Abstract
Activation of the Go/i-coupled cannabinoid 1 receptor (CB1R) has been shown to induce neurite outgrowth in Neuro2A cells through activation of Src kinase and STAT3 transcription factor. Signaling by the interleukin 6 receptor (IL-6R) also activates STAT3 through Jak kinase. We studied if signals from the two pathways could be integrated in a synergistic manner to trigger neurite outgrowth in Neuro2A cells. At low concentrations, when agonist at either receptor by itself has no effect, we found that CB1R and IL-6R stimulation together induced synergistic neurite outgrowth. Signal integration requires activation of transcription factors by Src, Jak, and mitogen-activated protein kinases. Mitogen-activated protein kinase can be activated by both receptors and shows enhanced early activation in the presence of both ligands. CREB and STAT3 transcription factors are required for synergy and show enhanced DNA-binding activity when both receptors are activated. STAT3 plays a critical role in integration of the signals downstream of the two receptors. When both pathways are activated, STAT3 phosphorylation is sustained for 6 h. This prolonged activation of STAT3 requires deactivation of SHP2 phosphatase. Reduction of SHP2 levels by RNA interference results in greater synergy in neurite outgrowth. Simultaneous knockdown of both SHP2 and STAT3 blocks the synergistic triggering of neurite outgrowth, indicating that STAT3 is downstream of SHP2. CB1R and IL-6R co-stimulation enhanced the differentiation of rat cortical neuron primary cultures. These results provide a mechanism where multiple protein kinases and transcription factors interact to integrate signals from G protein-coupled and cytokine receptor to evoke neurite outgrowth in Neuro2A cells.
Introduction
Cells must integrate the signals that they receive from multiple pathways in order to respond effectively to environmental cues. Neurons in particular have to be able to respond to signals not only biochemically but also morphologically, in order to properly develop and grow through their environment. One of the early stages of neuronal differentiation involves the process of neurite outgrowth, during which neurons begin to extend processes, which will later become dendrites and axons (1). Neurite outgrowth is a tightly controlled process that is known to be regulated by multiple pathways, controlled by growth factor receptors (2), cytokine receptors (3), and G-protein-coupled receptor pathways (4–6). All neurites have the potential to develop into an axon (7–9) and keep their ability to differentiate even after they take on a specific function, suggesting a role in neuronal regeneration.
Interleukin-6 (IL-6)3 belongs to a group of structurally related cytokines, called neuropoietic cytokines, which have all been reported to promote neuronal survival and differentiation and to prevent neuronal apoptosis in central nervous system in the event of injury (10, 11). In an intact and healthy brain, IL-6 levels usually remain low, but expression of IL-6 increases dramatically in various neurological diseases (12, 13) and acute brain injury (14). Although cerebral overexpression of IL-6 can lead to neurodegeneration, gliosis, microglial activation, and vascular proliferation (15), when used in moderate amounts, IL-6 enhances the differentiation and survival of neuronal cell lines and primary neurons (10, 16). IL-6 has also been found to have neuroprotective effects in neurons exposed to glutamate excitotoxicity (17) as well as in sensory neuron development in vivo (18). Evidence suggests that IL-6 exerts its neuroprotective effects by binding to the IL-6 receptor (IL-6R)-gp130 complex and activating the Jak/STAT pathway. Phosphorylation of STAT3 results in its nuclear translocation, which has been reported to stimulate neuronal cell survival (19). Importantly, IL-6R activation has been found to lead to axonal regeneration after spinal cord injury in vivo (20).
As an important process in neuronal regeneration, neurite outgrowth has been extensively studied in multiple cell lines of neuronal origin, such as PC12 cells (a rat pheochromocytoma cell line) and Neuro2A (N2A) cells (mouse neuroblastoma cell line). IL-6 in particular has been demonstrated to play an important role in the induction of neuronal differentiation in PC12 cells (3, 21, 22). Although IL-6 signaling seemed to be insufficient to induce neurite outgrowth on its own, it was able to enhance neuronal differentiation in nerve growth factor-stimulated PC12 cells (3).
Another signaling pathway that has been previously demonstrated to play an important role in neurite outgrowth is the Gαo/i- coupled cannabinoid 1 receptor (CB1R) pathway. Our laboratory has previously shown that CB1R activates the Src/STAT3 pathway through Gαo/i (4). Activation of Gαo/i generates a signaling cascade that includes Rap1, Ral, Src, Rac, and JNK. He et al. (4) have demonstrated that each one of these components in the signaling network plays a critical role in regulating CB1R/Gαo/i-induced neurite outgrowth. A more recent study from our laboratory illustrated that CB1R-stimulated neurite outgrowth may be regulated by a much larger signaling network, allowing for many potential points of signal integration (23). For integration of IL-6R and CB1R signals, STAT3 appears to be a logical choice, since it is activated by both pathways.
STAT3 activation has been shown to play an important role in neurite outgrowth. Wu and Bradshaw (24) found that STAT3 tyrosine phosphorylation increased in nerve growth factor-primed cells after IL-6 application. They also saw that IL-6 was able to induce a dose-dependent neurite outgrowth in primed PC12 cells. He et al. (4) observed that although activation of the CB1R pathway caused STAT3 phosphorylation and induced neurite outgrowth, dominant negative STAT3 inhibited HU-210-induced neurite outgrowth in a dose-dependent manner. Both IL-6R and CB1R pathways have STAT3 as a common signal transducer, which becomes phosphorylated, homodimerizes, translocates into the nucleus, and induces gene expression. Because STAT3 provides a possible point of convergence of two signaling pathways, we reasoned that its activation from these two signaling pathways may induce a synergistic enhancement of neurite outgrowth. Using Neuro2A cells, we examined if signals from the CB1R and IL-6 receptors could be integrated and investigated the mechanism underlying signal integration.
In this study, we show that concurrent activation of the endogenous CB1R and IL-6R signaling pathways leads to synergistic neurite outgrowth in Neuro2A cells. STAT3 serves as a major point of convergence for signals from CB1R and IL-6R, and signal integration at STAT3 leads to synergistic triggering of neurite outgrowth. The underlying mechanism involves sustained STAT3 activity through deactivation of SHP2 (protein tyrosine phosphatase, non-receptor type 2) at later times. The MAPK/CREB signaling cascade also shows enhanced activation when both receptors are stimulated and is required for synergistic neurite outgrowth. These results show that MAPK, STAT3, and SHP2 promote signal integration between G-protein-coupled receptors and cytokine receptors.
EXPERIMENTAL PROCEDURES
Materials and Antibodies
HU-210 was obtained from Sigma, and mouse IL-6 was from Invitrogen. PP2, PD98059, and LY294002 were from Calbiochem. Polyclonal rabbit antibodies directed against phospho-STAT3, total STAT3, phospho-SHP2, total SHP2, MAP2, phospho-MAPK1/2, total MAPK1/2, phospho-Erk5, total Erk5, phospho-Akt, total Akt, and monoclonal mouse antibodies directed against Tau were from Cell Signaling Technology Inc. Monoclonal mouse antibodies against β-tubulin were from Sigma.
Neuro2A Cell Culture, Transfection, and Immunoblotting
N2A cells were cultured in minimal essential medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, 1% l-glutamine, and 1% sodium pyruvate. For phosphorylation time course experiments, cells were serum-starved in minimal essential medium supplemented with 0.1% bovine serum albumin for 24 h. DNA transfections were performed when cells were ∼75% confluent using Lipofectamine 2000® (Invitrogen) according to the manufacturer's instructions. For all DNA transfections, cells were co-transfected with green fluorescent protein to identify transfected cells during neurite outgrowth assays. Dominant negative mutants of STAT3 contain a mutation of tyrosine 705 to phenylalanine (DN STAT3 (Y)) or mutation of the DNA-binding domain (DN STAT3 (VVV)) (25). The dominant negative mutant of Jak1 contains a 3-amino acid change in the conserved region VIII of the kinase domain that impairs its catalytic function (26). Dominant negative CREB (KCREB) contains a mutation in its DNA-binding domain. Twenty-four hours after transfection, cells were plated into 35-mm Mattek® dishes and treated with the indicated concentrations of IL-6 and/or HU-210, as described for neurite outgrowth assays. Control, SHP2, and STAT3 on-target plus smart-pool siRNAs were obtained from Dharmacon, and siRNA transfections were performed according to the manufacturer's instructions using the Dharmafect 2 reagent (Dharmacon). Cells were transfected with siRNA for 48 h, split into Mattek® dishes at 45,000 cells per dish, and subjected to neurite outgrowth assays as described below.
For immunoblotting, cells were serum-starved for 24 h, treated with IL-6, HU-210, or both, and lysed in radioimmunoprecipitation assay buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 1 mm EDTA) supplemented with protease and phosphatase inhibitors. After determination of protein concentration, the cell lysates were subjected to immunoblot analysis using standard procedures and visualized by enhanced chemiluminescence.
Gel Shift Assay
Electrophoretic mobility shift assays were performed using the LightShift chemiluminescent electrophoretic mobility shift assay kit (Pierce). Consensus sequences for STAT3 (5′-TGCATTTCCCGTAAATCT-3′ and its complement) and CREB (5′-TCAAATTGACGTCATGGTAA-3′ and its complement) were biotinylated with the Biotin 3′-end labeling kit (Pierce) and then annealed. Biotinylated consensus sequences were then incubated with 10 μg of nuclear extracts for 20 min at room temperature and then loaded on a 0.5× TBE 5% non-denaturing polyacrylamide gel. The DNA-protein complexes were transferred to a Biodyne B membrane (Pall), incubated with streptavidin-horseradish peroxidase, and visualized by enhanced chemiluminescence according to the manufacturer's instructions.
Neurite Outgrowth Assays
N2A cells were plated in 35-mm Mattek® dishes at 45,000 cells/plate. The cells were serum-starved for 1 h and then treated with IL-6, HU-210, or both for 18 h. For Src, MAPK, and PI3K inhibition experiments, the cells were serum-starved for 1 h, treated with the corresponding inhibitor for an indicated period of time, and then treated with IL-6 and HU-210 as above. Calcein AM (Invitrogen) was then added to visualize live cells, and cells were imaged with a Zeiss 510 Meta confocal microscope at ×20 magnification. For DNA transfections, green fluorescent protein expression was used for imaging of transfected cells. Pictures of 5–10 random fields/condition were taken per dish. The cells with projections of a length at least 2 times greater than the cell diameter were scored as positive for neurite outgrowth (4, 23). Neurite outgrowth was quantified as the percentage of cells with neurites of total cells in the fields of corresponding condition.
Data Analysis
All N2A experiments were carried out at least three times, and the indicated values are represented as mean ± S.E. Statistically significant differences were determined by unpaired Student's t test. Significance was defined as p < 0.05.
Rat Primary Cortical Neuron Cultures
Rat primary cortical cultures were dissected from postnatal day 1 rat brains. Cortices were incubated with trypsin and DNase for 15 min at 37 °C, and cells were dissociated by trituration. Cells were plated on glass coverslips in 12-well dishes coated with 1 μg/μl poly-l-lysine (Sigma) at ∼6000 cells/well in plating medium (minimal essential medium supplemented with 10% horse serum, 0.6% glucose, and 1 mm sodium pyruvate). After cells adhered (∼4 h later), cultures were grown in Neurobasal medium (Invitrogen) supplemented with B27 and 1 mm sodium pyruvate. At 1 day in vitro (DIV), cultures were treated with 5 μm AraC to prevent glial growth. At 2 DIV, cultures were treated with PBS as a control, IL-6, HU-210, or IL-6 and HU-210. After 24 h, the cells were fixed for 10 min at room temperature in 4% para-formaldehyde, 4% sucrose in PBS and then permeabilized in 0.25% Triton X-100 in PBS for 5 min. After blocking in 10% bovine serum albumin in PBS for 1 h, cells were incubated with antibodies directed against MAP2 (rabbit) and Tau (mouse) in 1% bovine serum albumin, 0.01% Triton X-100 overnight at 4 °C in a humidified chamber. Cells were then washed three times with PBS and incubated with AlexaFluor-568-conjugated goat anti-rabbit and Oregon Green-488-conjugated goat anti-mouse secondary antibodies (Invitrogen) for 1 h at room temperature in 1% bovine serum albumin in PBS. Neurons were washed with PBS, incubated with Hoechst dye, and mounted on microscope slides in ProLong Gold (Invitrogen). Images were acquired using a Zeiss 510 Meta confocal microscope at ×40 magnification. For each experiment 30–50 cells were imaged per condition, and experiments were repeated four times. The length of the longest neurite of each cell was measured morphometrically using the Zeiss LSM image browser.
RESULTS
Stimulation of N2A Cells with Low Concentrations of IL-6 and CB1R Agonist HU-210 Results in Synergistic Neurite Outgrowth
Our laboratory has previously shown that treatment with CB1R agonist HU-210 induces neurite outgrowth in N2A cells through activation of STAT3 (4, 23). Since IL-6 signaling also signals through STAT3 and has been shown to enhance nerve growth factor-induced neurite outgrowth (24), we decided to test whether activation of both CB1R and IL-6R pathways may result in synergistic neurite outgrowth due to two sources of STAT3 activation. We began this study by testing a range of IL-6 concentrations for the ability to cause STAT3 phosphorylation through activation of IL-6R-gp130 and the Janus kinase (Jak/STAT pathway). N2A cells were treated with a range of IL-6 concentrations for 5 min, and protein lysates were subjected to immunoblot analysis. As shown in supplemental Fig. 1S, treatment of N2A cells with IL-6 led to a dose-dependent increase in STAT3 phosphorylation. Doses of HU-210 used in this study were based on previously published work from our laboratory (4).
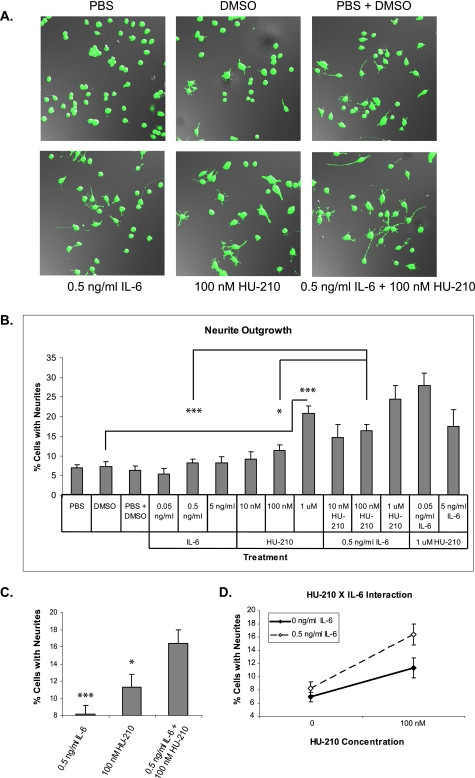
In order to test whether CB1R and IL-6R activation would cause a synergistic effect on neuronal differentiation, we performed neurite outgrowth assays. N2A cells were plated at low density, serum-starved for 1 h, and treated with a range of IL-6 and HU-210 concentrations as well as combinatorial treatments as indicated in Fig. 1, A and B. Treatment with IL-6 did not induce neurite outgrowth, whereas treatment with HU-210 resulted in low levels of neurite outgrowth in a dose-dependent manner. When 0.5 ng/ml IL-6 was combined with a range of HU-210 concentrations, combinations of low doses resulted in synergistic neurite outgrowth (Fig. 1, C and D, and supplemental Table 1S). Fig. 1C represents an inset from Fig. 1B with basal levels of neurite outgrowth subtracted, demonstrating that application of 0.5 ng/ml IL-6 and 100 nm HU-210 results in synergistic neurite outgrowth. Fig. 1D illustrates the same data, emphasizing that the addition of IL-6 results in a greater enhancement of neurite outgrowth in the presence of HU-210. Based on the finding that 0.5 ng/ml IL-6 and 100 nm HU-210 resulted in the greatest synergy, we used these ligand concentrations to examine the mechanism for synergy.
FIGURE 1.
Stimulation of CB1R and IL-6R results in synergistic neurite outgrowth. A, N2A cells were serum-starved for 1 h and treated with the indicated concentrations of the CB1R agonist (HU-210) and IL-6 cytokine for 18 h. Calcein AM was then added to cells, and live cells were imaged via confocal microscopy. B, the bar graph shows mean ± S.E. for percentages of cells with neurites. Quantifications were performed from at least four independent experiments. Statistical differences were calculated using a two-tailed t test; *, p < 0.05, ***, p < 0.0005. See supplemental Table 1S for statistical significance comparisons of all conditions. C, inset of data from B with basal levels of neurite outgrowth removed. Application of both ligands produced an enhancement in neurite outgrowth. D, two-way analysis of variance suggests a possible interaction between 0.5 ng/ml IL-6 and 100 nm HU-210, where the presence of IL-6 enhances the effect of 100 nm HU-210 on neurite outgrowth.
Regulation by Protein Kinases MAPK (p42/p44), Src, and Jak
Activation of Kinases Downstream of both IL-6R and CB1R Pathways Is Necessary for Synergistic Neurite Outgrowth
Our laboratory has previously reported that stimulation of the CB1R pathway in N2A cells activates MAPK and Src kinases (4, 23). MAPK may also be activated by IL-6R through recruitment of the Grb2-SOS complex, followed by activation of the Ras-Raf-MAPK cascade (27). It is also well known that IL-6R activates several Jak family member kinases, including Jak1 (28, 29).
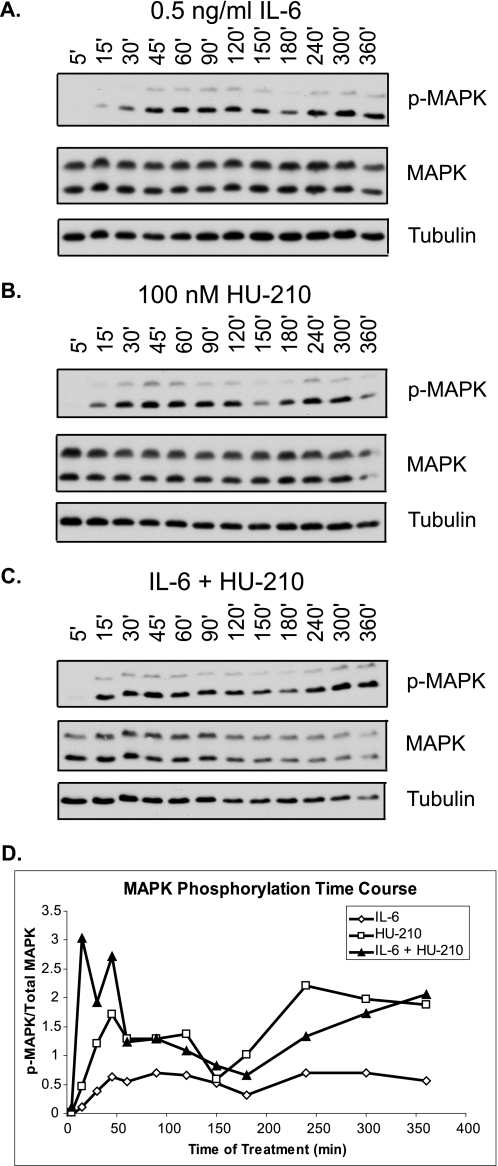
Because MAPK may be a potential point of signal convergence at the level of kinase regulation, IL-6, HU-210, and combined treatment may result in differential MAPK signaling. Therefore, we examined the time course response of MAPK phosphorylation to individual and combined treatments. N2A cells were serum-starved for 24 h and treated with 0.5 ng/ml IL-6, 100 nm HU-210, or both. The cells were lysed at a number of time points after treatment ranging from 5 min to 6 h. Treatment of cells with both ligands resulted in enhanced early MAPK phosphorylation at 15, 30, and 45 min post-treatment (Fig. 2, A–D), supporting its potential role in synergistic neurite outgrowth.
FIGURE 2.
Stimulation of CB1R and IL-6R results in enhanced early MAPK phosphorylation. N2A cells were serum-starved for 24 h and treated with 0.5 ng/ml IL-6 (A), 100 nm HU-210 (B), or both (C) for the indicated periods of time. The cells were lysed and subjected to immunoblot analysis for phospho-MAPK, total MAPK, and tubulin. The blots shown are representative of three independent experiments. D, quantification of MAPK phosphorylation represented as phospho-MAPK (p-MAPK)/total MAPK ratio over time.
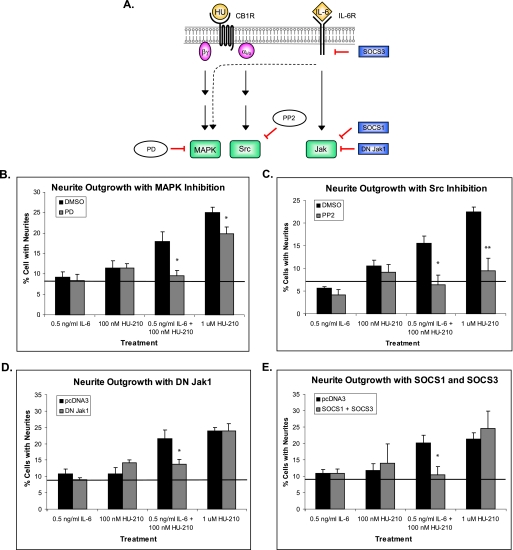
To examine the contribution of MAPK, Src, and Jak protein kinases activated by CB1R and IL-6R to synergistic neurite outgrowth, we individually inhibited the activity of these kinases and performed neurite outgrowth assays (Fig. 3A). To inhibit MAPK signaling, N2A cells were pretreated with MEK inhibitor PD98059 (PD) for 4 h prior to stimulation of cells with IL-6 and HU-210. Treatment with 20 μm PD did not inhibit Erk5 activity (supplemental Fig. 14S). To inhibit Src activity, N2A cells were treated with PP2 for 1 h prior to stimulation. For Jak1 inhibition, the cells were transfected with a dominant negative Jak1 kinase containing a mutation in its kinase domain (DN Jak1). Neurite outgrowth assays were then performed as described under “Experimental Procedures.”
FIGURE 3.
Activation of kinases downstream of both CB1R and IL-6R is necessary for synergistic neurite outgrowth. A, schematic representation of individual kinase inhibition during synergistic neurite outgrowth. The dashed line represents indirect activation of MAPK by IL-6R through recruitment of the Grb2-SOS complex. N2A cells were pretreated for 4 h with PD (MEK inhibitor PD98059) (B), pretreated for 1 h with PP2 (Src inhibitor) (C), or transfected with DN Jak1 (D) or SOCS1 and SOCS3 (E) proteins. The cells were then treated with the indicated concentrations of IL-6 and HU-210 for 18 h and imaged, and neurite outgrowth was quantified. The bar graphs show mean ± S.E. for percentages of cells with neurites. For each graph, quantifications were performed from at least three independent experiments. The solid line indicates the average of PBS, DMSO, and PBS + DMSO treatments. Statistical differences were calculated using a one-tailed t test. See supplemental Figs. 2S, 3S, 4S, and 5S for statistical significance comparisons of all conditions. *, significance compared with DMSO pretreatment or pcDNA3 transfection within the same ligand treatment; *, p < 0.05; **, p < 0.005.
We found that MAPK inhibition abolished synergistic neurite outgrowth in the presence of low concentrations of IL-6 and HU-210 (Fig. 3B and supplemental Fig. 2S). Pretreatment with PD also resulted in partial inhibition of neurite outgrowth in the presence of high HU-210 (1 μm), consistent with our previous findings (23).
We found that Src inhibition strongly blocked neurite outgrowth in response to all treatments by reducing the percentage of cells with neurites to the level of negative control (Fig. 3C and supplemental Fig. 3S). These results are consistent with previous findings from our laboratory (4, 23) and indicate that Src signaling emanating from CB1R plays a primary role in neurite outgrowth induced by activation of the two pathways and is essential for synergy.
Cells that were transfected with DN Jak1 showed a minor reduction in neurite outgrowth in response to the IL-6 and HU-210 combinatorial treatment (Fig. 3D and supplemental Fig. 4S). As expected, DN Jak1 did not affect the response of cells to HU-210 treatments. It is also important to note that cells that were treated with IL-6 alone were not affected by DN Jak1 transfection, because IL-6 treatment alone does not induce neurite outgrowth above basal level. The fact that DN Jak1 did not completely abolish neurite outgrowth in response to the combinatorial treatment suggests that Jak1 may not be the only Janus kinase activated by IL-6 receptor.
In order to inhibit IL-6R-induced kinase activity more globally, we transfected cells with SOCS1 and SOCS3 proteins, which provide a negative feedback loop in IL-6R/Jak signaling. SOCS1 inhibits IL-6 signaling by binding to the activation loop of Jak proteins via its Src homology 2 domain (27), and SOCS3 negatively regulates IL-6R activity (27) by binding to the IL-6 signal-transducing receptor subunit gp130 at Tyr759, which has been identified as being involved in negative regulation of IL-6 signaling (30). Overexpression of SOCS proteins did not result in a differential response to single treatments but completely abolished neurite outgrowth in response to the double treatment (Fig. 3E and supplemental Fig. 5S). The effects of SOCS3 could in part be due to the MAPK pathway (see “Regulation by SHP2 Phosphatase”). Taken together, these results indicate that activation of MAPK, Src, and Jak signaling is necessary for synergistic neurite outgrowth.
PI3K/Akt Signaling Is Generally Required for Neurite Outgrowth
In addition to Src and MAPK, the PI3K/Akt signaling cascade has previously been described as necessary for CB1R-induced neurite outgrowth (23). PI3K may also be activated by IL-6R signaling (31, 32). We examined its role in synergistic neurite outgrowth in response to CB1R/IL-6R activation. We found that inhibition of PI3K signaling with LY294002 (LY) resulted in loss of synergistic neurite outgrowth (supplemental Fig 12S, A and B). However, time course analysis of Akt phosphorylation demonstrated equivalent activation at 30 and 60 min irrespective of ligand treatment (supplemental Fig. 12SC). Therefore, PI3K/Akt signaling may be necessary for general neurite outgrowth but may not contribute to the synergistic effect.
Regulation by Transcription Factors CREB and STAT3
Activation of CREB and STAT3 Transcription Factors Is Required for Synergistic Neurite Outgrowth
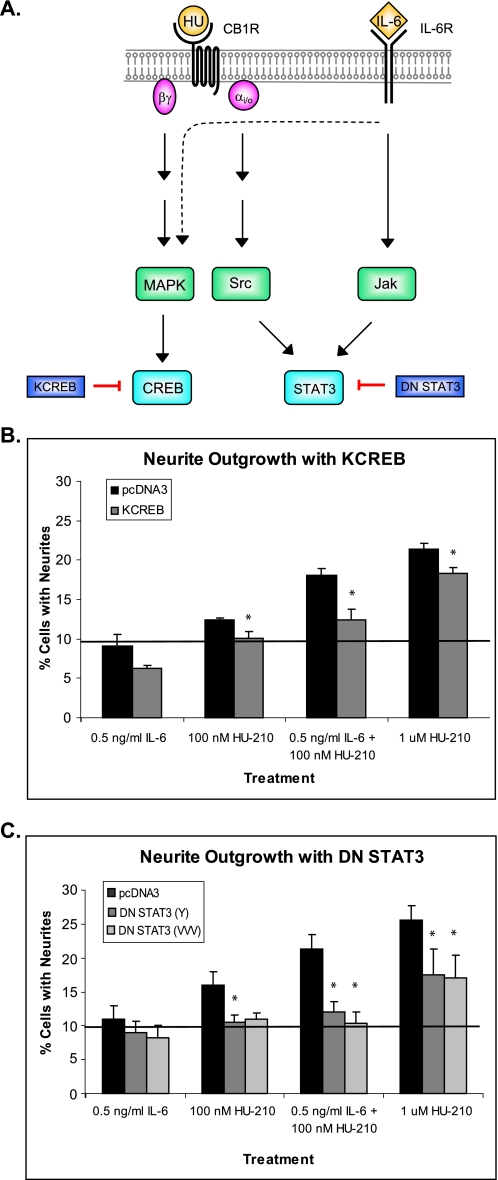
Our laboratory has previously demonstrated that activation of MAPK and Src kinases by CB1R leads to phosphorylation of transcription factors CREB and STAT3, respectively (23). STAT3 can also be activated by IL-6R through Jak, thereby providing a potential mechanism for synergistic neurite outgrowth (Fig. 4A).
FIGURE 4.
Activation of CREB and STAT3 transcription factors is required for synergistic neurite outgrowth. A, schematic representation of individual transcription factor inhibition during synergistic neurite outgrowth. N2A cells were transfected with pcDNA3, KCREB (dominant negative CREB) (B), DN STAT3 (Y) (C), or DN STAT3 (VVV) (C). All cells were co-transfected with green fluorescent protein. Neurite outgrowth of green fluorescent protein-expressing cells was scored after 18 h of the indicated treatments. The bar graph shows mean ± S.E. for percentages of cells with neurites. For each graph, quantifications were performed from three independent experiments. The solid line indicates the average of PBS, DMSO, and PBS + DMSO treatments. Statistical differences were calculated using a one-tailed t test. See supplemental Figs. 6S and 7S for statistical significance comparisons of all conditions. Within a given ligand treatment, asterisks indicate significance of inhibition of neurite outgrowth compared with pcDNA3-transfected cells; *, p < 0.05.
To determine whether MAPK signaling exerts its actions through CREB transcription factor, N2A cells were transfected with dominant negative CREB (KCREB), which contains a mutation in its DNA-binding domain. Twenty-four hours after transfection, cells were plated in Mattek® dishes, and neurite outgrowth assays were carried out. Transfection of cells with KCREB resulted in complete abolishment of neurite outgrowth in the presence of low concentrations of IL-6 and HU-210 and partial inhibition in the presence of high HU-210, supporting our hypothesis that MAPK signaling positively regulates neurite outgrowth through the transcription factor CREB (Fig. 4B and supplemental Fig. 6S).
In order to test whether STAT3 is necessary for synergistic neurite outgrowth, N2A cells were transfected with two forms of dominant negative STAT3. The dominant negative mutants of STAT3 contained a mutation in either the tyrosine 705 phosphorylation site (DN STAT3 (Y)) or the DNA binding site (DN STAT3 (VVV)) (25). Twenty-four hours after transfection, cells were plated into 35-mm Mattek® dishes and stimulated with low concentrations of IL-6 and/or HU-210. Both DN STAT3 (Y) and DN STAT3 (VVV) abolished synergy in response to combinatorial treatment and significantly reduced neurite outgrowth to basal level in response to single treatments (Fig. 4C and supplemental Fig. 7S). These results suggest that STAT3 is important for transmitting the signals from both pathways and plays a critical role in synergistic neurite outgrowth by providing a potential point of integration between the two pathways.
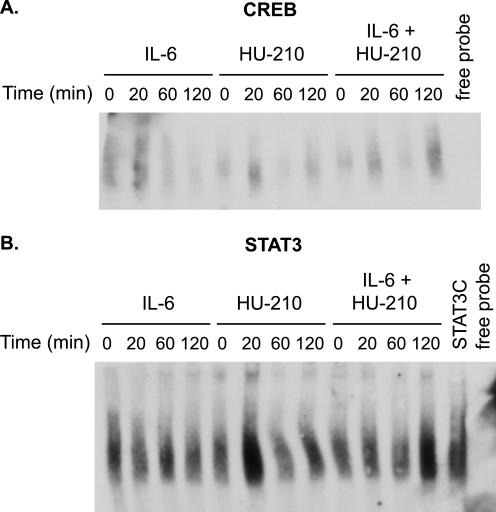
To further examine the transcriptional activation of STAT3 and CREB during synergistic neurite outgrowth, we performed gel shift assays. N2A cells were treated with 0.5 ng/ml IL-6, 100 nm HU-210, or both over a period of 0–120 min, and nuclear extracts were incubated with biotinylated consensus sequences for CREB and STAT3. Both transcription factors showed a marked increase in DNA binding at 120 min in the presence of both stimulatory ligands as compared with either ligand alone (Fig. 5, A and B, and supplemental Fig. 13S).
FIGURE 5.
Time course gel shift assays of CREB and STAT3 DNA-binding activity. N2A cells were serum-starved for 24 h and treated with 0.5 ng/ml IL-6, 100 nm HU-210, or both for the indicated periods of time. Nuclear extracts were prepared, and gel shift assays were performed using consensus sequences for CREB (A) and STAT3 (B). N2A cells were transfected with constitutively active STAT3 (STAT3C) as a positive control for gel shift. The blots shown are representative of two independent experiments.
Although both transcription factors showed enhanced DNA binding after co-stimulation of CB1R and IL-6R, we were unable to detect a difference in CREB phosphorylation levels in response to single and double treatments. Because we used suboptimal concentrations of ligands in this study, it appears that the level of CREB phosphorylation may be sufficient for biological effects but below the limits of detection by immunoblotting.
Combined Treatment with IL-6 and HU-210 Causes Sustained Phosphorylation of STAT3 in N2A Cells
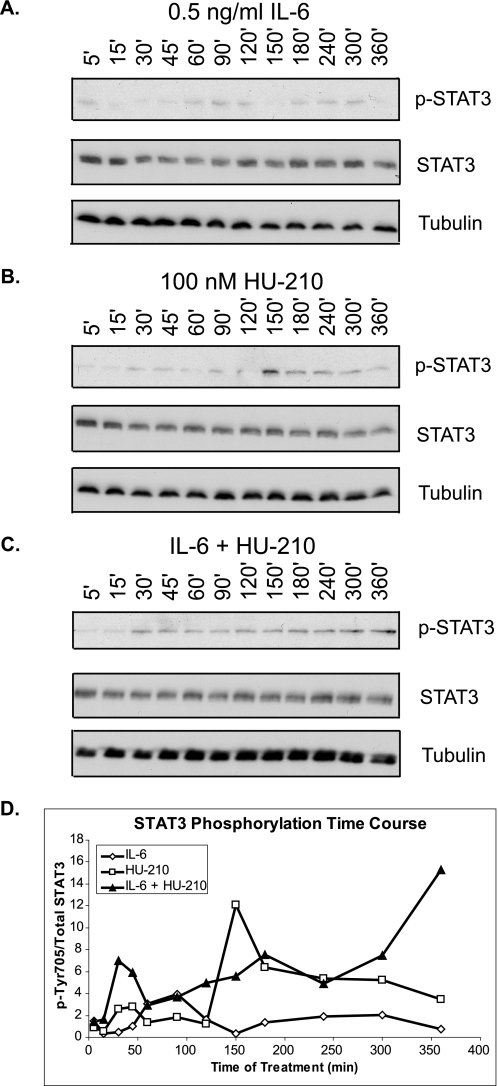
Because STAT3 can be activated by kinases downstream of both CB1R and IL-6R (Fig. 4A), we investigated how differential STAT3 activation may result in synergistic neurite outgrowth. Therefore, we assessed the time course response of STAT3 phosphorylation to individual and combined pathway activation. We hypothesized that combinatorial treatment may cause a higher level of STAT3 activation or allow for STAT3 activity to be sustained over a longer period of time. N2A cells were serum-starved for 24 h and received individual and combinatorial treatments of 0.5 ng/ml IL-6 and 100 nm HU-210. The cells were lysed at a number of time points after treatment ranging from 5 min to 6 h. Treatment of cells with both ligands resulted in sustained STAT3 phosphorylation for up to 6 h. In contrast, treatment with either ligand singly induced transient phosphorylation of STAT3 (Fig. 6, A–D).
FIGURE 6.
Activation of CB1R and IL-6R results in sustained STAT3 phosphorylation. N2A cells were serum-starved for 24 h and treated with 0.5 ng/ml IL-6 (A), 100 nm HU-210 (B), or both (C) for the indicated periods of time. The cells were lysed and subjected to immunoblot analysis for phospho-STAT3 (p-STAT; phospho-Tyr705), total STAT3, and tubulin. The blots shown are representative of three independent experiments. D, quantification of STAT3 phosphorylation represented as phospho-Tyr705/STAT3 ratio over time.
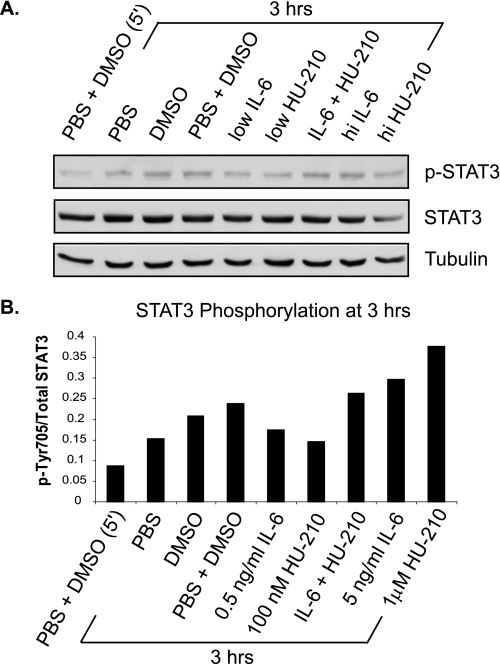
Based on the STAT3 phosphorylation time course, we chose 3 h post-treatment as the point in time when STAT3 phosphorylation keeps increasing only in the presence of both IL-6 and HU-210. N2A cells were treated with low and high concentrations of IL-6 and HU-210 and a combination of low doses for 3 h. The cells were lysed and subjected to immunoblot analysis for phospho-STAT3. As shown in Fig. 7, STAT3 phosphorylation was enhanced in the presence of both ligands at low concentrations, compared with either ligand alone. Treatment with a high concentration of HU-210 (1 μm) resulted in increased STAT3 phosphorylation (Fig. 7, A and B), correlating with its ability to induce neurite outgrowth (Fig. 1B). High IL-6 (5 ng/ml) also resulted in increased STAT3 phosphorylation at 3 h in the blot shown in Fig. 7, A and B, but this increase was less consistent across the three experiments. The high dose of IL-6 was also insufficient to induce neurite outgrowth (Fig. 1B), supporting the hypothesis of its secondary role in synergistic neurite outgrowth. Similarly, treatment with DMSO (with or without PBS) raised STAT3 activity, indicating a possible biochemical stress response. No stimulation of neurite outgrowth was seen with DMSO.
FIGURE 7.
Activation of CB1R and IL-6R results in enhanced STAT3 phosphorylation at 3 h of treatment. N2A cells were serum-starved for 24 h and treated with 0.5 ng/ml IL-6 (low), 5 ng/ml IL-6 (hi), 100 nm HU-210 (low), 1 μm HU-210 (hi), and a combination of low doses for 3 h. The cells were lysed and subjected to immunoblot analysis for phospho-STAT3 (phospho-Tyr705), total STAT3, and tubulin. A, the blot shown is representative of three independent experiments. B, quantification of STAT3 phosphorylation represented as phospho-Tyr705/STAT3 ratio.
Regulation by SHP2 Phosphatase
Stimulation of CB1R and IL-6R Results in Down-regulation of SHP2 Negative Feedback Activity
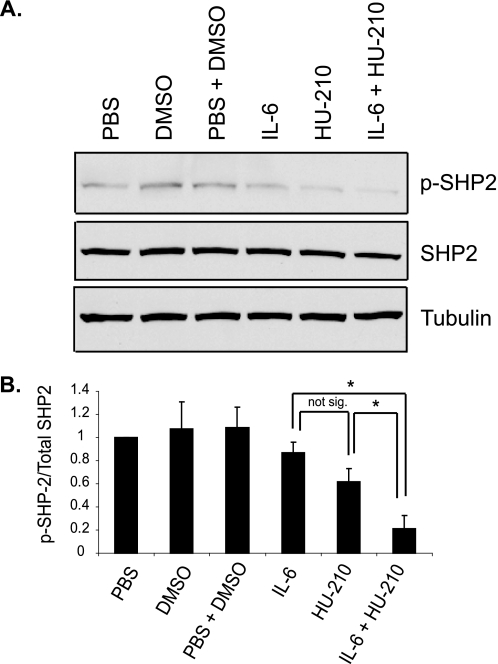
STAT3 activity is negatively regulated by SHP2 phosphatase, which binds and dephosphorylates the gp130 receptors, Jaks and STATs (27) (see Fig. 9A). In the absence of a tyrosine-phosphorylated binding partner, SHP2 has a low level of basal activity as its N-terminal Src homology 2 domain folds over the active site and inhibits its enzymatic activity (33). Binding of the Src homology 2 domains of SHP2 to phosphotyrosine motifs of receptors or adaptors unfolds the protein, leading to phosphorylation of tyrosine residue 542 or 580 within the C-terminal part of the enzyme, and results in enzymatic activity (34–37). In order to investigate the mechanism by which STAT3 activation is sustained in the presence of both CB1R and IL-6R ligands, we examined the activation of SHP2 at 6 h of treatment, when STAT3 phosphorylation was highest. We found that SHP2 phosphorylation was slightly reduced by application of either ligand alone, whereas a more marked reduction was observed in response to combinatorial treatment (Fig. 8, A and B), suggesting that negative feedback to STAT3 is down-regulated.
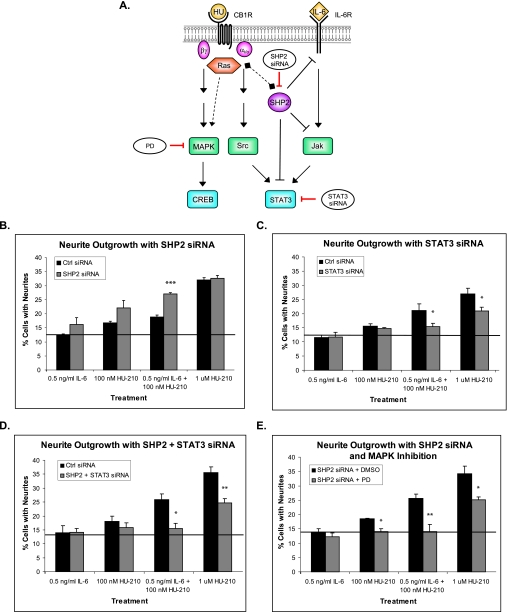
FIGURE 9.
SHP2 negative feedback acts upstream of STAT3 and MAPK/CREB signaling. A, schematic representation of SHP2 negative feedback signaling. The dashed lines represent recruitment of Ras by SHP2 through the Grb2-SOS scaffold complex, followed by activation of Ras-Raf-MAPK cascade. N2A cells were transfected with scrambled control siRNA, SHP2 siRNA (B), STAT3 siRNA (C), or both SHP2 and STAT3 siRNAs (D), and neurite outgrowth assays were carried out. E, N2A cells were transfected with SHP2 siRNA and then treated with PD for 4 h followed by neurite outgrowth assays. The bar graphs show mean ± S.E. For each graph, neurite outgrowth quantifications were performed from three independent experiments. Statistical differences were calculated using a one-tailed t test. See supplemental Figs. 8S, 9S, 10S, and 11S for statistical significance comparisons of all conditions. *, significance compared with control (Ctrl) siRNA or DMSO pretreatment within the same ligand treatment; *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
FIGURE 8.
SHP2 negative feedback is down-regulated in the presence of IL-6 and HU-210. A, N2A cells were serum-starved for 24 h and treated with 0.5 ng/ml IL-6, 100 nm HU-210, or both ligands for 6 h. The cells were lysed and subjected to immunoblot analysis for phospho-SHP2 (p-SHP2), total SHP2, and tubulin. The bar graph (B) shows mean ± S.E. from three independent experiments. Statistical differences were calculated using a one-tail t test. *, p < 0.05.
In order to examine the role of SHP2 in STAT3-induced neurite outgrowth, we transfected N2A cells with SHP2 siRNA and performed neurite outgrowth assays. Consistent with the decrease in SHP2 phosphorylation in Fig. 8, knockdown of SHP2 expression caused a significant enhancement in neurite outgrowth in response to the combinatorial treatment (Fig. 9B and supplemental Fig. 8S). Taken together, these data suggest that inactivation of SHP2 results in sustained STAT3 activation.
Previous studies (4, 23) and the findings in this study indicate that STAT3 is a key transcriptional regulator leading to neurite outgrowth. If so, SHP2 would be upstream of STAT3, and transfection with STAT3 siRNA should block neurite outgrowth, irrespective of the effects of SHP2 siRNA. Transfection with STAT3 siRNA inhibited neurite outgrowth (Fig. 9C and supplemental Fig. 9S), similar to the effect of DN STAT3 in Fig. 4C. Neurite outgrowth assays of cells co-transfected with STAT3 siRNA and SHP2 siRNA also resulted in abolishment of synergistic neurite outgrowth (Fig. 9D and supplemental Fig. 10S), supporting the hypothesis that STAT3 is a major downstream target necessary for neurite outgrowth and CB1R and IL-6R pathway integration.
SHP2 has previously been suggested to act as an adaptor in recruitment and activation of Ras by the IL-6 receptor, leading to the Ras-Raf-MAPK signaling cascade (27, 38). We investigated if MAPK/CREB signaling acts downstream of SHP2. N2A cells were transfected with SHP2 siRNA, pretreated with PD for 4 h (at 48 h post-transfection), and subjected to neurite outgrowth assays. Pretreatment with PD in the presence of SHP2 siRNA resulted in abolishment of synergistic neurite outgrowth in response to low concentrations of IL-6 and HU-210 and partial inhibition in response to high HU-210 (Fig. 9E and supplemental Fig. 11S). Therefore, MAPK signaling appears to act downstream of SHP2 and to be necessary for synergistic neurite outgrowth.
Stimulation of CB1R and IL-6R Results in Enhanced Neurite Outgrowth in Primary Cortical Neurons
To determine if co-stimulation of CB1R and IL-6R regulates neuronal differentiation in primary neurons, we stimulated rat cortical neuron primary cultures with HU-210 and IL-6 at 2 DIV. After 24 h of stimulation, the cultures were fixed and stained with antibodies against MAP2 and Tau as markers of developing dendrites and axons, respectively. Stimulation with HU-210 and IL-6 resulted in enhanced development of neurons, compared with single treatments (Fig. 10A). In studies of axonal regeneration in vitro, the length of the longest neurite is often used as a read-out because the longest neurite is most likely to later differentiate into an axon (20, 39). Due to skewed distributions of lengths of longest neurite under each analyzed condition, we chose the distribution-free Wilcoxon Mann-Whitney test to compare treatments. We found that stimulation of both signaling pathways resulted in a 53% increase in median length compared with IL-6 treatment and a 20% increase compared with HU-210 treatment (Fig. 10B).
FIGURE 10.
Stimulation of CB1R and IL-6R results in enhanced neurite outgrowth in primary rat cortical neurons. Rat primary cortical neuron cultures were treated with IL-6 and/or HU-210 at 2 DIV and fixed after 24 h. The cells were labeled for MAP2 (red) and Tau (green) to identify developing neurites. A, representative confocal images of cells under each condition are shown. B, the lengths of the longest neurite of each cell were measured morphometrically. The bar graph shows mean ± S.E. for average neurite length and median neurite length for each treatment. Statistical differences were calculated using the one-tailed Wilcoxon Mann-Whitney test for medians; *, p < 0.05; ***, p < 0.0005. See supplemental Table 2S for statistical significance comparisons of all conditions.
DISCUSSION
Neuronal differentiation is a complex process that integrates many signals to drive electrophysiological, morphological, and transcriptional changes (1, 41–43). This process is characterized by the initial formation of immature neurites, commonly referred to as neurite outgrowth. A number of studies from our laboratory have previously shown the importance of the CB1R signaling pathway in neurite outgrowth regulation through activation of the transcription factors CREB (23) and STAT3 (4, 23). Other studies have suggested that IL-6 cytokine signaling may also be an important component of signaling in neurite outgrowth (24, 44) by signaling through the Jak/STAT3 pathway. Here we show how CB1R and IL-6R signaling induce synergistic neurite outgrowth in Neuro2A cells through a downstream signaling network that converges on STAT3 and necessitates MAPK/CREB activation. We found that simultaneous activation of the two receptors results in synergistic neurite outgrowth.
In order to confirm that inputs from both receptors are important for synergistic neurite outgrowth, we individually blocked the kinases that activate downstream transcription factors from either arm. Consistent with previous studies (4), complete abolishment of neurite outgrowth in response to Src inhibition supports the assumption that the CB1R pathway plays a central role in neurite outgrowth. However, IL-6R signaling through the Jak/STAT pathway is still necessary for synergistic neurite outgrowth. Although the IL-6R-gp130 complex can activate a number of tyrosine kinases of the Jak family (Jak1, Jak2, and Tyk2) (27), Jak1 plays an essential role in STAT3 activation (29), and in cells lacking Jak1, IL-6 signal transduction is greatly impaired (28, 29, 45). We found that expression of dominant negative Jak1 markedly inhibited the synergistic effect of combinatorial treatment but did not bring neurite outgrowth back to unstimulated levels. These results suggested that despite being a necessary component for activation of STAT3, Jak1 may not be the only Jak involved in this signaling network. Because SOCS proteins provide a negative feedback loop to Jak signaling and IL-6R activation (27), overexpression of these proteins allowed us to inhibit Jak signaling more globally and completely block synergistic neurite outgrowth.
Our laboratory has recently demonstrated that MAPK signaling is necessary for CB1R-induced neurite outgrowth (23). MAPK can also be activated by IL-6R through recruitment of SHP2 and the Grb2-SOS complex, followed by activation of Ras-Raf-MAPK cascade (27). We found that inhibition of MAPK abolished synergistic neurite outgrowth in response to co-stimulation of CB1R and IL-6R. Since MAPK is the only kinase receiving direct activation from both receptors, we examined its activation in more detail. We found that in the presence of both ligands, MAPK has an enhanced early wave of phosphorylation compared with either treatment alone. Taken together, these data indicate that MAPK signaling is required for synergistic neurite outgrowth.
MAPK, Src, and Jak kinases phosphorylate STAT3 and CREB transcription factors. We found that dominant negative forms of STAT3 and CREB block neurite outgrowth in N2A cells. These results are consistent with previous studies from our laboratory, in which dominant negative STAT3 (4, 23) and dominant negative CREB (23) significantly inhibited HU-210-induced neurite outgrowth. Although CREB can be activated only by MAPK, STAT3 may be phosphorylated by both Src and Jak kinases. This suggests that STAT3 may play a more central role in signal integration and synergy.
To examine the mechanism of synergistic neurite outgrowth through STAT3 activation more closely, we performed a 6-h time course of STAT3 phosphorylation. We found that unlike individual treatments, simultaneous activation of both receptors results in prolonged STAT3 (Tyr705) phosphorylation. STAT3 phosphorylation has been shown to play an important role in neurite outgrowth. Wu and Bradshaw (24) found that STAT3 tyrosine phosphorylation increased in nerve growth factor-primed cells after IL-6 application, resulting in dose-dependent neurite outgrowth in primed PC12 cells. Similarly, He et al. (4) observed that activation of the CB1R pathway caused STAT3 phosphorylation and induced neurite outgrowth. Because STAT3 acts as a transcription factor, its sustained activity suggests that there may be a prolonged period of transcription of genes necessary for neurite outgrowth, providing a possible mechanism for synergistic neurite outgrowth. STAT3 also showed sustained DNA binding at 120 min only after treatment with both ligands, thereby supporting this hypothesis.
Previous studies have shown that in response to IL-6 application, STAT3 either goes through a transient wave of tyrosine phosphorylation, which goes back to basal levels after about 2 h (46, 47), or may exhibit two sequential waves of tyrosine phosphorylation over a longer period of time (24). Stimulation of N2A cells with HU-210 also causes a transient wave of STAT3 phosphorylation that lasts for about 30–60 min (4). Our observation of a prolonged state of Tyr705 phosphorylation suggests that in the presence of both IL-6 and HU-210 ligands, a negative regulator of STAT3 may be inactivated. One such protein is SHP2, which is a ubiquitously expressed cytoplasmic protein-tyrosine phosphatase, containing two N-terminal Src homology 2 domains and a catalytic phosphatase domain in the C-terminal half of the protein (27). SHP2 can be activated by downstream kinases of both IL-6R and CB1R pathways and can negatively regulate gp130, Jaks, and STATs (27). Previous studies have shown that prevention of SHP2 activation causes an enhanced and prolonged phosphorylation of gp130, Jak1, Jak2, and STAT3 as well as extended DNA binding by STAT3 (30, 48).
In its inactive state, SHP2 folds onto itself and autoinhibits, whereas phosphorylation of tyrosine 542 or 580 releases the block and activates SHP2 phosphatase activity (34). Therefore, we tested how SHP2 phosphorylation is affected by activation of IL-6R and CB1R signaling pathways. We found that after 6 h of treatment, which corresponds to the highest level of STAT3 phosphorylation, SHP2 phosphorylation is slightly reduced by each individual treatment but shows a much greater reduction in the presence of both ligands. These data support our hypothesis that sustained STAT3 phosphorylation results from a down-regulation of the autonegative feedback loop in the signaling network. We went on to confirm the negative regulatory role of SHP2 in neurite outgrowth assays and found that RNA interference of SHP2 resulted in enhanced neurite outgrowth, providing further evidence for the negative feedback hypothesis.
The mechanism of SHP2 inactivation is still unclear and requires further investigation, and there are several possibilities. The autoinhibited, “closed” conformation is highly favored under basal conditions, resulting in very low basal catalytic activity of SHP2. Moreover, SHP2 has been suggested to dephosphorylate itself in solution (49), corresponding to the observation of a very transient wave of SHP2 phosphorylation in response to IL-6 treatment in cell culture (45, 46). Since SHP2 can be phosphorylated by both Src (50) and Jaks (51), simultaneous activation of the two pathways may enhance early catalytic activity of SHP2, which may then lead to greater autoinhibition. Such a mechanism would suggest that early autoinhibition would leave less active SHP2 available to dephosphorylate STAT3.
Additionally, SHP2 has been shown to dephosphorylate Src on an inhibitory tyrosine, thereby releasing its inhibition (52, 53). Such dephosphorylation causes up-regulation of Src kinase activity. Since both IL-6R and CB1R pathways can activate SHP2, SHP2 may provide a positive feedback loop by activating more Src, thereby offering another mechanism for sustained phosphorylation of STAT3. Finally, both receptors activated in the network have been shown to activate Ras, either through the Grb2-SOS complex (54) or through Src activation of RasGRF (55, 56). Among its multitude of functions, Ras has been implicated in regulating the production of intracellular reactive oxygen species (ROS) (57), which can modulate and inhibit the activity of low molecular weight protein-tyrosine phosphatases (58). Rac proteins also function as a necessary switch for ROS generation (40, 57), and Rac has previously been demonstrated to be activated by the CB1R pathway via Src (4). Therefore, activation of Ras and Rac through the IL-6R and CB1R pathways could result in up-regulation of ROS, ultimately leading to inhibition of SHP2 negative feedback on STAT3.
A model for the mechanism of CB1R and IL-6R signal integration based on these findings is depicted in Fig. 11. STAT3 receives positive signals from CB1R and IL-6R, through Src and Jak kinases, respectively. SHP2 can also be activated by these two kinases and in presence of both ligands over time may become more inactivated either due to its transient activity (46) and highly favored closed conformation (33) or from the generation of ROS through activation of Ras by both pathways (57). Additionally, SHP2 may further activate Src at an early time after stimulation, providing a positive feedback loop for sustained STAT3 activity. Further studies are necessary to determine the mechanism of late SHP2 inactivation.
FIGURE 11.
Model for mechanism of CB1R and IL-6R signal integration. During activation of CB1R and IL-6R, STAT3 becomes activated by two sources, Src and Jak. SHP2 is also activated by both receptors but shows greater inactivation in the presence of both ligands, due to either greater autoinhibition or Ras-dependent ROS production. Additionally, SHP2 activates Src, which may provide another mechanism of positive feedback for sustained STAT3 activation. The MAPK/CREB signaling cascade is also necessary for synergistic neurite outgrowth and acts downstream of SHP2. The solid and dashed lines represent direct and indirect interactions, respectively. The black lines correspond to previously reported data for CB1R signaling, and brown lines correspond to new data suggesting CB1R/IL-6R signal integration. See under “Discussion” for more details.
In conclusion, we demonstrated that parallel activation of CB1R and IL-6R pathways results in synergistic neurite outgrowth in N2A cells and primary cortical neurons, and activation of both signaling pathways is necessary for this effect to take place. Synergistic neurite outgrowth can be explained by sustained activation of STAT3, which may allow a longer time period for gene transcription. STAT3 phosphorylation is sustained for at least 6 h after stimulation, due to down-regulation of a negative feedback loop of SHP2 phosphatase. Finally, since CB1R and IL-6R co-stimulation enhanced differentiation in rat cortical neurons, the integration of these pathways during neural development merits further investigation.
Supplementary Material
Acknowledgment
We thank Dr. Lu-Hai Wang for the generous gift of DN Jak1, SOCS1, and SOCS3 plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants GM5408, P50GM071558, and T32 CA88796 (to K. D. B.). This work was also supported by an American Cancer Society Spirit of Birmingham and Johnson Memorial Postdoctoral Fellowship Award.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1S and 2S and Figs. 1S–14S.
- IL-6
- interleukin-6
- IL-6R
- interleukin-6 receptor
- CB1R
- cannabinoid 1 receptor
- Jak
- Janus kinase
- STAT
- signal transducer and activator of transcription
- DIV
- day(s) in vitro
- ROS
- reactive oxygen species
- N2A
- Neuro2A
- MAPK
- mitogen-activated protein kinase
- CREB
- cAMP-response element-binding protein
- DN
- dominant negative
- siRNA
- small interfering RNA
- PI3K
- phosphatidylinositol 3-kinase
- PBS
- phosphate-buffered saline
- PD
- PD98059
- LY
- LY294002.
REFERENCES
- 1.Govek E. E., Newey S. E., Van Aelst L. (2005) Genes Dev. 19, 1–49 [DOI] [PubMed] [Google Scholar]
- 2.Mocchetti I., Wrathall J. R. (1995) J. Neurotrauma 12, 853–870 [DOI] [PubMed] [Google Scholar]
- 3.Ihara S., Iwamatsu A., Fujiyoshi T., Komi A., Yamori T., Fukui Y. (1996) J. Biochem. 120, 865–868 [DOI] [PubMed] [Google Scholar]
- 4.He J. C., Gomes I., Nguyen T., Jayaram G., Ram P. T., Devi L. A., Iyengar R. (2005) J. Biol. Chem. 280, 33426–33434 [DOI] [PubMed] [Google Scholar]
- 5.Jordan J. D., He J. C., Eungdamrong N. J., Gomes I., Ali W., Nguyen T., Bivona T. G., Philips M. R., Devi L. A., Iyengar R. (2005) J. Biol. Chem. 280, 11413–11421 [DOI] [PubMed] [Google Scholar]
- 6.Arthur D. B., Akassoglou K., Insel P. A. (2006) Biochem. Biophys. Res. Commun. 347, 678–682 [DOI] [PubMed] [Google Scholar]
- 7.Dotti C. G., Banker G. A. (1987) Nature 330, 254–256 [DOI] [PubMed] [Google Scholar]
- 8.Goslin K., Banker G. (1989) J. Cell Biol. 108, 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradke F., Dotti C. G. (2000) Curr. Biol. 10, 1467–1470 [DOI] [PubMed] [Google Scholar]
- 10.Gadient R. A., Otten U. H. (1997) Prog. Neurobiol. 52, 379–390 [DOI] [PubMed] [Google Scholar]
- 11.Nakashima K., Wiese S., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Yoshida K., Kishimoto T., Sendtner M., Taga T. (1999) J. Neurosci. 19, 5429–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strauss S., Bauer J., Ganter U., Jonas U., Berger M., Volk B. (1992) Lab. Invest. 66, 223–230 [PubMed] [Google Scholar]
- 13.Müller T., Blum-Degen D., Przuntek H., Kuhn W. (1998) Acta Neurol. Scand. 98, 142–144 [DOI] [PubMed] [Google Scholar]
- 14.Hans V. H., Kossmann T., Lenzlinger P. M., Probstmeier R., Imhof H. G., Trentz O., Morganti-Kossmann M. C. (1999) J. Cereb. Blood Flow Metab. 19, 184–194 [DOI] [PubMed] [Google Scholar]
- 15.Campbell I. L., Abraham C. R., Masliah E., Kemper P., Inglis J. D., Oldstone M. B., Mucke L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10061–10065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton A. R., Barlett P. F., Pennica D., Davies A. M. (1998) Eur. J. Neurosci. 10, 673–679 [DOI] [PubMed] [Google Scholar]
- 17.Ali C., Nicole O., Docagne F., Lesne S., MacKenzie E. T., Nouvelot A., Buisson A., Vivien D. (2000) J. Cereb. Blood Flow Metab. 20, 956–966 [DOI] [PubMed] [Google Scholar]
- 18.Zhong J., Dietzel I. D., Wahle P., Kopf M., Heumann R. (1999) J. Neurosci. 19, 4305–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunioku H., Inoue K., Tomida M. (2001) Neurosci. Lett. 309, 13–16 [DOI] [PubMed] [Google Scholar]
- 20.Cao Z., Gao Y., Bryson J. B., Hou J., Chaudhry N., Siddiq M., Martinez J., Spencer T., Carmel J., Hart R. B., Filbin M. T. (2006) J. Neurosci. 26, 5565–5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satoh T., Nakamura S., Taga T., Matsuda T., Hirano T., Kishimoto T., Kaziro Y. (1988) Mol. Cell Biol. 8, 3546–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y. Y., Bradshaw R. A. (1996) J. Biol. Chem. 271, 13033–13039 [DOI] [PubMed] [Google Scholar]
- 23.Bromberg K. D., Ma'ayan A., Neves S. R., Iyengar R. (2008) Science 320, 903–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y. Y., Bradshaw R. A. (1996) J. Biol. Chem. 271, 13023–13032 [DOI] [PubMed] [Google Scholar]
- 25.Ram P. T., Horvath C. M., Iyengar R. (2000) Science 287, 142–144 [DOI] [PubMed] [Google Scholar]
- 26.Zong C. S., Chan J., Levy D. E., Horvath C., Sadowski H. B., Wang L. H. (2000) J. Biol. Chem. 275, 15099–15105 [DOI] [PubMed] [Google Scholar]
- 27.Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., Schaper F. (2003) Biochem. J. 374, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guschin D., Rogers N., Briscoe J., Witthuhn B., Watling D., Horn F., Pellegrini S., Yasukawa K., Heinrich P., Stark G. R. (1995) EMBO J. 14, 1421–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuroda H., Sugimoto T., Horii Y., Sawada T. (2001) Med. Pediatr. Oncol. 36, 118–121 [DOI] [PubMed] [Google Scholar]
- 30.Lehmann U., Schmitz J., Weissenbach M., Sobota R. M., Hortner M., Friederichs K., Behrmann I., Tsiaris W., Sasaki A., Schneider-Mergener J., Yoshimura A., Neel B. G., Heinrich P. C., Schaper F. (2003) J. Biol. Chem. 278, 661–671 [DOI] [PubMed] [Google Scholar]
- 31.Wegiel B., Bjartell A., Culig Z, Persson J. L. (2008) Int. J. Cancer 122, 1521–1529 [DOI] [PubMed] [Google Scholar]
- 32.Morris V. A., Punjabi A. S., Lagunoff M. (2008) J. Virol. 82, 8771–8779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hof P., Pluskey S., Dhe-Paganon S., Eck M. J., Shoelson S. E. (1998) Cell 92, 441–450 [DOI] [PubMed] [Google Scholar]
- 34.Lu W., Gong D., Bar-Sagi D., Cole P. A. (2001) Mol. Cell 8, 759–769 [DOI] [PubMed] [Google Scholar]
- 35.Pluskey S., Wandless T. J., Walsh C. T., Shoelson S. E. (1995) J. Biol. Chem. 270, 2897–2900 [DOI] [PubMed] [Google Scholar]
- 36.Sugimoto S., Wandless T. J., Shoelson S. E., Neel B. G., Walsh C. T. (1994) J. Biol. Chem. 269, 13614–13622 [PubMed] [Google Scholar]
- 37.Lechleider R. J., Sugimoto S., Bennett A. M., Kashishian A. S., Cooper J. A., Shoelson S. E., Walsh C. T., Neel B. G. (1993) J. Biol. Chem. 268, 21478–21481 [PubMed] [Google Scholar]
- 38.Dance M., Montagner A., Salles J. P., Yart A., Raynal P. (2008) Cell. Signal. 20, 453–459 [DOI] [PubMed] [Google Scholar]
- 39.Cai D., Shen Y., De Bellard M., Tang S., Filbin M. T. (1999) Neuron 22, 89–101 [DOI] [PubMed] [Google Scholar]
- 40.Groemping Y., Rittinger K. (2005) Biochem. J. 386, 401–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohnuma S., Harris W. A. (2003) Neuron 40, 199–208 [DOI] [PubMed] [Google Scholar]
- 42.da Silva J. S., Dotti C. G. (2002) Nat. Rev. Neurosci. 3, 694–704 [DOI] [PubMed] [Google Scholar]
- 43.Diez del Corral R., Storey K. G. (2001) Nat. Rev. Neurosci. 2, 835–839 [DOI] [PubMed] [Google Scholar]
- 44.Wu Y. Y., Bradshaw R. A. (2000) J. Biol. Chem. 275, 2147–2156 [DOI] [PubMed] [Google Scholar]
- 45.Rodig S. J., Meraz M. A., White J. M., Lampe P. A., Riley J. K., Arthur C. D., King K. L., Sheehan K. C., Yin L., Pennica D., Johnson E. M., Jr., Schreiber R. D. (1998) Cell 93, 373–383 [DOI] [PubMed] [Google Scholar]
- 46.Schaper F., Gendo C., Eck M., Schmitz J., Grimm C., Anhuf D., Kerr I. M., Heinrich P. C. (1998) Biochem. J. 335, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Croker B. A., Krebs D. L., Zhang J. G., Wormald S., Willson T. A., Stanley E. G., Robb L., Greenhalgh C. J., Förster I., Clausen B. E., Nicola N. A., Metcalf D., Hilton D. J., Roberts A. W., Alexander W. S. (2003) Nat. Immunol. 4, 540–545 [DOI] [PubMed] [Google Scholar]
- 48.Kim H., Hawley T. S., Hawley R. G., Baumann H. (1998) Mol. Cell Biol. 18, 1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barua D., Faeder J. R., Haugh J. M. (2007) Biophys. J. 92, 2290–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng G. S., Hui C. C., Pawson T. (1993) Science 259, 1607–1611 [DOI] [PubMed] [Google Scholar]
- 51.Yin T., Shen R., Feng G. S., Yang Y. C. (1997) J. Biol. Chem. 272, 1032–1037 [DOI] [PubMed] [Google Scholar]
- 52.Walter A. O., Peng Z. Y., Cartwright C. A. (1999) Oncogene 18, 1911–1920 [DOI] [PubMed] [Google Scholar]
- 53.Peng Z. Y., Cartwright C. A. (1995) Oncogene 11, 1955–1962 [PubMed] [Google Scholar]
- 54.Schlessinger J. (2000) Cell 103, 211–225 [DOI] [PubMed] [Google Scholar]
- 55.Kiyono M., Kaziro Y., Satoh T. (2000) J. Biol. Chem. 275, 5441–5446 [DOI] [PubMed] [Google Scholar]
- 56.Wei W., Das B., Park W., Broek D. (1994) Gene 151, 279–284 [DOI] [PubMed] [Google Scholar]
- 57.Sundaresan M., Yu Z. X., Ferrans V. J., Sulciner D. J., Gutkind J. S., Irani K., Goldschmidt-Clermont P. J., Finkel T. (1996) Biochem. J. 318, 379–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nimnual A. S., Taylor L. J., Bar-Sagi D. (2003) Nat. Cell Biol. 5, 236–241 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.