPC) (22). It is possible to restore locomotor activity when epSPC, activated by the injured tissue (epSPCi), are ectopically transplanted (23). Altering the fate of engrafted or endogenous epSPCi, to restrict differentiation to oligodendrocytes or a neuronal lineage, would replace the loss of functional units and would delay the demyelination process (23). In the present study, we identified and characterized a new chemical entity, FM19G11, which inhibits the expression and transcriptional activity of HIFα isoforms and their corresponding target genes, including the HIF2α-mediated regulation of Sox2, newly characterized here. The specific inhibition of HIFα proteins by FM19G11 reduces the transcriptional activation of the expression of pluripotency markers Sox2 and Oct4 and the corresponding target genes Tgf-α and Nanog in epSPC, thus driving cell differentiation to oligodendrocytes in a process that may favor the design of pharmacological strategies for spinal cord regeneration.
EXPERIMENTAL PROCEDURES
Plasmids and Treatments
The plasmid 9x-HRE-Luc with a luciferase reporter gene was kindly provided by Dr. M. O. Landazuri. This plasmid containing the neomycin resistance gene was used to generate a stably transduced HeLa cell line (HeLa-9x). Plasmids pCMV-TRE (12-O-tetradecanoylphorbol-13-acetate-responsive element) and pGL2-CRE containing a firefly luciferase gene, and the plasmids containing the cDNAs of ATF2, JunB, c-FOS, and c-JUN, kindly provided by Dr. R. Farrás, were used for stable and transient transfection in the HEK293T cell line (the 293-TRE and 293-CRE cell lines were created).
For Sox2 promoter transcriptional activity analysis, a reporter construct was created in pGL3-basic, including the region of the mouse Sox2 promoter sequence −392/−1725 upstream of start codon (pGL3-mpSox2). The in silico search of HRE sequences in the mouse sequence showed the presence of two sites, −725 and −1320, and both were point-mutated by PCR (mpSox2Δ) using the following primers: −1320 HRE FW: CCTATTTGTAACGGAAATGGGGCTGTGGCTC, RV_5′-GAGCCACAGCCCCATTTCCGTTACAAATAGG; −725 HRE FW_5′-GAATTAGGGGTTGAGGACAAATGCTGCGGTTCCTTGAGC and RV_GCTCAAGGAACCGCAGCATTTGTCCTCAACCCCTAATTC.
Luciferase Reporter Activity Assays
105 HEK293T, HeLa-9x, 293-TRE, or 293-CRE cells per well were seeded onto white 96-well plates in quadruplicate 24 h prior to assay. For transient overexpression, 0.05 μg/well in 96-well plates of each plasmid, ATF2, JunB, c-JUN, c-FOS, PGL3-basic, pGL3-mpSox2, pGL3, and mpSox2Δ were transfected with FuGENE6 HD (3:6) 24 h before stimulation. Serial dilutions of FM19G11 from 0 (containing DMSO as a control) up to 1 μm were added immediately before hypoxic stimulation in 1% O2 atmospheres created by the In vivo2 400 chamber (Ruskinn Life Sciences). 6 h after stimulation, luciferase activity was quantified by addition of an equal volume of Bright-Glo Luciferase Reagent (Promega) and detected in the VICTOR3 luminometer (PerkinElmer Life Sciences).
Cytotoxicity Assay
Cell viability was measured by following the CellTiter 96® Aqueous Non-radioactive Cell Proliferation Assay instructions (Promega). 5 × 104 HeLa cells per well were seeded onto 96-well plates 24 h before assay. Serial dilutions of FM19G11 from 0 (containing DMSO as a control) up to 100 μm were used to stimulate the cells for 72 h under standard oxygen conditions (∼20% O2) or hypoxic atmosphere (1% O2).
Chemical Synthesis of FM19G11
The detailed protocols for the synthesis of FM19G11 and its precursors are included in supplemental data I.
Ependymal/Progenitor Cell Isolation and Culture
epSPC were harvested from adult female Sprague-Dawley rats (∼200 g), isolated, and cultured as described elsewhere (23).
Oligodendrocyte-directed Differentiation
Differentiation was performed as previously described (23). Briefly, epSPCs were cultured with glial restriction medium: Dulbecco's modified Eagle's medium, F-12, B27 supplement (Invitrogen), 25 μg/ml of insulin, 6.3 ng/ml of progesterone, 10 μg/ml of putrescine, 50 ng/ml of sodium selenite, 50 μg/ml of holotransferrin, 40 ng/ml of tri-iodothyroidin, supplemented with 4 ng/ml of basic fibroblast growth factor and 10 ng/ml of EGF (Sigma) for 1 day. Subsequently, cells were incubated with 20 ng/ml of EGF and 10 μm of all-trans-retinoic acid for 1 week. All-trans-retinoic acid was then removed and the cells were exposed to glial restriction medium supplemented with 20 ng/ml of EGF for 25 days. At day 28, the spheres were plated in Petri dishes coated with 1:30 Matrigel for 1 week and cultured on glial restriction medium supplemented with 20 ng/ml of EGF. For terminal differentiation, at day 35, oligodendrocyte precursor cells were seeded on poly-l-lysine and human laminin (Sigma)-coated slides. At days 0 and 35, the cells were incubated under hypoxic conditions (1% O2) in the In vivo2 400 chamber (Ruskinn Life Sciences) for 72 h with 500 nm FM19G11 or its DMSO vehicle as control. Then, the cells were harvested for total RNA or immunocytochemical staining.
hESC Culture
Primary human embryonic stem cell (hESC) colonies from the H9 line (WiCell Inc., Madison, WI) were cultured as described elsewhere (24). Briefly, hESC were mechanically dispersed into several small clumps and cultured on fresh, commercially available human foreskin fibroblasts (American Type Culture Collection, Manassas, VA), inactivated by mitomycin C in ES medium containing knock-out Dulbecco's modified Eagle's medium, 1 mm l-glutamine, 100 mm non-essential amino acids, 20% serum replacement, 1% penicillin/streptomycin, 8 ng/ml of basic fibroblast growth factor (Invitrogen), and 100 mm β-mercaptoethanol (Sigma). ES medium was changed every second day. Human embryonic stem cells were passaged by mechanical dissociation and then removed to a freshly prepared human foreskin fibroblast layer.
RNA Isolation, Semi- and Quantitative Reverse Transcription-PCR
One microgram of total RNA, extracted by using the RNeasy Mini-kit (Qiagen, Germany), was reverse transcribed in a total reaction volume of 50 μl by means of incubation at 42 °C for 30 min using random hexamer primers. The primer sequences for semiquantitative PCR are detailed in supplemental data II. The target gene value was normalized to the expression of an endogenous reference (GAPDH).
For quantitative PCR, mRNAs were amplified and quantified by SYBR Green or TaqMan probes (Applied Biosystems) (supplemental data II). As template, 40 ng of cDNA from the target and housekeeping gene (GAPDH) were prepared in separate tubes for each primer master mixture reaction. The comparative threshold cycle (CT) method was used to calculate the relative expression (25).
DNA Microarray Analysis
epSPC isolation and DNA microarray hybridization was performed as described elsewhere (23). The gene profile was sorted by differential expression levels between the two experimental conditions (epSPC 48 h in hypoxia with FM19G11 versus DMSO) and clustered into biological functional profiles by FatiGO application (26).
Western Blot Analysis
Cells were collected and washed with cold phosphate-buffered saline. Total cell protein extracts were isolated by use of 2% SDS Tris-Cl lysis buffer plus proteinase inhibitors. Subcellular fractionation was performed in two steps, by using hypotonic and hypertonic buffers for cytoplasm and nuclear fraction isolation, respectively. SDS-PAGE and hybridization steps were carried out as previously described (23), with antibodies against HIF1α (a kind gift from Dr. Berra), HIF2α, PHD3, Sox2, Oct4, Notch1 (Abcam, UK), RIP, NG2, Nestin, and glial fibrillary acidic protein (GFAP) (Chemicon) at 1:1000 dilution. β-Actin at 1:5000 dilution (Sigma) was used as loading control. The resulting bands were densitometrically analyzed by ImageJ software.
Immunocytochemistry
Fixed and permeabilized cells (0.05% Triton X-100), after blocking (1% fetal bovine serum), were incubated overnight at 4 °C with the primary mouse antibodies (1:200), α-RIP, α-O4 (Chemicon), α-HIF-1α (BD Bioscience), and rabbit antibodies, α-NG2 (Chemicon) and α-Sox2 (Abcam). For detection, Texas Red dye-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories) and Oregon Green 488 goat anti-mouse IgG at 1:400 (Invitrogen) were used. Signals were viewed by confocal microscopy (Leica).
RNA Interference by Small Interfering RNA (siRNA) Duplex Transfection for HIF1α and HIF2α
Annealed siRNA duplexes were purchased from Applied Biosystems. The siRNA sequences targeting rat HIF1α (accession number NM_0243591.1) and HIF2α (accession number NM_023090.1) corresponded to catalog numbers 4390816_s131713 and 4390816_s131443, respectively (Applied Biosystems). 500 nm siRNA were used for transfection.
ChIP Analysis
Chromatin immunoprecipitation analysis used the LowCell ChIP kit (Diagenode), following the manufacturer's instructions, as described elsewhere (27). Samples were incubated with 10 μg of anti-HIF2α or AcH3 antibodies (Abcam). An isotype-matched antibody was used as control for nonspecific binding. The rat Sox2 promoter region was analyzed in silico using the Genomatix bioinformatics software portal.
Statistical Analysis
Statistical comparisons were assessed by the Student's t test. All p values were derived from a two-tailed statistical test using the SPSS 11.5 software. A p value <0.05 was considered statistically significant.
RESULTS
FM19G11, a New HIFα Inhibitor
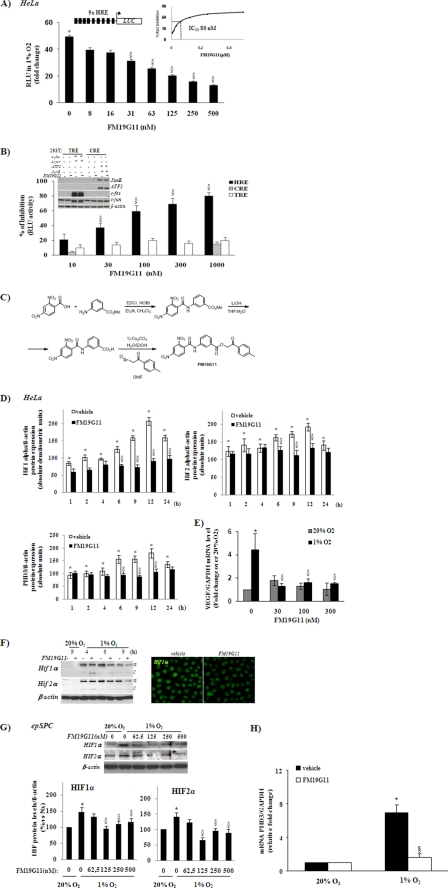
To identify novel molecules targeting the HIF pathway, we used a stable luciferase reporter gene-based screen containing 9 repetitions of the HRE 5′ upstream of the start codon in the active promoter region constitutively expressed in the HeLa cell line (HeLa-9x-HRE-Luc). These cells displayed more than 100-fold higher luciferase activity after incubation in hypoxia (1% O2). We tested the HeLa-9x-HRE-Luc screen against a compound bank, containing more than 12,000 compounds, chosen as representatives of the total chemical space. FM19G11 reduced hypoxia-induced luciferase activity by 50% (IC50) at 80 ± 5 nm concentration (Fig. 1A, inset). FM19G11 showed significant relative luciferase unit (RLU) inhibition from 30 nm, with steady reduction in a characteristic dose-dependent manner (Fig. 1A) and reaching 80% inhibition at 1 μm (Fig. 1B). To test the specificity of FM19G11 for HRE binding sites, we reproduced the luciferase-reporter gene assay by stable overexpression of CRE or TRE reporter constructs. ATF2 and JunB transient overexpression was used to induce CRE-mediated reporter gene activity, and c-fos and c-jun overexpression, for TRE induction (Fig. 1B, inset). No significant RLU inhibition of TRE or CRE transcriptional activity was found in the presence of any tested concentration of FM19G11 (Fig. 1B). It is worth mentioning that no cytotoxicity for concentrations of FM19G11 lower than 30 μm, in standard oxygen tension, or 50 μm, under hypoxic conditions, was observed on the HeLa cell line (supplemental Fig. S1).
FIGURE 1.
Synthesis and analysis of FM19G11 activity in HIFα proteins and their target genes under hypoxia. A, HeLa-9x-HRE-Luc cells treated with different concentrations of FM19G11 (0–0.5 μm) were exposed to 1% O2 for 6 h. FM19G11 inhibits in a dose-responsive way the HIF-translational activity in the luciferase reporter assay. Inset, calculation of IC50 by converting the RLU into % of RLU inhibition. B, 293-TRE or 293-CRE, transiently overexpressing c-fos and c-jun or ATF2 and JunB, respectively, were treated with 0.3 μm FM19G11 for Western blot analysis (inset) or with 0–1 μm for 6 h in the luciferase reporter assay. Luciferase activity in vehicle-treated cells was taken as 0% of inhibition. C, chemical synthesis of FM19G11 in three steps. The reaction of 2,4-dinitrobenzoic acid with methyl 3-aminobenzoate was followed by methyl ester hydrolysis and coupling with (bromomethyl)p-tolylketone to form the target compound. D, intracellular levels of HIF1 and -2α and PHD3 proteins were assayed after HeLa exposition to 1% O2 for 1–24 h with 300 nm FM19G11 (+) or its vehicle (−). 24 h exposition at 20% O2 in the absence of FM19G11 (−) was taken as the basal condition. β-Actin served as loading control. The graphs represent the mean of absolute densitometry values of each condition from three independent experiments. E, TaqMan® real time-PCR analysis of VEGF in HeLa cells treated for 6 h with FM19G11 (0–300 nm) at 1 or 20% O2. F, left panel, representative example of nuclear (n) and cytoplasm (c) fractionated cell proteins assayed for HIFα protein expression. HeLa cells were treated with 300 nm FM19G11 (+) or its vehicle (−) for 4–9 h at 1 or 20% O2. Right panel, immunostaining for HIF1α of HeLa cells treated with FM19G11 or its vehicle (DMSO) under hypoxia for 6 h. G, epSPC, upper panel: representative experiment of the dose-response of FM19G11, showing the effect on HIFα protein expression after 48 h in hypoxia (1% O2). Lower panel, mean of densitometry values of HIFα protein expression analysis. Values are shown as a percentage of the control (20% O2). H, epSPC: TaqMan real time-PCR analysis of PHD3 relative expression levels with vehicle or FM19G11 (500 nm) treated for 48 h under normoxia (20% O2) or hypoxia (1% O2). Results were standardized by the housekeeping gene GAPDH. mRNA levels were calculated by the 2ΔΔCT method. Results were obtained from three independent experiments. Error bars represent S.D. *, p < 0.05 versus 20% O2; §, p < 0.05 versus vehicle at 1% O2 determined by Student's t test.
Chemical Synthesis of FM19G11
The FM19G11 compound was chemically re-synthesized for extensive evaluation. As shown in Fig. 1C, reaction of 2,4-dinitrobenzoic acid with methyl 3-aminobenzoate was followed by methyl ester hydrolysis and coupling with (bromomethyl)p-tolylketone to afford the target compound. FM19G11 was purified by flash column chromatography.
FM19G11 Inhibits HIFα Proteins in Human Tumor Cell Lines
We evaluated the effect of FM19G11 on total protein levels and on nuclear- and cytoplasm-fractionated extracts of HIF1α and -2α (Fig. 1, D and F), as well as the effect on the expression of their target genes PHD3 (Fig. 1D) and VEGF (Fig. 1E) in the HeLa cell line. Simultaneous incubation at 300 nm FM19G11 from 1 to 12 h of hypoxia exposure (1% O2) prevented HIF1α and HIF2α accumulation (this effect was extended to 24 h for HIF1α accumulation). Although most HIFα protein was found in the nucleus in all cases, a detectable amount of both isoforms (1 and 2α) was found in the cytosolic fraction in the presence of FM19G11 after 4 h of incubation at 1% O2. However, no significant changes in the subcellular location of HIFα proteins were observed at any other tested time (Fig. 1F). The prolyl hydroxylases (PHDs/EGLNs) are the central regulators of the molecular responses to oxygen availability (28) and PHD3 is also directly regulated by both α proteins (29). FM19G11 significantly inhibited PHD3 protein levels (Fig. 1D). In addition, the hypoxic transcriptional induction of VEGF, a well known target gene of HIFα proteins (30), was significantly blocked by all tested doses of FM19G11 (Fig. 1E). Because the HIF promoter region contains multiple HRE, indicating a self-regulating mechanism (31), we also analyzed both HIFα isoforms at the mRNA levels in the presence of FM19G11. The hypoxia-dependent induction of HIF1 and 2α mRNA was significantly lower in the presence of FM19G11 after 6 h of incubation (data not shown), coinciding with the significant reduction of the protein levels induced by the compound. However, no significant changes in protein levels, in comparison with vehicle-treated cells, were obtained at shorter (1 or 3 h) or longer (9 or 12 h) hypoxic exposition times (data not shown).
To investigate whether HIF1α protein inhibition by FM19G11 was mediated by promoting the activation of the proteasomal system, we performed an experiment in the presence of the proteasomal inhibitor MG132 in normoxia. Interestingly, MG132 did not affect any isoform, HIF1, or 2α accumulation by FM19G11, suggesting a proteasome-independent mechanism on HIFα inhibition (data not shown). The action of FM19G11 on HIFα proteins was not restricted to HeLa cells, because it was also observed in adult human cell lines derived from various tissue types, including colon HT-29 and the breast cancer cell line MDA-MB-435-S (data not shown).
FM19G11 Inhibits HIFα Protein Accumulation in Adult and Embryonic Stem Cells from Rodents and Humans, Respectively
Interestingly, the compound FM19G11 had a similar effect on HIF regulation in stem cells. We analyzed the effect of the compound on adult rat epSPC and on hESC. Fig. 1G shows the dose-dependent inhibitory effect of FM19G11 on both HIFα proteins in epSPC. Transcriptional repression of Phd3 was also observed in the rat epSPC (Fig. 1H) and hESC (data not shown) treated with 500 nm FM19G11 for 48 h under hypoxia.
FM19G11 Regulates Oct4 and Sox2 Pluripotency Markers
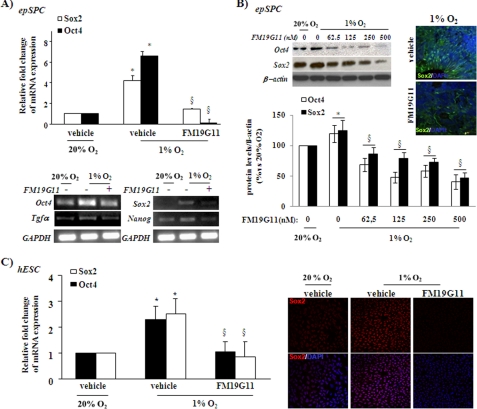
HIF2α transcription factor directly regulates the expression of Oct4, indicating a function of this HIF protein in the self-renewal and differentiation of stem cell properties (8). We observed by real time PCR that FM19G11 abrogates the increment associated with hypoxia of the transcriptional expression of Oct4 in both tested stem cell types, rat epSPC (Fig. 2, A and B) and human ESC (Fig. 2C). In confirmation of recently published results (32), this transcriptional inhibition of Oct4 by FM19G11 was observed in parallel with the down-regulation in both stem cell types of Sox2, another important player in stemness maintenance (33) (Fig. 2). In addition, epSPC treated with FM19G11 under hypoxia showed lower mRNA levels of Tgf-α and Nanog, target genes of Oct4 and Sox2, respectively (Fig. 2A, lower panel). The regulation of the two proteins, Oct4 and Sox2, by the compound showed a dose-dependent inhibitory pattern. The time and dose-response experiments carried out indicated that the hypoxia-induced expression of Oct4 and Sox2 was more efficiently reduced at 500 nm FM19G11 after 48 h in 1% O2 (Fig. 2B, upper and left panel). The immunocytochemical studies confirmed the diminished expression of Sox2 in cells treated with FM19G11, with no apparent alterations in protein location (Fig. 2, B and C) in relation to the controls.
FIGURE 2.
FM19G11 regulates Oct4 and Sox2 pluripotency markers in rodent and human stem cells. epSPC (A and B) and hESC (C): A, upper panel: real time SYBR Green PCR analysis of rat Sox2 and Oct4 relative expression levels. Lower panel, semi-quantitative PCR of Oct4, Sox2, and their direct target genes, Tgfα and Nanog, respectively. Cells were incubated with 500 nm FM19G11 (+) or its vehicle (−) for 48 h under hypoxia (1% O2). 20% O2 was taken as the basal condition. GAPDH served as a loading control. B, left panels: representative Western blot (upper panels) and densitometry analysis of three independent experiments (lower panels). FM19G11 dose-dependent effect on Oct4 and Sox2 protein expression after 48 h of hypoxia exposure. Values are shown as a percentage of the control (20% O2). Right panels: representative immunostaining of the Sox2 protein in undifferentiated neurospheres treated for 48 h in 1% O2 with vehicle or 500 nm FM19G11. C, left panel: TaqMan real time-PCR analysis of human Sox2 and Oct4 relative expression levels in hESC treated with 500 nm FM19G11 or its vehicle; right panel: qualitative immunostaining analysis of human Sox2 protein expression in undifferentiated hESC colonies treated for 48 h in 1% O2 with DMSO (vehicle) or 500 nm FM19G11. 20% O2 condition served as the basal control. Results were obtained from three independent experiments. Error bars represent S.D. *, p < 0.05 versus 20% O2; §, p < 0.05 versus vehicle at 1% O2, determined by Student's t test.
HIF2α Regulates Sox2 Expression
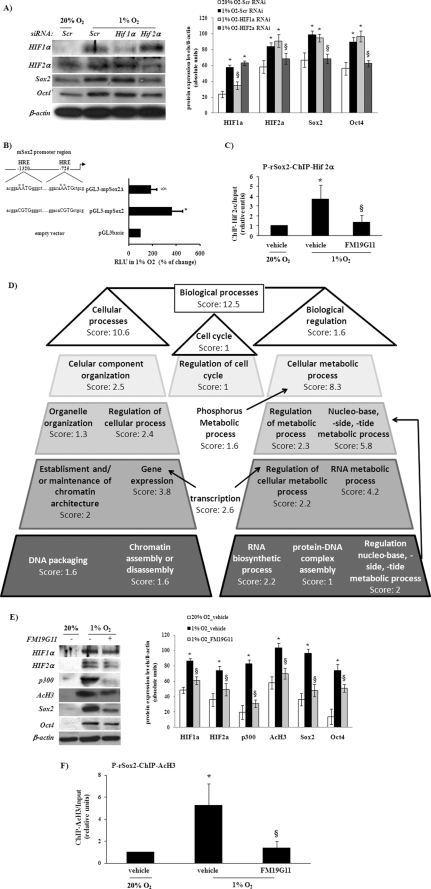
The role of Sox2 in pluripotency is mostly based on its function of maintaining Oct4 levels and the consequent expression of many pluripotency associated genes, e.g. Fgf4, Lefty1, and Nanog, which are tightly regulated by an enhancer containing Oct4 and Sox2 binding motifs (18). Oct4 was reported to be a direct target of HIF2α (8). Recently McCord et al. (32), using the siRNA approach, reported for the first time experimental evidence indicating that Sox2 have HIF-dependent regulation. Here, we validated these results, by using specific siRNA oligos, and the knockdown expression of HIF2α, but not of HIF1α, which blocks hypoxia-induced expression of both Oct4 and Sox2 in epSPC (Fig. 3A). These results strongly indicate the direct involvement of HIF2α in the positive regulation of both pluripotency markers, Oct4 and Sox2. However, to provide further and new data that may indicate the direct connection between HIF2α and Sox2 regulation, we searched for putative HRE-binding sequences within the promoter region of the rodent Sox2 gene. The in silico-predicted occupancy of HIF for HRE binding sites, over the 5-kb promoter region immediately upstream of the transcription start signal of Sox2, was first analyzed by a reporter-based screen including the promoter sequence containing two HRE sites (pGL3-mpSox2) and after the performance of point mutation (pGL3-mpSox2Δ) to inactivate both sites. Transcriptional activation under hypoxia, by using the wild-type sequence (pGL3-mpSox2) rather than the empty vector, was significantly abolished by using the mutated sequence (pGL3-mpSox2Δ; Fig. 3B). Finally, to disclose a specific role for HIF2α in Sox2 promoter activity depending on HRE activation, a ChIP assay was performed. The specific ChIP signals obtained after immunoprecipitation with anti-HIF2α antibody showed, first, more amplified immunoprecipitated sequences after hypoxia stimulation and, then, reduced binding of HIF2α within the Sox2 promoter when epSPC were treated with FM19G11 (Fig. 3C).
FIGURE 3.
Hif2α regulates Sox2 expression and influences the epigenetic mechanisms. A, epSPC were exposed to 1 or 20% O2 for 48 h. 500 nm of each siRNA duplex, scramble (Scr, nonspecific probe), HIF1α, or HIF2α-specific rat probes were transfected 24 h before oxygen-dependent stimulation. The Western blot assay showed that only HIF2α knockdown also reduced the protein levels of Sox2 and Oct4. β-Actin served as a loading control. *, when compared with scramble at 20% O2; §, when compared with scramble at 1% O2; p < 0.05 was determined by Student's t test. B, ChIP analysis within the rat Sox2 promoter. There were a significantly higher number of copies of the Sox2 promoter by real time-PCR amplification after chromatin immunoprecipitation assays using specific antibody for HIF2α. The presence of FM19G11 significantly inhibited hypoxia-dependent induction. *, p < 0.05 versus 20% O2; §, p < 0.05 versus vehicle at 1% O2 determined by Student's t test. C, luciferase reporter assay under hypoxia for 6 h. HEK293T cells transiently transfected with pGL3-basic, empty vector, including the wild-type mouse Sox2 promoter sequence (pGL3-mpSox2) or point mutated at both HRE sites (pGL3-mpSox2Δ) (see diagram on the right) 24 h before hypoxic stimulus. D, FatiGO analysis of epSPC treated with 500 nm FM19G11 was compared with vehicle alone for 48 h in hypoxia. Two biological functional groups were overrepresented in the FM19G11-treated sample after hierarchical clustering. E, left panel: representative Western blot assay for epSPC treated (+) or not (−) with 500 nm FM19G11, exposed for 48 h at 1 or 20% O2; right panel: densitometry analysis of three independent experiments. F, ChIP analysis within the rat Sox2 promoter by using AcH3 antibody for chromatin immunoprecipitation. Error bars represent S.D. *, versus 20% O2; §, versus vehicle at 1% O2; p < 0.05 was determined by Student's t test.
Epigenetic Influences on FM19G11-dependent Sox2 Regulation
To reveal the extended mechanism involved in FM19G11-dependent regulation of Sox2 expression, we performed a DNA microarray analysis, comparing the gene expression profile of epSPC when treated under hypoxia with the compound FM19G11 or its vehicle. The differentially expressed genes were organized according to gene ontology (GO) by using the corresponding gene-GO association table to obtain FatiGO-implemented analysis. As shown in Figs. 3D or supplemental S2 (for more details), two main groups after biological function clustering were overrepresented in the presence of FM19G11. The first group was related to chromatin assembly; and the second, to transcriptional regulation. Taken together, they may indicate that FM19G11 activity in transcriptional regulation is mediated throughout alteration of epigenetic events by chromatin modifications. Stabilized HIFα proteins bind with the ARNT subunit and recruit the p300·CBP complex, two coactivators with histone acetyltransferase activity (34). Based on the transcription profile obtained in the presence of FM19G11, we tested whether FM19G11 would behave as a histone deacetylase inhibitor, but no positive results were obtained (data not shown). However, the presence of FM19G11 inhibited the hypoxia-induced expression levels of the histone 3-acetylated form (AcH3), as well as the levels of total p300, which act as acetyltransferase (Fig. 3E). These results were obtained in close association with the repression of HIF targets, including pluripotency markers. Furthermore, the ChIP signals obtained after immunoprecipitation with anti-AcH3 antibody revealed a rich acetylated region in the Sox2 promoter when the epSPC were exposed to hypoxia. In contrast, a significant reduction in AcH3 signals was seen in epSPC maintained under hypoxic conditions when FM19G11 was present (Fig. 3F).
FM19G11 Promotes Oligodendrocyte Differentiation under Hypoxia
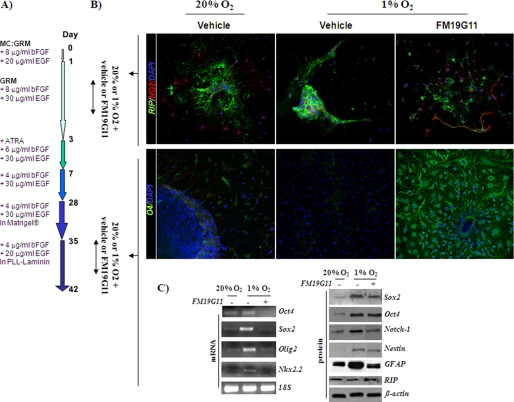
Hypoxia blocks stem cell differentiation; and HIF expression is widely accepted to be associated with stemness. Here, we showed the effect of FM19G11, an HIFα inhibitor, on oligodendrocyte differentiation under hypoxia. An oligodendrocyte differentiation protocol from undifferentiated epSPC was induced until day 42, in line with our earlier research (23), by following the steps described in Fig. 4A. The process of differentiation with epSPC occurred in parallel, in the presence and absence of FM19G11, in an atmosphere of 1% O2, and for early and late differentiation stages from day 1 to 3 and from day 35 to 37, respectively (Fig. 4). At both stages, the hypoxic conditions blocked the differentiation process, according to cell markers RIP and NG2 at early stage (day 1–3) and the mature oligodendrocyte markers 04 and RIP at late stage (day 35–37) (Fig. 4, B and C, for RIP expression). In all cases, the addition of FM19G11 rescued the expression of the above mentioned cell markers (Fig. 4, B and C, for RIP expression). It is also important to mention the poor migration from the epSPC neurospheres into the matrix, under low oxygen concentration and at an early stage of the differentiation protocol (Fig. 4B). At day 37, the cells were also harvested for reverse transcription-PCR and Western blot analysis (Fig. 4C). From day 35 of the differentiation protocol, the precursors were forced to a definitive maturation by culturing in a laminin matrix (Fig. 4A). Sox2, Oct4, Notch1, and Nestin, typically expressed in undifferentiated progenitor cells, were at this late stage up-regulated in hypoxia, in comparison with normoxic conditions (Fig. 4C). Olig2 and Nkx2.2, homeodomain transcription factors, are linked to oligodendrocyte early specification during spinal cord development, gradually reducing in mature cells. The exposure to low oxygen concentration induced the expression of these early specific oligodendrocyte markers, Olig2 and Nkx2.2 (Fig. 4C), but in the presence of FM19G11 the cells recovered the low expression levels of these transcription factors at late differentiation stages. The expression of the astrocytic marker GFAP diminished from day 3 of the differentiation protocol after all-trans-retinoic acid addition (23). Hypoxia significantly induced the expression of GFAP, indicating lower oligodendrocyte specification in the epSPC culture. This induction was abolished by FM19G11 treatment (Fig. 4C). Considering all of the above, the hypoxia-induced delay in directed oligodendrocyte differentiation was aborted by FM19G11 treatment.
FIGURE 4.
FM19G11 favors oligodendrocyte cell differentiation of epSPC under hypoxia. A, diagram of differentiation protocol. B, immunostaining assay for oligodendrocyte cell markers: RIP and NG2 (upper panels) and O4 (lower panels). Upper panels, cells treated with 500 nm FM19G11 or DMSO (vehicle) from day 1 to 3 of the differentiation protocol; lower panels, cells treated during day 35 to 37 of the differentiation protocol under both normoxic (20% O2) and hypoxic (1% O2) conditions. C, extended analysis at day 37 of the differentiation process. epSPC were cultured under normoxic (20% O2) and hypoxic (1% O2) atmospheres and treated with 500 nm FM19G11 (+) and vehicle alone (−). The undifferentiated stage (Oct4, Sox2, Olig2, Nkx2.2, Notch1, Nestin, and GFAP) and the oligodendrocyte-specific fate cell marker (RIP) were assayed by Western blot (right panel) and/or PCR (left panel). 18S and β-actin expression served as loading controls for PCR and Western blot, respectively.
DISCUSSION
Over the last 20 years, major efforts have gone into the search for HIFα inhibitors for use in new drugs (35). Although a wide range of diverse molecules have been found to inhibit the HIF pathway, these molecules often have other actions that indirectly cause lower HIF protein levels. At present, none of the reported HIF inhibitors have met the pharmacokinetic requirements for human therapeutic use. Here we demonstrate that the new chemical entity FM19G11 acts as a potent inhibitor of HIFα proteins in hypoxia, giving high selectivity against other transcription factors of the AP-1 complex used during the screening campaign. Furthermore, we show that this molecule represses the target genes of both HIF proteins, 1α and 2α, in cancer cell lines of various tissues showing lower transcript and protein levels in rat epSPC and human ESC, which suggests a steady mechanism of action for this new drug. The complete eradication of what are known as cancer stem cells might be crucial in curing cancer; reduction of HIF activity may promote their differentiation and decrease their ability to repopulate tumors after chemo- and radiotherapy (6, 36). The low toxicity of this small molecule, no cytotoxicity was observed at concentrations a thousand times higher than the IC50 even in a hypoxic atmosphere, permitted its safe use in a wide variety of live-cell assays, including immuno-based determinations and long-lasting experiments in stem cell differentiation.
Although hypoxia is widely linked to many pathological procedures (1–3), it is also a controller of major physiological processes, such as differentiation status during embryogenesis and in adulthood (1, 6, 8). Hypoxia is associated with the undifferentiated status of stem cells; and the function of HIFα proteins in maintaining multipotency was only found quite recently. The real mechanisms by which the HIF pathway interacts with other pathways to keep stemness are still largely unknown, despite a great many publications in the last few years (6, 36). First, HIF1α was shown to block neuronal and myogenic differentiation in a Notch-dependent manner (5) and, more recently, OCT4 was identified as a HIF2α-specific target gene (8) controlled by Sox2 (18). First of all, Keith and Simon (36) elegantly hypothesized that SOX2 and KLF4 might also be HIF targets and recently McCord et al. (32) validated this hypothesis based on the inhibition by a siRNA of the 2α isoform. However, McCord et al. (32) do not show whether Sox2 is solely under the control of HIF2α or if there is an overlap with the 1α isoform. A reporter assay, based on the promoter region of Sox2 containing two HRE sites, a HIF2α ChIP experiment, and the use of siRNA experiments leading to HIF2α knockdown cells strongly demonstrated that Sox2 is a direct target of the HIFα proteins, and, in particular, that its regulation resides specifically in the 2α isoform. Complementary information that reinforces the role of HIF2α in the direct control of Sox2 was provided by ChIP experiments carried out in the presence of the inhibitor FM19G11. All of the above clearly point to the utility of this small molecule, at present seen just as a tool compound, to clarify the hierarchy of HIF2α in the control of two key genetic factors that govern pluripotency.
Microenvironment influence on chromatin assembly and accessibility and/or dynamic interplay of certain transcription factors determines the stem cell differentiating status (27, 37–39). In fact, the Oct4 locus adopts a closed conformation in differentiating embryonic somatic cells, making it refractory to regulation by HIF2α (8). Here, we confirmed the direct association between the HIF2α-positive transcriptional regulation of Sox2 and the open chromatin conformation of its promoter. FM19G11 prevented the general H3 acetylation induced by hypoxia in epSPC and reduced the expression of p300, the main co-activator for transcriptional activation of HIFα proteins with histone acetyltransferase activity. ChIP analysis by AcH3 immunoprecipitation showed direct involvement of the acetylation mechanism in hypoxia and FM19G11 regulation over the Sox2 transcriptional activity. Although p300 immunoprecipitation experiments proved a Sox2 interaction,4 no evidence linking Sox2 and p300 transcriptional regulation on maintaining the undifferentiated stage was found, as was previously described in the case of Notch1 (40). The inhibitory activity of FM19G11 on Oct4 and Sox2, Notch, and Nanog and transforming growth factor-α opened up new approaches to its use in cell reprogramming experiments with neural progenitor cells for the SCI regeneration model in the rat. Therefore, loss of myelinating oligodendrocytes or oligodendrocyte progenitor cells is a feature of many central nervous system injury and disease states. Moreover, due to secondary damage after SCI, the ischemic environment does not allow re-myelinization, partly because there is an arrest of oligodendrocyte lineage maturation (41). Indeed, when undifferentiated progenitors are transplanted into an ischemic environment, no significant cell differentiation occurs (23, 42). The cell fate modulation of transplanted or endogenous stem cells by forcing the generation of oligodendrocytes to re-myelinate spared axons in the vicinity of the lesion would be a powerful therapeutic approach for SCI regeneration (23, 43, 44). As mentioned above, FM19G11 did repress a variety of key genes involved in stemness, and our reprogramming experiments showed that the inhibitor favors oligodendrocyte differentiation, possibly through modulation of Sox2 and Oct4 expression and by allowing neural stem and/or precursor cells to differentiate. Sox-2 was shown to be the key player in cell fate control, regulating Oct4 and, combined with a few other factors (c-myc and/or Klf4), confers ES-like properties on mature murine fibroblasts (20). However, given the results reported here, it should be emphasized that HIF2α is now positioned in the upper hierarchy of cell fate. All in all, the low toxicity profile of this drug favors pharmacological approaches and enables it to act on SCI regeneration in rigorously defined models.
Supplementary Material
Acknowledgments
We are especially grateful to Dr. M. O. Landazuri and Dr. R. Farras for the HeLa-9x cell line and AP1 related plasmid, respectively. We gratefully acknowledge Dr. Maria Teresa Calvo and Raquel Garijo Fernádez for excellent technical support. We also thank the Confocal Microscopy and Genomic Services of the Centro de Investigación Príncipe Felipe (Valencia, Spain).
This work was supported by the Fondo de Investigaciones Sanitarias, the Instituto de Salud Carlos III (Spain), the Ministerio de Educación, Ciencia y Tecnología, and Generalitat Valenciana (Spain) Projects PI051973 RD06/0010/1006, SAF2007-63714, and GVRE/2008/254.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3, Tables S1–S3, and data I and II.
V. Moreno-Manzano, F. J. Rodríguez-Jiménez, and J. M. Sánchez-Puelles, unpublished data.
- HIF
- hypoxia-inducible transcription factors
- HRE
- hypoxia-responsive element
- PHD
- prolyl-hydroxylase
- ChIP
- chromatin immunoprecipitation
- RLU
- relative luciferase unit
- CRE
- cAMP-response element
- GO
- gene ontology
- AcH3
- histone 3-acetylated form
- DMSO
- dimethyl sulfoxide
- epSPC
- ependymal progenitor stem cell
- SCI
- spinal cord injury
- EGF
- epidermal growth factor
- hESC
- human embryonic stem cell
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- siRNA
- small interfering RNA
- TRE
- 12-O-tetradecanoylphorbol-13-acetate-responsive element
- GFAP
- glial fibrillary acidic protein.
REFERENCES
- 1.Folkman J. (2007) Nat. Rev. Drug Discov. 6, 273–286 [DOI] [PubMed] [Google Scholar]
- 2.Pouysségur J., Dayan F., Mazure N. M. (2006) Nature 441, 437–443 [DOI] [PubMed] [Google Scholar]
- 3.Bertout J. A., Patel S. A., Simon M. C. (2008) Nat. Rev. Cancer 8, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rankin E. B., Giaccia A. J. (2008) Cell Death Differ. 15, 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gustafsson M. V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J., Ruas J. L., Poellinger L., Lendahl U., Bondesson M. (2005) Dev. Cell 9, 617–628 [DOI] [PubMed] [Google Scholar]
- 6.Simon M. C., Keith B. (2008) Nat. Rev. Mol. Cell Biol. 9, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raval R. R., Lau K. W., Tran M. G., Sowter H. M., Mandriota S. J., Li J. L., Pugh C. W., Maxwell P. H., Harris A. L., Ratcliffe P. J. (2005) Mol. Cell. Biol. 25, 5675–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covello K. L., Kehler J., Yu H., Gordan J. D., Arsham A. M., Hu C. J., Labosky P. A., Simon M. C., Keith B. (2006) Genes Dev. 20, 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Covello K. L., Simon M. C., Keith B. (2005) Cancer Res. 65, 2277–2286 [DOI] [PubMed] [Google Scholar]
- 10.Hu Y., Leaver S. G., Plant G. W., Hendriks W. T., Niclou S. P., Verhaagen J., Harvey A. R., Cui Q. (2005) Mol. Ther. 11, 906–915 [DOI] [PubMed] [Google Scholar]
- 11.Dutta D., Ray S., Vivian J. L., Paul S. (2008) J. Biol. Chem. 283, 25404–25413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruber M., Hu C. J., Johnson R. S., Brown E. J., Keith B., Simon M. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maynard M. A., Evans A. J., Shi W., Kim W. Y., Liu F. F., Ohh M. (2007) Cell Cycle 6, 2810–2816 [DOI] [PubMed] [Google Scholar]
- 14.Metzen E., Zhou J., Jelkmann W., Fandrey J., Brüne B. (2003) Mol. Biol. Cell 14, 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Studer L., Csete M., Lee S. H., Kabbani N., Walikonis J., Wold B., McKay R. (2000) J. Neurosci. 20, 7377–7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C. P., Zhu L. L., Zhao T., Zhao H., Huang X., Ma X., Wang H., Fan M. (2006) Neurosignals 15, 259–265 [DOI] [PubMed] [Google Scholar]
- 17.Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A. A., Ko M. S., Niwa H. (2007) Nat. Cell Biol. 9, 625–635 [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 21.Gordan J. D., Bertout J. A., Hu C. J., Diehl J. A., Simon M. C. (2007) Cancer Cell 11, 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson C. B., Momma S., Clarke D. L., Risling M., Lendahl U., Frisén J. (1999) Cell 96, 25–34 [DOI] [PubMed] [Google Scholar]
- 23.Moreno-Manzano V., Rodríguez-Jiménez F. J., García-Roselló M., Laínez S., Erceg S., Calvo M. T., Ronaghi M., Lloret M., Planells-Cases R., Sánchez-Puelles J. M., Stojkovic M. (2009) Stem Cells 27, 733–743 [DOI] [PubMed] [Google Scholar]
- 24.Erceg S., Laínez S., Ronaghi M., Stojkovic P., Pérez-Aragó M. A., Moreno-Manzano V., Moreno-Palanques R., Planells-Cases R., Stojkovic M. (2008) PLoS ONE 3, e2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 26.Al-Shahrour F., Minguez P., Vaquerizas J. M., Conde L., Dopazo J. (2005) Nucleic Acids Res. 33, W460–W464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Jiménez F. J., Moreno-Manzano V., Lucas-Dominguez R., Sánchez-Puelles J. M. (2008) Stem Cells 26, 2052–2062 [DOI] [PubMed] [Google Scholar]
- 28.Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. (2004) J. Biol. Chem. 279, 38458–38465 [DOI] [PubMed] [Google Scholar]
- 29.Pescador N., Cuevas Y., Naranjo S., Alcaide M., Villar D., Landázuri M. O., Del Peso L. (2005) Biochem. J. 390, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitz L. J., Morris J. C., Towler P., Long A., Burgess P., Greco R., Wang J., Gassaway R., Nickbarg E., Kovacic S., Ciarletta A., Giannotti J., Finnerty H., Zollner R., Beier D. R., Leak L. V., Turner K. J., Wood C. R. (1997) Oncogene 15, 613–618 [DOI] [PubMed] [Google Scholar]
- 31.Koh M. Y., Spivak-Kroizman T., Venturini S., Welsh S., Williams R. R., Kirkpatrick D. L., Powis G. (2008) Mol. Cancer Ther. 7, 90–100 [DOI] [PubMed] [Google Scholar]
- 32.McCord A. M., Jamal M., Shankavaram U. T., Lang F. F., Camphausen K., Tofilon P. J. (2009) Mol. Cancer Res. 7, 489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J. B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., Araúzo-Bravo M. J., Ruau D., Han D. W., Zenke M., Schöler H. R. (2008) Nature 454, 646–650 [DOI] [PubMed] [Google Scholar]
- 34.Fedele A. O., Whitelaw M. L., Peet D. J. (2002) Mol. Interv. 2, 229–243 [DOI] [PubMed] [Google Scholar]
- 35.Giaccia A., Siim B. G., Johnson R. S. (2003) Nat. Rev. Drug Discov. 2, 803–811 [DOI] [PubMed] [Google Scholar]
- 36.Keith B., Simon M. C. (2007) Cell 129, 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruau D., Ensenat-Waser R., Dinger T. C., Vallabhapurapu D. S., Rolletschek A., Hacker C., Hieronymus T., Wobus A. M., Müller A. M., Zenke M. (2008) Stem Cells 26, 920–926 [DOI] [PubMed] [Google Scholar]
- 38.Maltepe E., Krampitz G. W., Okazaki K. M., Red-Horse K., Mak W., Simon M. C., Fisher S. J. (2005) Development 132, 3393–3403 [DOI] [PubMed] [Google Scholar]
- 39.Lyssiotis C. A., Walker J., Wu C., Kondo T., Schultz P. G., Wu X. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14982–14987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallberg A. E., Pedersen K., Lendahl U., Roeder R. G. (2002) Mol. Cell. Biol. 22, 7812–7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segovia K. N., McClure M., Moravec M., Luo N. L., Wan Y., Gong X., Riddle A., Craig A., Struve J., Sherman L. S., Back S. A. (2008) Ann. Neurol. 63, 520–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parr A. M., Kulbatski I., Zahir T., Wang X., Yue C., Keating A., Tator C. H. (2008) Neuroscience 155, 760–770 [DOI] [PubMed] [Google Scholar]
- 43.Meletis K., Barnabé-Heider F., Carlén M., Evergren E., Tomilin N., Shupliakov O., Frisén J. (2008) PLoS Biol. 6, e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohori Y., Yamamoto S., Nagao M., Sugimori M., Yamamoto N., Nakamura K., Nakafuku M. (2006) J. Neurosci. 26, 11948–11960 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.