Abstract
Autophagosomes and their precursors are best defined by electron microscopy but may also be traced in living cells based on the distribution of specific autophagy molecules. LC3, the most commonly examined autophagy marker in mammalian cells, labels structures that are frequently manifested as dots or rings using light microscopy; however, the nature of these structures is not entirely clear. We reported here a novel approach to examine the LC3-positive compartment in cell-free lysates, which revealed that they were actually tubulovesicular structures with considerable heterogeneity. Using affinity purification, we isolated these membranes for electron microscopy, which indicated that they possessed ultrastructural features consistent with autophagosomal membranes at various maturation stages. Further biochemical and proteomics analyses demonstrated the presence of multiple autophagy-related and other functional molecules. The different distribution patterns of Atg5, Atg16, Atg9, and p62/SQSTM1 on the LC3-positive compartment provided new clues on how these molecules might be involved in the dynamics of the autophagosomal membranes. Finally, several morphologically unique groups of LC3-positive membranes were categorized. Their topological configurations suggested that double-membrane vesicles could be derived from single membrane compartments via different means, including tubule-to-vesicle conversion, whose presence was supported by live cell imaging. These findings thus provide new information on the dynamics of the autophagosomal compartment.
Keywords: Bacterial Transcription, Homeobox, Monoglyceride, Protein Export, Radiation Inactivation, Respiratory Burst, Autophagosome, Autophagy
Introduction
Macroautophagy, hereafter referred to as autophagy, constitutes a major catabolic mechanism in cells (1–3). Thirty two autophagy-related (Atg) molecules have been identified in yeast, and many are evolutionarily conserved in mammalian cells (4–7). How molecular events lead to the formation of autophagosomes is not known, although several hypotheses have been proposed based on electron microscopic studies (4, 5, 8).
Typical autophagosomes revealed by EM are vesicles delimited by double membranes or, in some cases, multiple membranes (1, 9–12). Various cytosolic contents could be found within the cisternal lumen of autophagosomes. Fusion of autophagosomes with lysosomes forms the single-membrane autolysosomes with increased electron density. EM studies had led to the hypothesis that the formation of autophagosomes might start with a crescent-shaped membrane cisterna, also called the isolation membrane or phagophore (4, 5, 8). The various components in the autophagosomal compartment could be recognized by the presence of specific autophagy molecules (9, 10, 13, 14). Thus, Atg16L1 and Atg5 were mainly present in the phagophore, whereas LC3, a mammalian Atg8 homologue, labels isolation membranes, matured autophagosomes, and autolysosomes, suggesting that different Atg molecules may participate in autophagosome biogenesis at different stages. However, cells processed for traditional EM could not be subjected to further investigation to test these hypotheses.
Consequently, various Atg markers have been examined in living cells to monitor and to understand the biological process. Atg16L1 and LC3 are diffusively present in the cytoplasm of cells under basal conditions but accumulate at discrete spots following autophagic stimulation. These puncta are thought to represent the same autophagosomal structures as those identified by EM at various maturation stages (9, 10). However, direct evidence has yet to be sought to document that these puncta indeed represent such structures even though their formation seems to be highly specific. One hurdle is that few morphological features, other than being described as dots or rings, could be ascribed to these puncta. Another is that no specific methods have been developed to probe these structures from living cells in greater detail.
We have attempted to overcome these hurdles by examining the LC3-labeled puncta in cell-free lysate and purifying them for EM and biochemical analysis. The results not only confirmed their autophagosomal nature, but defined new molecular, morphological, and topological features that could be important for understanding autophagosome membrane dynamics.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment
HEK-293 and HCT-116 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and other standard supplements. LC3 (specifically LC3B), Atg16L1, Atg5, and Atg9L1 were isolated from human cDNA and cloned to pEGFP or pDsRed/monomer vector (Clontech) at their N terminus. Stable cell lines with GFP-LC3 were created with standard methodology. Transient transfection was conducted using Lipofectamine 2000 (Invitrogen). Cells were treated 48 h later as indicated.
Starvation was induced by replacing the complete medium with Earle's balanced salt solution (EBSS).2 Autophagy induction by calcium phosphate precipitate (CPP) was conducted as described previously (15). Briefly, cells were cultured at the density of 2 × 105/ml/per well in 12-well plates overnight. Right before the treatment, CPP solution was prepared by mixing Buffer A (50 mm HEPES, pH 7.05, 3 mm Na2HPO4) and calcium chloride (256 mm) in equal volumes. For each milliliter of culture, 100 μl of CPP solution was added dropwise. Cells were then analyzed within 2–6 h.
To prepare cell lysates for the observation of the LC3-positive structure, cells cultured in 6-well plates were washed with cold PBS and then homogenized in 2 ml of cold Buffer B (0.25 m sucrose, 1 mm EDTA, 20 mm HEPES, pH 7.4) by passing 10–15 times through a 22-gauge needle (for HEK-293 cells) or a 28-gauge needle (for HCT116 cells). The lysates were centrifuged at 800 × g for 10 min. The post-nuclear fraction (10 μl) was loaded onto a glass slide and covered with a glass coverslip for immediate fluorescence microscopic examination.
Fluorescence and Electron Microscopy
Cells cultured in plastic plates were directly imaged using a Nikon TE 200 inverted epifluorescence microscope (Melville NY) equipped with a digital camera (CoolSNAP HQ2; Photometrics, Inc.). Time-lapse images of LC3-positive structures in cell lysates were acquired under a 60×/1.40 oil objective and assembled using NIS-Elements AR3.0 software (Nikon, Melville, NY) at 2–5-s intervals for 60–180 s.
Live cell confocal imaging was performed on cells grown in glass bottom culture dishes (Mattek), which were maintained in a temperature-controlled, humidified environmental chamber (Tokai Hit, Tokyo Japan), using a Nikon 200E inverted microscope and Yokagawa (Tokyo, Japan) scan head. Metamorph (Molecular Devices, Downingtown, PA) was used to control the microscope. Single midplane images were collected from multiple different stage positions once every 3 min. Following collection, images were deconvolved using a Weiner deconvolution filter with Nikon Elements software.
For electron microscopy, cells or affinity-purified LC3-TVS were fixed in 2.5% glutaraldehyde in 0.1 m PBS, pH 7.4, for 1 h and post-fixed in aqueous 1% OsO4, 1% K3Fe(CN)6 for 1 h. Following three PBS washes, the pellet was dehydrated through a graded series of 30–100% ethanol, 100% propylene oxide and then infiltrated in 1:1 mixture of propylene oxide/Polybed 812 epoxy resin (Polysciences, Warrington, PA) for 1 h. After several changes of 100% resin over 24 h, the pellet was embedded in molds, cured at 37 °C overnight, followed by additional hardening at 65 °C for 2 more days. Ultrathin (60 nm) sections were collected on 100 mesh copper grids, stained with 2% uranyl acetate in 50% methanol for 10 min, followed by 1% lead citrate for 7 min. Sections were imaged using a JEOL JEM 1011 transmission electron microscope (Peabody, MA) at 80 kV fitted with a bottom mount AMT 2k digital camera (Advanced Microscopy Techniques, Danvers, MA).
Isolation of the GFP-LC3-positive Subcellular Structure
HEK-293 cells stably expressing GFP-LC3 were treated with CPP for 6 h. Cells were then washed in PBS and disrupted in Buffer B. The post-nuclear supernatant was further centrifuged at 10,000 × g for 20 min. The pellets were washed twice in the washing buffer (PBS, pH 7.4, 0.1% bovine serum albumin, 2 mm EDTA) to remove residual cytosolic GFP-LC3. The pellets were finally suspended in PBS with 2 mm EDTA and 3% bovine serum albumin and incubated with an anti-GFP antibody (GFP, B2, Santa Cruz Biotechnology) at 1:25 dilution overnight at 4 °C. The unbound antibodies were removed by centrifugation. The pellet was washed and resuspended in PBS buffer, followed by incubation with the prewashed BioMag goat anti-mouse IgG beads (Qiagen) for 1 h at 4 °C. The magnetic beads were separated from the solution using a magnetic separator. After washing, the preparation was fixed in 2.5% glutaraldehyde for EM study or resuspended in RIPA buffer for biochemical analysis.
Immunoblot Analysis
Lysates from cells or the isolated LC3-positive membranes (10 μg) were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Antibodies against the following molecules were used: GFP, p62/SQSTM1, Rab5, Rab7, and Rabaptin-5 (Santa Cruz Biotechnology); Beclin-1, Rab4, Rab11, Rabex-5, and EEA-1 (BD Biosciences); Atg5, Atg9L1, Atg16L1, UVRAG, and VPS34 (Abgent, Inc.); clathrin and HSP70 (Cell Signaling Technology); β-actin (Sigma); glyceraldehyde-3-phosphate dehydrogenase (Novus Biologicals); LAMP1 (clone H4A3) and LAMP2 (clone ABL-93) (Developmental Studies Hybridoma Bank, University of Iowa); and Rab33 (16). The rabbit polyclonal anti-LC3B antibody was made using a peptide representing the N-terminal 14 amino acids of human LC3B and an additional cysteine (PSEKTFKQRRTFEQC).
Proteomics Studies
LC3-positive membranes on the magnetic beads were washed three times with ice-cold PBS. The beads were then suspended in RIPA and/or urea lysis buffer (9.5 m urea, 2% CHAPS, 0.8% ampholytes, and 1% dithiothreitol and protease inhibitors) for 30 min at room temperature. The supernatants were then collected by centrifugation for 15 min at 20,000 × g at 4 °C. After the non-protein elements were removed with the 2-D Clean-up kit (GE Healthcare), the protein precipitates were dissolved in 9.5 m urea lysis buffer containing 0.5% v/v IPG buffer (pH 3–10, nonlinear, GE Healthcare) and quantified for protein concentration using a Bradford-based protein assay (Bio-Rad). The two-dimensional electrophoresis was performed as described previously (17). Briefly, 80 μg of protein was separated by isoelectric focusing using IPG strips (11 cm, pH 3–10 nonlinear) with Protean IEF Cell System (Bio-Rad) for the first dimension and a vertical gel electrophoresis apparatus system (Invitrogen) for the second dimension. Gels were stained with Coomassie Blue as reported (18). The protein spots on the gels were digitalized at 400 d.p.i. resolution using a Kodak Image Station 4000MM (Carestream Health).
Protein spots were then excised and subjected to tryptic in-gel digestion and matrix-assisted laser desorption ionization time-of-flight analysis using Voyager-DE PRO mass spectrometer (Applied Biosystems, Inc.) as described previously (17). Results were analyzed using 4000 Series Explorer software version 4.0 (Applied Biosystems) to obtain accurate masses (±50 ppm) for all the peptides in the tryptic digest. The resulting peptide mass fingerprints, together with the pI and relative molecular mass values (estimated from two-dimensional gels), were used to search the Swiss-Prot or NCBInr protein data bases, MS-FIT from Protein Prospector version 5.0, which compares the experimentally determined tryptic peptide masses with theoretical peptide masses calculated for proteins contained in the Swiss-Prot or NCBInr protein data bases. Taxonomy was set to human, and trypsin and keratin peaks were excluded. Protein identifications with a score greater than 1000, with sequence coverage more than 10% and at least four matched peptides, were considered significant. Search parameters were ±50–100 ppm peptide mass tolerance and one maximum missed cleavage. The identified proteins were classified in terms of their physiological functions and localization using information from PubMed (www.ncbi.nlm.nih) and the Swiss-Prot/TrEMBL protein knowledge base.
RESULTS
Tubulovesicular Morphology of LC3-labeled Membrane Compartment
GFP-LC3 has been widely used in tracking autophagosome dynamics in mammalian cells (9, 20). GFP-LC3 labels punctated structures in cells under autophagy stimulation. Immuno-EM has confirmed that these structures represent autophagosomes at various maturation stages (9). At the cellular level, the GFP-LC3 puncta seem to possess different morphologies such as dots, rings, and even tubules (9, 14, 19), of which the dynamic nature, the relationship with autophagosome maturation, and the biological significance were not clear.
To better understand these structures, we developed a simple but revealing approach to examine the GFP-LC3 puncta in cell-free lysates. This approach gave rise to an unprecedented level of detail about these structures and further allowed their purification for biochemical analysis.
HEK-293 cells and HCT-116 cells, stably expressing GFP-LC3, were subjected to starvation in EBSS. As anticipated, GFP-LC3 puncta was induced (Fig. 1A). The morphology of the GFP-LC3 puncta under this stimulation was mostly fine dots with little definition. However, when the cells were disrupted and the lysates were observed using fluorescence microscopy, we found that these structures possessed very diverse morphology with many different configurations (Fig. 2, A and B). GFP-LC3-labeled membranes were found to be both vesicular and tubular. Tubules and vesicles could be linked together with vesicles more frequently at the end of the tubules. The cisternal structure could be dilated, twisted, and even composed of additional internal elements, which were also GFP-LC3 positive. The size of these structures was larger than would be anticipated based on the size of the puncta at the cellular level, suggesting that the puncta might represent a more packed configuration, which was relaxed upon cell lysis.
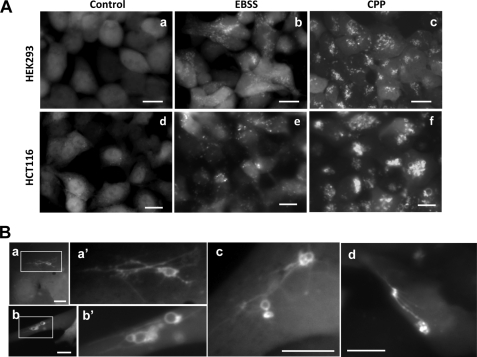
FIGURE 1.
GFP-LC3-positive tubulovesicular structures in cells. A, HEK-293 cells (panels a–c) and HCT-116 cells (panels d–f) stably expressing GFP-LC3 were treated with medium (panels a and d), EBSS (panels b and e), or CPP (panels c and f) for 4 h. LC3 punctation was observed under a fluorescence microscope. Scale bar, 10 μm. B, HEK-293-GFP-LC3 cells (panels a–c) or HCT-116-GFP-LC3 cells (panel d) were treated with CPP for 2 or 4 h, respectively. The details of the GFP-LC3-labeled structures were examined. Panels a′ and b′ are enlarged from the boxed area in panels a and b, respectively. Scale bar, 10 μm.
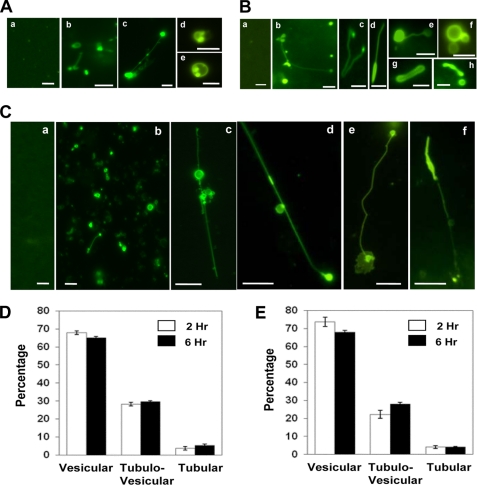
FIGURE 2.
GFP-LC3-positive tubulovesicular structures in cell lysates. A and B, HEK-293-GFP-LC3 (A) or HCT-116-GFP-LC3 (B) cells were treated with normal medium (panels a in A and B) or EBSS (rest of the panels) for 4 h. Cells were then lysed, and LC3-TVS were identified. Representative LC3-TVS are shown. Scale bar, 2 μm. C, HEK-293-GFP-LC3 cells treated with medium (panel a) or CPP for 3 h (panels b–f) were lysed and observed. Representative LC3-TVS are shown. Scale bar, 5 μm. D and E, HEK-293-GFP-LC3 cells were treated with CPP (D) or EBSS (E) for 2 or 6 h as indicated before being lysed. Three types of LC3-TVS were quantified in the lysates in randomly selected fields using fluorescence microscopy, and the percentages (mean ± S.D.) of each type of LC3-TVS were determined. Vesicular LC3-TVS are single ring-like structures (e.g. A, panels d and e); tubular LC3-TVS are tubules only (e.g. B, panel g); and tubulovesicular LC3-TVS are the combination of vesicular and tubular structures (e.g., A, panel c; B, panel b; B, panel d; B, panel e; and C, panels c–f).
We had also observed these structures, now termed as LC3-TVS, in two other types of cells, A549 and a murine embryonic fibroblast line following starvation (data not shown). To examine whether LC3-TVS could be induced by a different autophagic stimulus, we used CPP because it induces a strong autophagic response (15). Compared with starvation, the CPP-induced LC3 puncta were more prominent in terms of size and number (Fig. 1A). In addition to the smaller puncta, CPP also induced the formation of large sized ring-shaped LC3-positive structures (Fig. 1B). Interestingly, we observed that these dots or rings could be linked by LC3-positive tubules, readily visible at the cellular level (Fig. 1B).
LC3-TVS were prominently present in the lysate prepared from CPP-treated cells (Fig. 2C). LC3-labeled tubules tended to be longer than those found in the lysates prepared from starved cells, consistent with the observations made at the cellular level (Fig. 1B). The LC3-labeled tubules could span more than 10 μm, with vesicles on either the end or the middle part of the tubules.
However, in either starvation or CPP treatment, about two-thirds of LC3-TVS were vesicular, and the rest were either tubular or tubulovesicular (Fig. 2, D and E). Tubules accounted for less than 10% of LC3-TVS, suggesting that this configuration was relatively rare. There were no apparent differences in the distribution pattern between cells treated for 2 h and cells treated for 6 h, indicating a steady turnover of these various LC3-TVS.
Thus LC3-TVS were induced in multiple cell lines following autophagic stimulation, but not in cells without autophagic stimulation (Fig. 2, A, panel a, B, panel a, and C, panel a). The presence of LC3-TVS in the lysate was entirely consistent with the appearance of LC3 puncta at the cellular level. In addition, LC3-TVS were morphologically consistent with the LC3 tubules and vesicles observed within cells, such as in the case of CPP treatment, suggesting that they were not the result of the cell lysis procedure.
LC3-TVS Demonstrate Ultrastructural Features Consistent with Autophagosomal Membranes
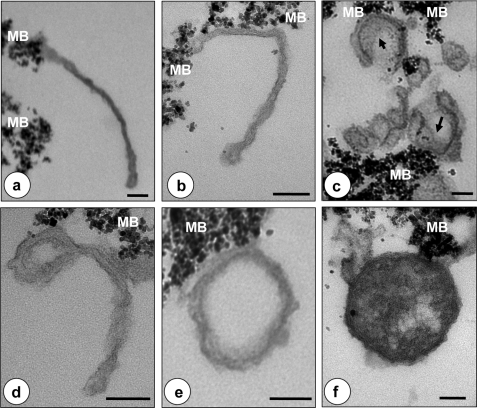
To determine the relationship of LC3-TVS with the autophagosomal vesicles identified by EM in intact cells, we treated HEK-293 cells, stably expressing GFP-LC3, with CPP and isolated LC3-TVS from the lysates by anti-GFP affinity purification. EM examination of the isolated materials confirmed the pleomorphic tubulovesicular nature. Some of these LC3-positive tubules (Fig. 3, a–c) clearly resembled the phagophores as defined previously (10, 11). Electron-dense material, which likely represented engulfed cellular components, could be found at the lumen side of some crescent-like cisternae (Fig. 3c). Other vesicular structures looked like mature autophagosomes delimited by double membranes (Fig. 3, e and f). Interestingly, vesicles with tubular stems could be found to have double membranes along the vesicular portion of the organelle (Fig. 3d), suggesting a transition from the tubular to vesicular component (see below). Finally, multimembranous vesicles that might represent a special form of autophagosome (11) were also observed (Fig. 3f). Together, these findings indicated that LC3-TVS were quite heterogeneous, containing autophagosomal membranes at various maturation stages as observed previously using immuno-EM for LC3 localization (9, 10).
FIGURE 3.
LC3-TVS exhibits heterogeneous ultrastructural features consistent with autophagosomes at different maturation stages. LC3-TVS were isolated by affinity purification and subjected to EM analysis. a–c show various tubular LC3-TVS. Some resembled the crescent-like phagophores with dense materials (arrows) along the inner surface (c), which may represent engulfed cellular substances. e and f show vesicles that may represent completed autophagosomes. d shows a tubulovesicular structure that may represent an intermediate stage of tubule-to-vesicle conversion. The dense particles at the edge of the membranes are magnetic beads (MB). Scale bar, 100 nm.
Biochemical Analysis Supports the Autophagosomal Nature of LC3-TVS
We further sought evidence that LC3-TVS represent autophagosomes using biochemical analysis. Like other autophagic stimuli (20), CPP induced the rapid formation of LC3-II and the release of the GFP moiety from the GFP-LC3 molecule, indicating the formation of autophagosome and its degradation in the lysosome (15) (Fig. 4A). Biochemical analysis of the isolated LC3-TVS demonstrated the enrichment of membranous GFP-LC3-II, with little to no GFP-LC3-I or GFP alone (Fig. 4A). Importantly, the endogenous LC3-II was also enriched within the same GFP-LC3-targeted membranes (Fig. 4B). This was consistent with our previous findings that CPP-induced GFP-LC3 puncta formation depended on LC3 processing and key autophagy machinery (15). We detected several other Atg molecules in the isolated LC3-TVS, including Atg9, Atg16L1, and Atg5 in the conjugation form (with Atg12) (Fig. 4B). Intriguingly, there seemed to be an additional Atg16L1 band in the isolated LC3-TVS, which was not obviously detected in the total lysate. The nature of this more slowly migrating Atg16L1 species was not clear at this moment. Beclin 1 signal was weakly observed, whereas VPS34 was not detected with two different antibodies. Isolated materials from untreated samples did not reveal any Beclin 1 (data not shown). However, UVRAG was readily detected, which may suggest that most UVRAG was not associated with Beclin 1 in LC3-TVS prepared here. UVRAG could function with the class C Vps complex to promote Rab7 activity and autophagosome maturation, which can be independent of its interaction with Beclin 1 (21). In addition, we also detected the presence of p62/SQSTM1, which bridges the autophagosomal membranes with the autophagic substrates (22–24). Interestingly, most of these molecules were not particularly enriched in the isolated LC3-TVS, suggesting that only a small fraction of these molecules was recruited to these structures. Alternatively, they might have rapidly recycled off, as suggested for Atg9 (25), Atg5, and Atg16 (10, 14). The presence of these markers was not due to membrane contamination because they could be demonstrated to be directly present on LC3-TVS (see below). Certainly, these findings did not imply that all vesicles would express all the molecules to the same extent because the LC3-TVS compartment appeared to be heterogeneous with regard to the maturation stage (Fig. 3).
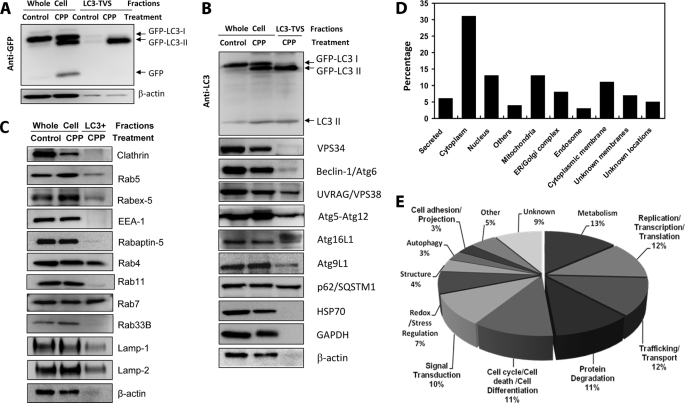
FIGURE 4.
Protein expression profile of LC3-TVS. A–C, total lysates from HEK-293-GFP-LC3 cells treated with control medium or CPP for 6 h (whole cells) or from isolated LC3-TVS were subjected to immunoblot assay with indicated antibodies. Equal amounts of proteins were loaded per lane. GFP-LC3 was detected using an anti-GFP antibody (A) or an anti-LC3 antibody (B). Endogenous LC3 was identified with the same anti-LC3 antibody (B). All other autophagy-related molecules (B) and endosomal related molecules (C) were detected using the corresponding antibodies except that the Atg5-Atg12 complex was identified using an anti-Atg5 antibody. D and E, total lysates of LC3-TVS were subjected to proteomics analysis. One hundred and one proteins were clearly identified and annotated. D shows the percentage of proteins at different subcellular locations. Others indicates locations at the proteasome, centrosome, or microtubules. Unknown membranes indicates that membrane location is predicted based on sequences, but the actual location is not known. E shows the potential functions of the identified proteins. Others indicates functions in immunity, angiogenesis, respiration, and microtubule nucleation. See supplemental Table S1 for details.
Because of the similarity of the tubulovesicular morphology of LC3-TVS to the endosomal membranes, we examined the presence of several endosomal markers that had been reported to be involved in the autophagy process (Fig. 4C) (26–29). We found that the isolated LC3-TVS contained the early endosomal markers, Rab5 and Rab4, but no significant amount of Rab11. They also contained the later endosomal markers, Rab7, LAMP1, and LAMP2. We also detected the GTP exchange factor for Rab5, Rabex-5. However, clathrin, an endocytic vesicle marker, as well as EEA-1 and Rabaptin-5, two of the Rab5 effector molecules, were nearly undetectable. We did not detect Rab33B, normally localized in the Golgi complex, but reported to interact with Atg16 and to affect LC3 lipidation (30), suggesting that the isolated compartment likely had matured beyond this stage or that the membranes positive for both Rab33B and LC3 were rare. In addition to the immunoblot analysis, we detected the presence of Rab5 or Rab7 on individual LC3-TVS by fluorescent microscopy (data not shown). Overall, the findings were consistent with the notion that the presence of these endosomal markers resulted from the fusion of autophagosomes with endosomal vesicles as indicated in earlier studies (28, 31–33). Alternatively, they might imply that the endosomal system could contribute to the autophagosome formation as one of the membrane sources.
The presence of other possible proteins in LC3-TVS was assessed by employing a proteomics approach. Among the 101 unique proteins that were clearly identified (supplemental Table S1), cytoplasmic proteins constituted the single largest group (31%). But a significant portion (42%) originated from various membrane compartments, including the endoplasmic reticulum, Golgi complex, mitochondria, endosomes, and cytoplasmic membranes (Fig. 4D).
Although most of the defined proteins did not seem to have a known function in autophagy (Fig. 4E and supplemental Table S1), we did detect the presence of LC3 and two other autophagy molecules, Atg7 and Atg9, with this approach as well. Other autophagy molecules were not detected perhaps due to lower abundance or detachment from the autophagosomes. We also detected a G protein α-subunit, Giα3, i.e. guanine nucleotide-binding protein G(k) subunit α, which had been associated with autophagy (34, 35). Furthermore, several proteins, such as peroxiredoxin 6, GRP-78, UMP-CMP kinase, and carbonic anhydrase, although not clearly known to be important for autophagy, had been found to be enriched in autophagosomal membranes in a previous proteomics study (36), which analyzed autophagosomes purified from the livers of starved rats using density gradient centrifugation. These results indicate the necessity for further investigation of these molecules and their roles in autophagy. We also realized that the presence of these markers could also be due to their engulfment by autophagosomes, in which case they would bear no direct function in autophagy.
Previous studies have demonstrated that Atg5, Atg12, and Atg16L1 are associated in a protein complex and suggested that Atg16L1 may be responsible for the complex to target to the phagophore, where it promotes the conjugation of LC3 to phosphatidylethanolamine (14, 37). Atg5 and Atg16L1 are not detected in completely delimited autophagosomes and are thus considered markers for the phagophore (9, 10, 13, 14). In our study, phagophore-like structures could be observed (Fig. 3, a–c), and endogenous Atg16L1 and Atg5 proteins could be detected in LC3-TVS (Fig. 4B). To further characterize the morphology of the phagophore, we cotransfected HEK-293 cells stably expressing RFP-LC3 with GFP-Atg16L1 and CFP-Atg5 and visualized the phagophore using fluorescence microscopy (Fig. 5A). Notably, unlike the crescent-like presentation observed by EM in intact cells, the Atg16L1/Atg5/LC3-positive phagophores were found to be tubules or vesicles or a combination of both. The EM presentation of phagophores might be affected by orientation within a given section. The morphology of the phagophore under the nonfixed and nonsectioned conditions would be a closer representation to the native state. This observation could affect the interpretation of how phagophores might evolve into autophagosomes (see below).
FIGURE 5.
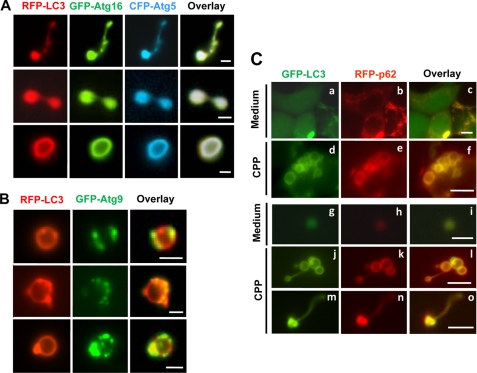
Colocalization of autophagy-related molecules on LC3-TVS. A and B, HEK-293-RFP-LC3 cells were transfected with GFP-Atg16L1 and CFP-Atg5 (A) or with GFP-Atg9 (B) overnight and then treated with CPP for 4 h. Cells were lysed, and LC3-TVS were identified that also contained GFP-Atg16L1 and CFP-Atg5 (A) or GFP-Atg9 (B). Three representative structures are shown for each combination. Note the different distribution pattern of Atg16L1 and Atg9 on LC3-TVS. Bar, 2 μm. C, HEK-293-GFP-LC3 transfected with RFP-p62 were cultured in medium (panels a–c and g–i) or treated with CPP for 4 h (panels d–f and j–o). The intact cells (panels a–f) or the lysates (panels g–o) were observed by fluorescence microscopy. Scale bars, 10 μm (panels a–f), 2 μm (panels g–l), and 5-μm (panels m–o). Low levels of LC3- and p62-positive smaller vesicles could be observed in control groups (panels a–c and g–i), although significantly more and larger LC3- and p62-positive vesicles were only seen in CPP-treated samples (panels d–f and j–o).
We next examined Atg9, which is another key molecule required for autophagy in both yeast (38) and mammalian cells (39, 40). It redistributes from the trans-Golgi network to the later endosomes and can colocalize with LC3 puncta upon autophagic stimulation (39, 40). We detected the endogenous Atg9 protein in lysates of isolated LC3-TVS (Fig. 4B) and observed colocalization of Atg9 on individual LC3-TVS (Fig. 5B), supporting the notion that Atg9 could be present in autophagosomal structures whose nature, however, was less clear in terms of its maturation stage. Interestingly, we noted that the distribution of Atg9 on LC3-TVS (Fig. 5B) was often asymmetric and tended to be located in certain areas of the membranes, in contrast to the even distribution of Atg16L1, Atg5, or LC3. The distribution pattern suggested that Atg9 might be associated with specific docking sites and unique membrane dynamics.
Finally, we examined p62/SQSTM1, a key adaptor molecule known to interact with LC3 and the ubiquitinated substrates of autophagy to facilitate their degradation (22–24). We confirmed that p62 could be specifically recruited to LC3-positive puncta following CPP treatment (Fig. 5C). In addition, the LC3G120A mutant, which cannot be processed by Atg4 for lipid conjugation, did not recruit p62 under the same conditions (data not shown). Furthermore, p62 was detected in LC3-TVS by Western blot (Fig. 4B) and by fluorescence microscopy (Fig. 5C). The presence of p62 in tubular LC3-TVS (Fig. 5C) might suggest that the recruitment of p62 by LC3 occurs before the closure of the autophagosomal membranes. This early recruitment of p62 could allow p62, and perhaps its associated proteins, to be enclosed in the autophagosome before the membrane fusion occurred.
Morphology Heterogeneity of LC3-TVS Could Suggest Diverse Membrane Dynamics
The morphology heterogeneity of LC3-TVS suggested that there might be dynamic conversions among the different forms, which could be associated with a maturation process. In the absence of additional molecular information, we explored the potential morphology relationships of various LC3-TVS and found that LC3-TVS could be categorized into several groups consisting of topologically related vesicles. By doing so, we noted that double-membrane vesicles could be derived from single-membrane vesicles by several different means (Figs. 6 and 7).
FIGURE 6.
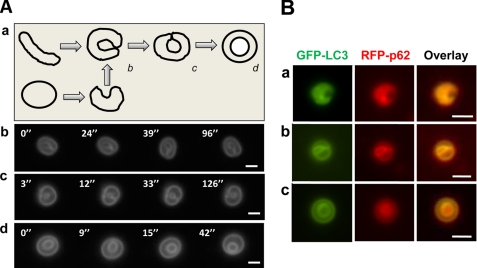
Topological configurations of LC3-TVS, group I. A, HEK-293-GFP-LC3 cells treated with CPP for 3 h were lysed. LC3-TVSs that seemed to be topologically related (b–d) were selected and imaged every 3–5 s for 180 s (see supplemental Videos 1–3, respectively) to visualize their three-dimensional configuration, which revealed that the inner ring was still attached to the outer ring in example c but was free in example d. Representative panels with the vesicles at different positions are shown. Panel a shows the model, suggesting that the vesicle in panel d could be topologically derived from the vesicle shown in panel c, which in turn could be derived from the vesicle shown in panel b. Although the vesicle in panel b could be derived from a tubule, it could be also topologically derived from another vesicle. Scale bar, 2 μm. B, HEK-293-GFP-LC3 cells were transfected with RFP-p62 and treated with CPP for 6 h. Three LC3-TVS that were also positive for RFP-p62 were found to be topologically related. Note the concentrated fluorescence signals of both LC3 and p62 in the inner surface (panel a), inner ring (panel b), or the lumen (panel c) of the vesicles. Scale bar, 1 μm.
FIGURE 7.
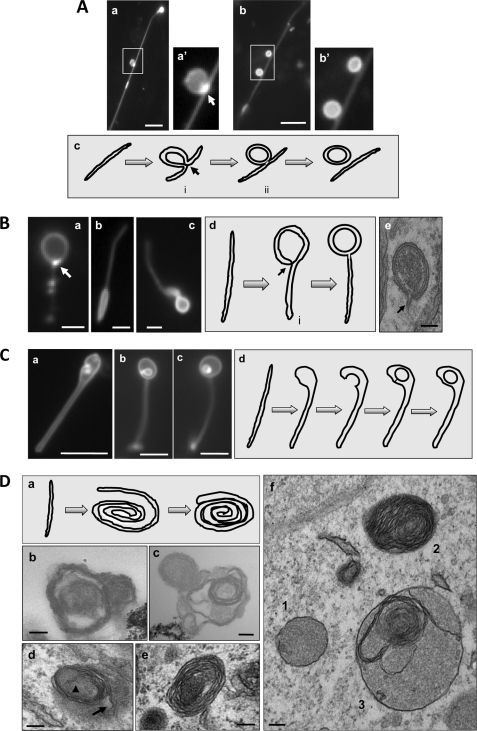
Topological configurations of LC3-TVS, groups II–V. A, group II. HEK-293-GFP-LC3 cells treated with CPP for 3 h were lysed. Two representative LC3-TVS that seemed to be topologically related (panels a and b) were presented. Boxed area in panels a and b are enlarged in panels a′ and b′, respectively. Panel a is the same as Fig. 2C, panel d. It is represented here to demonstrate the vesicle-tubule relationship. Panel c suggests a possible topological derivation of a vesicle from a tubular membrane with the intermediates i and ii resembling those shown in panels a and b, respectively. The arrow in panels a′ and c indicates the possible fusion site where the GFP-LC3 signal was particularly concentrated. Scale bar, 5 μm. B, group III. LC3-TVS identified in HEK-293 (panels a and c) or HCT-116 cells (panel b) treated with EBSS (panels a and b) or CPP (panel c) are presented to suggest a topological origin from a tubular membrane as illustrated in panel d. The intermediate vesicle (denoted with i in panel d) could be identified in EM (panel e and Fig. 3, panel d). The arrows in panels a and d indicate the possible fusion site where the GFP-LC3 signal was particularly concentrated. Panel e demonstrates a double-membrane autophagosome, which was identified in an intact HEK-293 cell treated with CPP. Note the tubular stem extending from the double-membrane vesicle (arrow). Scale bars, 1 μm (panels a and b), 2 μm (panel c), and 100 nm (panel e). C, group IV. Three LC3-TVS (panels a–c) from HEK-293-GFP-LC3 cells treated with CPP are presented, and their topological relationship could be as is illustrated in panel d, in which the tubule expands at one end, which is followed by invagination and internal vesicle formation. Scale bar, 5 μm. D, group V. Panel a shows a schematic representation of the formation of the roll-like vesicles. The tubular membrane may roll around the substrates to form a loose bundle, which may then be compacted and delimited by the outermost layer of the membrane. Electron microscopic examination of HEK-293-GFP-LC3 cells treated with CPP (panels d–f) or the LC3-TVS isolated from these cells (panels b and c) demonstrated the presence of multiple-membrane vesicles, which could be derived from the membrane rolling as illustrated in panel a. The arrowhead and arrow in panel d indicate the proximal and distal ends of the rolling tubule, respectively. Panel f also shows a vesicle (vesicle 3) with the combined morphology of vesicle 1 (double-membrane) and vesicle 2 (multimembrane). Scale bar, 100 nm.
LC3-TVS of group I (Fig. 6) would represent one such a process widely proposed in the literature (4, 5, 8). Here a single-membrane tubular sheet or vesicle may become curved with the leading edges contacting each other, resulting in membrane fusion and the formation of separated outer and inner membranes, i.e. a double-membrane structure (Fig. 6A, panel a). Electron microscopy has demonstrated the presence of the crescent-shaped phagophores that may represent the intermediate stage of the conversion in cells (9, 10) and in isolated LC3-TVS (Fig. 3c). We identified LC3-TVS that might represent additional intermediate stages (Fig. 6A, panels b–d), although such conversion had not been actually observed in real time fashion. These representative vesicles were tracked in vitro by time-lapse imaging for a better understanding of the spatial relationship of the outer and inner rings. One vesicle seemed to represent the stage where the curved leading edges contacted each other but had not been fused (Fig. 6A, panel b, and supplemental Video 1). In the other two vesicles where the fusion had occurred and thus the inner ring had formed, the video imaging showed that the inner ring could still be attached with the outer ring in one vesicle (Fig. 6A, panel c, supplemental Video 2) but completely detached from the outer ring in another (Fig. 6A, panel d, and supplemental Video 3). The former could represent an intermediate stage during the transition from the crescent-shaped single-membrane vesicle (Fig. 6A, panel b) to the completely delimited double-membrane vesicle (Fig. 6A, panel d).
To explore the potential biological relevance of these representative LC3-TVS, we examined whether and how the adaptor molecule p62 was present in these LC3-TVS. In HEK-293 cells expressing both GFP-LC3 and RFP-p62, CPP treatment led to the colocalization of p62 on LC3-TVS (Fig. 5C). Further examination of the lysates indicated that p62-positive LC3-TVS could possess various group I topological configurations. Interestingly, p62 seemed to be more concentrated in the inner surface of the “curving” membrane (Fig. 6B, panel a), the inner ring (Fig. 6B, panel b), or the lumen of the inner ring (Fig. 6B, panel c). Notably the distribution of GFP-LC3 seemed to follow the same pattern, with more in the inner ring. Because LC3-interacting p62 is important for bridging the autophagosome with its substrates during autophagy (22–24), the relative concentration of p62 in the inner part of the LC3-TVS supports the notion that these vesicles could have a dynamic relationship as depicted in Fig. 6A, panel a.
A significant fraction of LC3-TVS was composed of both tubular and vesicular parts (Fig. 2). For these LC3-TVS, it seemed that the vesicular part might be derived from the tubular membrane. In one possible scenario (group II), the middle part of the membrane budded out to form a vesicular compartment, followed by the detachment of the formed vesicles (Fig. 2C, panels c and d, and Fig. 7A). A key difference from the conventional membrane budding process might be that the budding in the present case would involve both sides of the tubule toward one direction, so the newly formed vesicle could be double-membraned (Fig. 7A, panel c). We noted that the GFP-LC3 signal was particularly concentrated at a site where the membrane fusion and vesicle detachment might occur (Fig. 7A, panels a and c), although GFP-LC3 signal was in general stronger in the budding vesicular part than in the tubular part in these LC3-TVS.
The group III LC3-TVS represented another topological variation in which a vesicular structure was associated with the end of a tubule (Fig. 2 and Fig. 7B, panels a–c). The vesicular part could be double-membraned as shown by EM (Fig. 3d). Topologically, one way that might explain the formation of a double-membrane vesicle, but not a single-membrane vesicle, at the end of a tubule is that the terminal part folded back toward the tubule, followed by fusion (Fig. 7B, panel d). As in group II conversion, we noted that GFP-LC3 signals at the possible joint site were particularly strong in some LC3-TVS (Fig. 7B, panel a), where the membrane fusion might occur. This process could have in vivo significance as autophagosomes with tubular stems that resembled the intermediate structure (Fig. 7B, panel d) could be observed in intact cells by EM (Fig. 7B, panel e).
Another possible means by which a double-membrane vesicle could be formed at the end of a tubule was the expansion of the lumen at that end, followed by invagination of a portion of the membrane, which is then detached to become the inner vesicle (group IV) (Fig. 7C). Interestingly, groups II–IV topological conversions could be found on single LC3-TVS (Fig. 2 and Fig. 7A, panel a).
A variation of group III conversion was the wrapping of the tubular membranes, perhaps around the substrates to be degraded, into multilayered vesicles (group V) (Fig. 7D, panel a). We could not discriminate such LC3-TVS by fluorescence microscopy, perhaps because such a configuration would render them to be manifested as strongly brightened dots. However, after purification, these multimembrane LC3-TVS could be identified by EM (Fig. 7D, panels b and c). Furthermore, multimembrane vesicles, alone or in fusion with the double-membrane vesicles, could be observed by EM in intact cells treated with CPP (Fig. 7D, panels d–f) or with other autophagic agents (11, 41). In some cases, the two ends of the rolling membrane could be clearly recognized (Fig. 7D, panel d), supporting the model of how they might be formed (Fig. 7D, panel a).
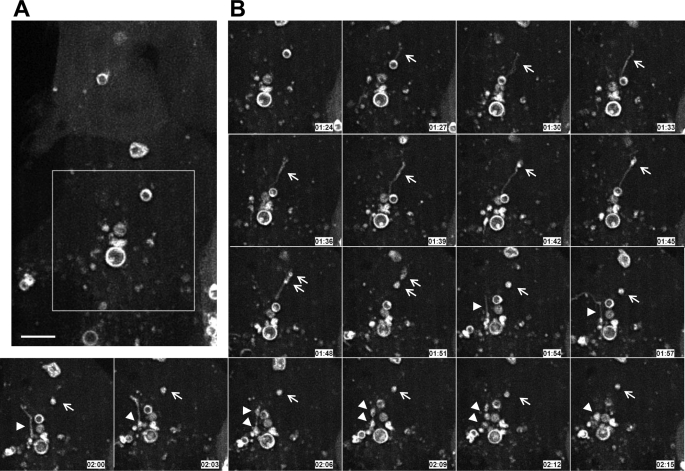
These models of tubule-to-vesicle conversion with the potential consequence of the formation of double-membrane structures were based on the morphology of LC3-TVS in lysates. In search for in vivo evidence, we monitored CPP-treated HEK-293 cells stably expressing GFP-LC3 by time-lapse confocal imaging. We could observe the stimulus-dependent appearance of GFP-LC3-positive tubules (Fig. 8 and supplemental Video 4). Subsequently, GFP-LC3 could become concentrated at a certain portion of the tubule where vesicles seemed to form. This process was relatively quick. In one example, it took about 24 min from the appearance of the tubule to its breakdown into the vesicles (Fig. 8B, arrows). In another example, the same process took about 15 min (Fig. 8B, arrowheads). Thus, time-lapse confocal imaging of live cells could detect sequences of membrane dynamics that would be consistent with the tubule-to-vesicle conversion concept of the proposed models (Fig. 7), although whether the process represents a maturation process has yet to be demonstrated.
FIGURE 8.
Dynamic conversion of LC3-TVS in live cells. HEK-293 cells stably expressing GFP-LC3 were grown in glass bottom culture dishes and treated with CPP for 2 h. Live cell confocal imaging was then performed in a temperature-controlled and humidified environmental chamber. Images were taken every 3 min (see supplemental Video 4). The left panel (A) is an overview of the field, where the boxed area is shown in panel B. Panel B shows two tubule-to-vesicle conversion events. In one event (indicated by the arrows), one GFP-LC3 positive tubule appeared at 1:27 and became two vesicles at 1:51. In another event (indicated by the arrowheads), one GFP-LC3 positive tubule appeared at 1:54 and became two vesicles at 2:09. Scale bar, 5 μm.
DISCUSSION
Autophagosomes are best characterized by electron microscopic examination of individual cells. They can also be isolated by gradient centrifugations from a given subcellular fraction (42), which are then subjected to electron microscopy or biochemical analysis. However, fixed samples cannot be subjected to dynamic analysis. The autophagosomal compartment can be tracked in living cells by tagging Atg molecules, among which LC3 is the most commonly used in mammalian cells (9, 20). Immunoelectron microscopy confirmed that GFP-LC3 can label autophagosomal membranes. However, at the light microscopic level, structures labeled by LC3 or other Atg molecules, such as Atg16L1 or Atg5, can often be only recognized as dots or, in some cases, rings. How these structures represent autophagosomes in the living cells and how they might evolve following autophagy stimulation is unclear because very little information is available other than that their formation is under autophagic control. We have thus developed a new approach to study the LC3-positive autophagosomal compartment under nonfixed conditions.
Lysis of cells that were undergoing autophagy resulted in the release of membrane structures labeled by GFP-LC3, which were then subjected to morphological and biochemical analyses. The conclusion that these structures were authentic rather than experimental artifacts is based on the following: 1) they could be observed not just in the cell lysate but also in intact cells without any physical disruption; 2) they were found only after autophagic stimulation but not in untreated condition; 3) they could be observed in different cell types following different autophagy stimulation, including the classical stimulation of starvation; and 4) they possessed the features of autophagosomal membranes.
Morphological and Molecular Features of LC3-TVS
Electron microscopy indicated that the isolated LC3-TVS assumed the same ultrastructural features as the previously defined autophagosomal membranes, including the phagophore and the delimited autophagosomes. The detection of Atg16L1 and Atg5 on LC3-TVS also supports that some LC3-TVS could represent the phagophore, which may now be better studied in the nonfixed status. Interestingly, the morphology of the phagophore under the nonfixed and nonsectioned conditions indicates that it could be vesicular and/or tubular. The vesicular presentation of phagophores was not expected based on the existing EM observation of strictly crescent-like tubular morphology, although the evidence for the presence of Atg5-positive vesicular phagophore could be found in earlier studies (10). Although the existing hypothesis (4, 5, 8) explains the formation of the autophagosome from the crescent-like tubular structure through membrane extension, curving, and fusion of the leading edges (Fig. 6A), it is also possible that the vesicular phagophores may evolve into the double-membrane autophagosomes via a different topological process, e.g. via invagination (Fig. 6A, panel a).
Isolation of LC3-TVS also allows the study of Atg molecules whose function is less clear, as in the case of the transmembrane protein, Atg9. In mammalian cells, autophagy stimulation promotes the transition of Atg9 from the trans-Golgi network to later endosomes (39, 40), which is controlled by ULK1, a mammalian homologue of Atg1 (40), and Atg13 (43). This redistribution leads to colocalization of at least some Atg9 with the LC3-positive puncta (39, 40). However, the nature of the relationship between Atg9 and the LC3-positive compartment and the significance of Atg9 redistribution is not clear, although it was hypothesized that Atg9 contributes to the membranes of autophagosomes (40) through vesicle transportation facilitated by this molecule (25). Atg9-positive LC3-TVS would likely represent the compartment where Atg9-positive vesicles fuse with the LC3-TVS or its precursor. A notable feature of this compartment is the asymmetric distribution of Atg9 (Fig. 5B), which was in stark contrast to the even distribution of Atg16L1, Atg5, or LC3. This feature may suggest that Atg9 is associated at some specific docking sites related to membrane fusion and/or topological changes that remodel the vesicles for the generation of mature autophagosomes. It would be interesting to identify these sites and the mechanisms involved in the interaction of Atg9 vesicles with the LC3-TVS.
The presence of Rab5, Rab4, and Rab7 in the LC3-TVS suggests that both early and later endosomes could contribute to the maturation of the autophagosome, a notion that has been well documented in the literature (12, 28, 31, 32). Fusion of the autophagosome with the endosomes, particularly the later endosomes, results in the generation of so-called amphisome (44). We have detected LC3-TVS that also express Rab5 or Rab7 (data now shown), which could be classified as amphisome by definition. However, they did not manifest a morphological feature significantly different from that of other LC3-TVS with fluorescent microscopy.
We conducted additional biochemical analysis of isolated LC3-TVS to search for novel molecular features. The nonbiased proteomics approach revealed mainly proteins that are not particularly known for autophagy. We cannot rule out that the presence of any of these proteins in the LC3-TVS was due to engulfment of organelles or cytoplasm by the autophagosomes. However, the possible involvement of some of the proteins in LC3-TVS biogenesis should be explored in future studies. Notably, several molecules, such as peroxiredoxin 6, GRP-78, UMP-CMP kinase, and carbonic anhydrase (supplemental Table S1), had also been found to be enriched in autophagosomal membranes in a previous proteomics study (36). The latter study used autophagosomes purified from the livers of starved rats using the classical density gradient centrifugation. The two studies shared the advantage of employing defined subcellular structures for proteomics investigation, thus reducing the nonspecific association and enhancing the relevance of the findings.
The presence of Giα3 in LC3-TVS was particularly interesting. This molecule has long been known to play a role in the regulation of autophagy (34) and could mediate the induction of autophagy. Giα3 has been shown to colocalize with LC3 in mouse hepatocytes upon starvation (35). Thus, in this regard, the finding of Giα3 in LC3-TVS may not be surprising. However, these observations raised questions regarding how Giα3 might mediate autophagy signaling. It would be tempting to speculate that this molecule may be targeted to the autophagosomal membranes where it functions.
Topology and Dynamic Conversions of LC3-TVS
LC3-TVS were heterogeneous with morphological features not obviously observed in intact cells. The definite biological relationship among LC3-TVS of various morphologies has yet to be determined, but the topological relationships defined in this study could suggest several possible membrane remodeling processes.
The group I conversion (Fig. 6A) would be most consistent with the current prevailing hypothesis that tubule-like membranes give rise to the double-membrane vesicles through membrane bending and enclosure (4, 5, 8). Our studies provide new pieces of evidence to support this hypothesis because we have defined LC3-TVS that could represent the intermediate status predicted by this hypothesis. Furthermore, the distribution of p62 with a relative concentration in the inner side of LC3-TVS corresponds well with the topology of these vesicles that would suggest a conversion into an autophagosome as predicted by the model. Future studies need to provide in vivo evidence to demonstrate the actual process of such a dynamic conversion.
The precursor membrane may not need to be strictly tubular but could also be vesicular (Fig. 6A, panel a), which can be remodeled to the crescent shape. This would be supported by the tubular or vesicular morphology of Atg16L1- and Atg5-positive LC3-TVS that likely represent phagophores (Fig. 5A).
Perhaps most revealing is that through similar analysis, we could identify additional patterns of topological conversion that may suggest how double-membrane vesicles could be formed from the single-membrane compartment by other means. In groups II–IV (Fig. 7, A–C), only a portion of a tubular membrane is likely involved in the generation of double-membrane vesicles through a budding-like process (group II), a folding-like process (group III), or a combination of lumen expansion and invagination (group IV). Although these processes were deduced from the morphology of LC3-TVS, we observed vesicles that may represent the intermediate stage of the conversion in autophagy-stimulated cells with conventional EM (Fig. 7B, panel d). However, these structures were not observed frequently, perhaps due to the rapid formation and turnover of autophagosomes. In addition, group II conversion would predict the generation of certain intermediate structures due to membrane budding (Fig. 7A, panels a and c). Coincidently, a membrane structure termed as omegasome with a similar morphology to these intermediates and positive for LC3 has been also proposed in another study (45). This omegasome structure was thought to be involved in autophagosome biogenesis. Certainly, the true correlation of the omegasome with the specific LC3-TVS defined here (Fig. 7A) has yet to be determined, but the topological conversion defined here could explain the formation of both structures.
LC3-TVS of group V indicated that vesicular structure could be formed through revolving of the tubular membranes. Multimembrane autophagosomes are not uncommon (Fig. 7D, panel f) (11, 41), but their biogenesis is rarely discussed. The proposed mechanism is based on topology possibilities with some morphological evidence (Fig. 7D, panel d). Clearly, this process could be intertwined with other topological changes, which leads to the generation of hybrid types of autophagosomes (Fig. 7D, panel f). It would be important to determine the condition under which and the mechanism by which this type of topological changes occur.
Overall, it is not clear how frequently these various membrane conversions could occur. Live cell imaging could detect the tubular LC3-positive structure being converted to vesicular structure (Fig. 8), although the resolution of the imaging did not allow us to depict the details of the conversion. It is thus unknown to what models the particular converting event fitted. In lysates prepared from cells treated with EBSS or CPP for 2 or 6 h, about two-thirds of LC3-TVS possessed vesicular morphology and one-third possessed tubulovesicular or tubular morphology (Fig. 2, D and E). This may suggest that the vesicular configuration is easier to form, relatively stable, or the most frequently targeted by GFP-LC3. Thus, the dynamics of LC3-TVS in vivo have yet to be fully explored.
How the tubule-to-vesicle conversion occurs is not clear. We observed that in LC3-TVS of groups II–IV, the LC3 signal is generally stronger in the vesicular part compared with the tubular part. Unless LC3 preferably targets to the vesicular portion, one has to assume that there might be a lateral movement of LC3 in the membrane toward the area of vesicle formation. Neither hypothesis has experimental support at the present. However, the latter hypothesis would be consistent with the proposed topological changes that allow the generation of double-membrane vesicles from the single-membrane tubular portion. There was increased concentration of LC3 on the vesicle near where it encountered the tubule (Fig. 7). The yeast counterpart of LC3, Atg8, is able to induce membrane hemifusion (46). Interestingly, Atg8 has been found to be mainly present at the site where membranes contact each other and hemifusion occurs, despite that it is apparently evenly distributed before fusion occurred. These findings allow us to hypothesize that localized concentration of LC3 in LC3-TVS through lateral movement may indicate a similar process and that the site where LC3 is most concentrated could be where membrane fusion occurs, leading to the closure of the vesicle during the formation.
We have to caution that the in vivo significance of LC3-TVS dynamics could be subjected to alternative interpretations. Because LC3 is present in autophagosomal membranes at various stages, a particular morphological feature of LC3-TVS may represent a type of membrane dynamics other than those depicted in our models (Figs. 6 and 7). These other events could possibly include fusions between LC3-TVS and other membrane compartments with or without significant autophagy consequences.
It also should be noted that CPP appear to be a particularly strong stimulus for the formation of LC3-TVS, although starvation could induce the same event (Figs. 1 and 2). Thus, it is possible that some of the membrane events may be more significantly amplified in CPP-treated cells, but may not necessarily be universally conserved in other autophagy conditions.
In summary, the most notable finding of this study was that the commonly observed LC3 dots or rings were actually quite pleomorphic and could be both tubules and vesicles. Both biochemical and ultrastructural analysis support that they represent the autophagosomal compartment with some components likely at the early stage of maturation. Under a given stimulation, LC3-TVS could be quite heterogeneous in size and in morphology. Their overall complexity revealed in this study would not be anticipated based on previous observations made at the intact cell level. As the result, further analysis of the morphology of the heterogeneous LC3-TVS population suggests that single-membrane compartment could be converted into double-membrane vesicles through several different means, although the biological significance and frequency of these events have yet to be fully determined.
Supplementary Material
Acknowledgments
We thank Ming Sun for EM sample preparation, Dr. Manimalha Balasubramani for proteomics assistance, and Dr. Gerald Apodaca for valuable comments. We thank Dr. Zhenyu Yue (Mount Sinai Medical Center) for the p62 construct.
This work was supported, in whole or in part, by National Institutes of Health Grants CA111456 and CA83817 (to X. M. Y.) and NS058463 (to Y. J. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Videos 1–4.
- EBSS
- Earle's balanced salt solution
- LC3-TVS
- LC3-positive tubular vesicular structures
- PBS
- phosphate-buffered saline
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- GFP
- green fluorescent protein
- CPP
- calcium phosphate precipitate.
REFERENCES
- 1.De Duve C., Wattiaux R. (1966) Annu. Rev. Physiol. 28, 435–492 [DOI] [PubMed] [Google Scholar]
- 2.Levine B., Klionsky D. J. (2004) Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 3.Kundu M., Thompson C. B. (2008) Annu. Rev. Pathol. 3, 427–455 [DOI] [PubMed] [Google Scholar]
- 4.Suzuki K., Ohsumi Y. (2007) FEBS Lett. 581, 2156–2161 [DOI] [PubMed] [Google Scholar]
- 5.Xie Z., Klionsky D. J. (2007) Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 6.Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. (2009) Dev. Cell 17, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009) Dev. Cell 17, 87–97 [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N. (2007) Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 9.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. (2001) J. Cell Biol. 152, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fengsrud M., Sneve M. L., Overbye A., Seglen P. O. (2004) in Autophagy (Klionsky D. J. ed) pp. 11–25, Landes Bioscience, Georgetown, TX [Google Scholar]
- 12.Eskelinen E. L. (2005) Autophagy 1, 1–10 [DOI] [PubMed] [Google Scholar]
- 13.Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. (2003) J. Cell Sci. 116, 1679–1688 [DOI] [PubMed] [Google Scholar]
- 15.Gao W., Ding W. X., Stolz D. B., Yin X. M. (2008) Autophagy 4, 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng J. Y., Koda T., Fujiwara T., Kishi M., Ikehara Y., Kakinuma M. (1998) J. Cell Sci. 111, 1061–1069 [DOI] [PubMed] [Google Scholar]
- 17.Kang J. H., Kim H. T., Choi M. S., Lee W. H., Huh T. L., Park Y. B., Moon B. J., Kwon O. S. (2006) Proteomics 6, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 18.Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988) Electrophoresis 9, 255–262 [DOI] [PubMed] [Google Scholar]
- 19.Köchl R., Hu X. W., Chan E. Y., Tooze S. A. (2006) Traffic 7, 129–145 [DOI] [PubMed] [Google Scholar]
- 20.Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang C., Lee J. S., Inn K. S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J. U. (2008) Nat. Cell Biol. 10, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 24.Yue Z. (2007) Autophagy 3, 139–141 [DOI] [PubMed] [Google Scholar]
- 25.He C., Baba M., Cao Y., Klionsky D. J. (2008) Mol. Biol. Cell 19, 5506–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutierrez M. G., Munafó D. B., Berón W., Colombo M. I. (2004) J. Cell Sci. 117, 2687–2697 [DOI] [PubMed] [Google Scholar]
- 27.Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E. L. (2004) J. Cell Sci. 117, 4837–4848 [DOI] [PubMed] [Google Scholar]
- 28.Fader C. M., Sánchez D., Furlán M., Colombo M. I. (2008) Traffic 9, 230–250 [DOI] [PubMed] [Google Scholar]
- 29.Ravikumar B., Imarisio S., Sarkar S., O'Kane C. J., Rubinsztein D. C. (2008) J. Cell Sci. 121, 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itoh T., Fujita N., Kanno E., Yamamoto A., Yoshimori T., Fukuda M. (2008) Mol. Biol. Cell 19, 2916–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon P. B., Seglen P. O. (1988) Biochem. Biophys. Res. Commun. 151, 40–47 [DOI] [PubMed] [Google Scholar]
- 32.Liou W., Geuze H. J., Geelen M. J., Slot J. W. (1997) J. Cell Biol. 136, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demarchi F., Bertoli C., Copetti T., Tanida I., Brancolini C., Eskelinen E. L., Schneider C. (2006) J. Cell Biol. 175, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meijer A. J., Codogno P. (2004) Int. J. Biochem. Cell Biol. 36, 2445–2462 [DOI] [PubMed] [Google Scholar]
- 35.Gohla A., Klement K., Piekorz R. P., Pexa K., vom Dahl S., Spicher K., Dreval V., Häussinger D., Birnbaumer L., Nürnberg B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3003–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Øverbye A., Fengsrud M., Seglen P. O. (2007) Autophagy 3, 300–322 [DOI] [PubMed] [Google Scholar]
- 37.Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. (2008) Mol. Biol. Cell 19, 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reggiori F., Shintani T., Nair U., Klionsky D. J. (2005) Autophagy 1, 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada T., Carson A. R., Caniggia I., Umebayashi K., Yoshimori T., Nakabayashi K., Scherer S. W. (2005) J. Biol. Chem. 280, 18283–18290 [DOI] [PubMed] [Google Scholar]
- 40.Young A. R., Chan E. Y., Hu X. W., Köchl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. (2006) J. Cell Sci. 119, 3888–3900 [DOI] [PubMed] [Google Scholar]
- 41.Ding W. X., Ni H. M., Gao W., Hou Y. F., Melan M. A., Chen X., Stolz D. B., Shao Z. M., Yin X. M. (2007) J. Biol. Chem. 282, 4702–4710 [DOI] [PubMed] [Google Scholar]
- 42.Strømhaug P. E., Berg T. O., Fengsrud M., Seglen P. O. (1998) Biochem. J. 335, 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. (2009) Mol. Cell. Biol. 29, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seglen P. O., Bohley P. (1992) Experientia 48, 158–172 [DOI] [PubMed] [Google Scholar]
- 45.Axe E. L., Walker S. A., Manifava M., Chandra P., Roderick H. L., Habermann A., Griffiths G., Ktistakis N. T. (2008) J. Cell Biol. 182, 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakatogawa H., Ichimura Y., Ohsumi Y. (2007) Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.