Abstract
Half of all human transcription factors use C2H2 zinc finger domains to specify site-specific DNA binding and yet very little is known about their role in gene regulation. Based on in vitro studies, a zinc finger code has been developed that predicts a binding motif for a particular zinc finger factor (ZNF). However, very few studies have performed genome-wide analyses of ZNF binding patterns, and thus, it is not clear if the binding code developed in vitro will be useful for identifying target genes of a particular ZNF. We performed genome-wide ChIP-seq for ZNF263, a C2H2 ZNF that contains 9 finger domains, a KRAB repression domain, and a SCAN domain and identified more than 5000 binding sites in K562 cells. Our results suggest that ZNF263 binds to a 24-nt site that differs from the motif predicted by the zinc finger code in several positions. Interestingly, many of the ZNF263 binding sites are located within the transcribed region of the target gene. Although ZNFs containing a KRAB domain are thought to function mainly as transcriptional repressors, many of the ZNF263 target genes are expressed at high levels. To address the biological role of ZNF263, we identified genes whose expression was altered by treatment of cells with ZNF263-specific small interfering RNAs. Our results suggest that ZNF263 can have both positive and negative effects on transcriptional regulation of its target genes.
Keywords: Chromatin/Epigenetics, DNA/Transcription, Transcription/Regulation, Transcription/Target genes, Transcription/Zinc Finger, KRAB domain, SCAN domain, ZNF263
Introduction
C2H2 zinc fingers comprise the largest class of site-specific DNA-binding proteins encoded in the human genome (1). Of the 2000 predicted DNA binding transcription factors, ∼900 contain C2H2 zinc finger domains. This abundance suggests that the C2H2 zinc finger factors (ZNFs)3 may be critical regulators of a large number of important biological networks. The high specificity and high affinity of zinc finger transcription factors has enabled many in vitro DNA-protein interaction studies such as CASTING and DNA affinity purification. In fact, a C2H2 zinc finger protein, SP1, was the first site-specific transcription factor purified based on its DNA binding properties (2). Decades of research on the in vitro DNA binding properties of the zinc finger proteins have provided critical insights into how transcription factors recognize their cognate sites.
C2H2 zinc finger transcription factors contain from 1 to more than 30 fingers. Based on the distribution of the fingers along the coding region, the proteins can be classified into triple C2H2, multiple adjacent C2H2, and separated paired C2H2 finger proteins (3). Each finger contains two-to-three B strands and one α-helix. DNA binding specificity is conferred by several amino acid residues in the α-helix of the finger with support provided by conserved linkers (TG(Q/E)KP) present between fingers. Triple C2H2 proteins, which include the well studied SP1 and early growth response (EGR) family members, use their three fingers to bind to G-rich consensus motifs (e.g. GGGGCGGGG for SP1 and GCGTGGGCG for EGR1). In contrast to the single, high specificity motif that characterizes binding of triple C2H2 factors, proteins with multiple adjacent zinc fingers are hypothesized to have the ability to bind to different motifs, depending upon which fingers are used for recognition of the DNA. However, of the hundreds of multiple adjacent C2H2 factors, very few have been extensively characterized.
Although there is a paucity of experimental analyses on the binding specificity of multiple adjacent C2H2 factors, previous work using EGR family members as a model system has resulted in the construction of a zinc finger code that allows binding site motif predictions to be made based on the amino acid sequence of the fingers (4–6). After the development of a putative binding motif for a particular zinc finger protein, a simple bioinformatics analysis could be used to predict the genomic locations of the binding sites for that ZNF. However, it is unlikely that every one of the sequences in the human genome having a good match to a predicted motif is in fact occupied in vivo (due to negative influences from repressive chromatin, nucleosomal positioning, overlap of binding sites with other factors, etc). It is also important to consider that the zinc finger code is based in large part on in vitro studies performed using isolated DNA fragments and purified proteins; very few analyses of the in vivo binding specificity of C2H2 zinc finger proteins have been performed. It is possible that in vivo binding specificities may follow a different code than that developed from in vitro studies, perhaps due to heterodimer formation of a C2H2 zinc finger factor with another partner. Finally, it is not clear if in vivo, ZNFs that have a large number of finger domains will bind to a large site encompassing the predicted binding specificity of all the fingers. It is also possible that they will bind to a consensus motif dictated by the set of three adjacent fingers having the highest DNA binding affinity or to multiple different consensus motifs by employing alternative different combinations of three adjacent fingers.
To address these questions, we have chosen to study ZNF263 (Fig. 1). This protein contains 9 C2H2 zinc finger domains in the C terminus of the protein (7). In addition, ZNF263 contains two other conserved domains associated with the C2H2 family, an N-terminal SCAN domain, and a Kruppel-associated box (KRAB) domain located between the SCAN domain and the finger domains (8). At least one-third of the C2H2 zinc finger proteins contain a KRAB domain, which is thought to mediate interactions with TRIM28 (KAP1), and members of this set of factors are termed KRAB-ZNFs. Recent curation of the human genome identified 423 KRAB-ZNF genes that have the potential through alternative transcripts to produce at least 742 distinct proteins (9). A small number of the KRAB-ZNF proteins (25 of the 423, including ZNF263) also contain a leucine-rich region called a SCAN domain. Many of the KRAB-ZNFs are primate-specific. For example, of the human 366 KRAB-ZNFs that lack a SCAN domain, 342 are present in other primates but only 76 are conserved in mouse. In contrast, the subset of SCAN-domaining containing KRAB-ZNFs tend to be highly conserved; of the 25 KRAB-ZNFs that contain a SCAN domain, 19 are conserved in mouse (9). Thus, ZNF263 is a multiple adjacent C2H2 factor that belongs to a large set of KRAB-ZNFs but is also contained within a much smaller subset of highly conserved KRAB-ZNFs that have a SCAN domain. We have analyzed the in vivo binding specificity of ZNF263 and have also addressed the mechanisms by which this KRAB-ZNF factor may regulate transcription.
FIGURE 1.
Schematic of the ZNF263 protein. The SCAN protein-protein interaction domain, the TRIM28 (KAP1) interaction domain, and the nine zinc fingers of ZNF263 are shown.
EXPERIMENTAL PROCEDURES
Cell Culture
Human chronic myelogenous leukemia cells (K562, ATCC #CCL-243) were grown in RPMI supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin-streptomycin. Human cervical carcinoma cells (HeLa-S3, ATCC CCL-2.2) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin-streptomycin.
Chromatin Immunoprecipitation (ChIP)
ZNF263 ChIP samples were prepared from K562 cells as follows. Cultures of 1 × 108 K562 cells were harvested at a density of 106 cells/ml cells and cross-linked with 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by the addition of glycine to 125 mm final concentration, and cells were washed twice with 1 × phosphate-buffered saline. The cell pellet was then resuspended in 2 ml of ChIP lysis buffer (50 mm Tris-Cl pH 8.0, 5 mm EDTA, 1% SDS, 1 complete protease inhibitor tablet (Roche Applied Science) and incubated on ice for 30 min. Samples were sonicated for 30 min with 30-s pulses and 1 min of resting using the Bioruptor sonicator (Diagenode) to produce chromatin fragments of 0.5 kb on average. After clarification by centrifugation, sonicated extracts were precleared with 20 μl of Staphylococcus aureus cells per 107 cells blocked with 10 mg/ml bovine serum albumin. The precleared extracts were diluted 1:10 with ChIP dilution buffer (1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-Cl pH 8.0, 5 mm phenylmethylsulfonyl fluoride). Anti-ZNF263 antibody (Novus Biologicals, catalog no. H00010127-A01) was added at a concentration of 2 μg/107 cells and incubated for 12 h at 4 °C. Complexes were recovered with S. aureus cells for 15 min at room temperature and washed 5 times with ice-cold radioimmune precipitation assay buffer. Precipitates were resuspended in 100 μl of ChIP elution buffer (1% SDS, 0.1 m NaHCO3), incubated at 65 °C for 12 h, and treated with 10 μg of RNaseA for 20 min at 37 °C. After pooling, the DNA was recovered from the eluate using the QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions.
Quantitative Real-time PCR
Quantitative real-time PCR was performed on a Bio-Rad DNA Engine Opticon real-time PCR system using SYBR® Green Master PCR Mix according to the manufacturer's instructions (Invitrogen). The -fold enrichment of each target site was calculated as 2 to the power of the cycle threshold (cT) difference between input chromatin and ChIP samples. We selected primer sets for five negative regions, all of which showed no enrichment. Ten other validation sites were chosen over a range of subgroups as defined in the results; primers are listed in supplemental Table S1.
ChIP-seq Library Construction and Quantitation
ChIP libraries were created according to Robertson et al. (10) using 15 cycles of amplification. Libraries were run on a 2% agarose gel, and the 150–450-bp fraction of the library was extracted and purified. To estimate the yield of library and its relative amplification value, library DNA was quantitated using a Nanodrop spectrometer, and serial dilutions of 1.25 nm library were compared with a reference library by real-time PCR using primers complementary to the library adapters. The amplification value relative to the reference library was used to estimate the flow cell loading concentration. The ChIP-seq libraries were run on an Illumina GA2 by the DNA Technologies Core Facility at the University of California-Davis or by the laboratory of Mike Snyder (Yale University) as part of the ENCODE Consortium. The ChIP-seq data (GSE19235) has been deposited in the NCBI Gene Expression Omnibus.
Processing ChIP-seq Data
A standard routine procedure for extracting image files, mapping the reads onto human genome, and filtering the mapped reads to unique reads was followed using the Solexa 1.6 pipeline. Only uniquely mapped reads with a length of 27 bp were then further used for determining the ZNF263 binding sites. We used the bin-based enrichment threshold level (BELT) program, which we developed for analyzing ChIP-seq data. In brief, the algorithm of the program is: 1) develop a bin-based method to obtain a sum of the uniquely mapped reads for each bin, providing the results in GFF- or BED-formatted files (the formatted GFF or BED files can be visualized on the University of California, Santa Cruz Genome Browser or using the Affymetrix Integrated Genome Browser (see Fig. 2A for an example of BED files and identified binding regions)); 2) apply a percentile rank statistic method to determine each level of scores from the Top 0.1% to the Top 10% level; 3) generate a background model of binding peaks by randomizing the data and identifying the number of the binding sites for each percentile rank in the randomized data; 4) estimate the false discovery rate (FDR) to measure the significance of identified target loci. A detailed description of the algorithm and how to determine the bin-size and thresholds can be found under supplemental Methods.
FIGURE 2.
Reproducibility of the ZNF263 ChIP-seq experiments. A, ChIP-seq profiles of two biologically independent replicate experiments (labeled RepA and RepB) using the ZNF263 antibody are shown for a region of chromosome 1. The peaks called for each replicate and the overlapping peaks are also shown. B, Venn diagrams indicate the number of peaks called at the 0.5% FDR level for each of two biologically independent ZNF263 ChIP-seq experiments and the overlap between the two replicates.
Identification of a ZNF263 Consensus Recruitment Motif
To identify a binding motif for ZNF263, we applied our de novo motif discovery approach ChIPMotifs developed in our previous study (11). Briefly, the ChIPMotifs approach incorporates a statistical bootstrap re-sampling method to identify a number of motifs detected from the set of high stringent 1473 ZNF263 binding regions obtained from the Top 0.1% level using ab initio motif-finding programs such as Weeder (12), MaMF (13), and MEME (14). A set of ∼24,000 human promoter sequences of 500 bp in length for each promoter from 1000 bp upstream to the 5′ transcription start site were selected as a negative control dataset. The identified significant motifs evaluated by the Fisher Test were then screened against the JASPAR (15) and TRANSFAC (16) databases using STAMP (17); all these programs are built into our ChIPMotifs program. A final novel ZNF263 motif was then determined. For those ZNF263 binding sites without a good match to the first identified novel ZNF263 motif, ChIPMotifs were further run on these sites, and other known or novel motifs were then determined.
To obtain a motif predicted for ZNF263 by the zinc finger code, we used a prediction program (5) that predicts binding sites for zinc finger domains (see supplemental Fig. S3). This program predicted motifs for fingers 2-3-4, 3-4-5, 6-7-8, and 7-8-9. We merged the individual triplet predictions to obtain a predicted WebLogo for fingers 2–9 (see Fig. 4 and supplemental Fig. S3). To search a set of genomic regions for the predicted motif, we adapted the WebLogo to create a nucleotide string; the sequence NNGGANGANGGANGGGANNANGGA was used as the predicted motif bound by fingers 2–9. However, because there is a gap between fingers 5 and 6 (see Fig. 1), we also made individual motifs for fingers 2–5 and 6–9; the sequence NGGGANNANGGA was used as the motif bound by fingers 2–5, and the sequence NNGGANGANGGA was used as the motif bound by fingers 6–9.
FIGURE 4.
Motif analysis of ZNF263 binding sites. A, a WebLogo representing the 24-nt experimentally derived ZNF263 binding site is shown. B, a WebLogo representing the ZNF263 binding site predicted using the zinc finger code is shown. ZNFs bind in the C-terminal to N-terminal orientation. Therefore, the first 12 nt in the motif are those predicted to be bound by fingers 9 to 6, and the second 12 nt in the motif are those predicted to be bound by fingers 5 to 2 (see supplemental Fig. S3). For searching of the ZNF263 binding sites for the predicted motif, the sequence NNGGANGANGGANGGGANNANGGA was used as the motif bound by fingers 2–9; the sequence NGGGANNANGGA was used as the motif bound by fingers 2–5, and the sequence NNGGANGANGGA was used as the motif bound by fingers 6–9.
Microarray Data Analysis
Affymetrix Human Exon 1.0 ST (HuEx-1_0-st-v2) arrays consisting of 1.4 million probe sets of clustering 1.0 million exon clusters from >5,500,000 features, were used for measuring basal level gene expression in K562 cells. A total of seven biological samples from seven different institutions (from seven ENCODE consortium groups) were performed on this array platform, and the generated quantified Affymetrix image files (seven *.CEL files) were analyzed simultaneously and normalized using the RMA algorithm provided in the Expression Console software developed by Affymetrix Inc. Probe sets were annotated as supported by RefSeq and full-length GenBankTM Transcripts, resulting in 21,924 annotated probe sets representing ∼18,000 annotated genes. A scale of LOG2 was used for calculating the abundance for each probe set gene for each sample. A mean value of seven replicates is computed for each core gene and used as a final value of the gene expression level.
siRNA Treatment and Illumina Expression Arrays
For ZNF263 knockdown RNA analysis, HeLa-S3 cells were transfected with ZNF263 siRNAs (SMARTpool; Dharmacon, catalog no. L-018336-01-0005) or si-GLO RISC-Free (Dharmacon, catalog no. D-001600-01) as a nonspecific control using Invitrogen Lipofectamine2000 according to the manufacturer's recommendations. Cells were harvested at 72 h; RNA was prepared using Invitrogen Trizol according to manufacturer's recommendations and then assayed using the Agilent Systems Bioanalyzer to ensure that high quality RNA was used for the array experiments. The Illumina TotalPrep RNA amplification kit from Ambion (AMIL1791) was used to generate biotinylated, amplified RNA for hybridization with the Illumina Sentrix Expression Beadchips, HumanHt-12. The Sentrix gene expression beadchips used for this study consisted of a 12-array, 2 stripe format comprising ∼48,000 probes/array. In this collection 24,000 probes were from refseq sequences and 24,000 from other GenBankTM sequences (see the Illumina website for more details). Arrays were processed as per the manufacturer's instructions, scanned at medium photomultiplier tube settings as recommended by the manufacturer, and analyzed using Bead Studio Software Version 2.3.41. The arrays were hybridized and processed by the University of California Davis Expression Analysis Core Facility. Data were normalized using the “average” method, which simply adjusts the intensities of two populations of gene expression values such that the means of the populations become equal. Differential expression was calculated for the control versus siZNF263 data sets using an algorithm provided by Bead Studio. -Fold enrichment values were used to obtain the list of candidates with greater than a two-fold change. The array data (GSE19146) has been deposited in the NCBI Gene Expression Omnibus.
RESULTS
Genome-wide Identification of ZNF263 Binding Sites Using ChIP-seq
As indicated above, ZNF263 contains 9 zinc fingers, with the conserved DNA binding specificity linker being present after fingers 2, 3, and 4 in the first cluster and after 6, 7, and 8 in the second cluster. Thus, ZNF263 is predicted to bind to DNA, with some combination of fingers 2–5 and fingers 6–9 (Fig. 1). Our goal was to use ChIP-seq to identify all genomic binding sites for ZNF263. However, before our studies binding of ZNF263 in human cells had not been investigated. We began by examining expression levels of ZNF263 in a series of human cell lines (supplemental Fig. S1A). Because ZNF263 is expressed in K562 cells at levels equal to or greater than in the other cells we tested and because we have found that K562 cells are very good for ChIP-seq (for a variety of factors we have observed less background and higher signals in these cells than in many other cell types), we decided to initially focus on the binding pattern of ZNF263 in K562 cells. We next showed that an antibody that recognizes the endogenous ZNF263 could immunoprecipitate the protein from K562 nuclear extract (supplemental Fig. S1B). Importantly, these experiments showed that the ZNF263 antibody is highly specific, recognizing only the correct size protein in nuclear extract prepared from K562 cells, and that the antibody can effectively immunoprecipitate the ZNF263 protein from nuclear extract. The ZNF263 antibody was then used in ChIP assays, and the resultant immunoprecipitated DNA was purified and used to prepare a library for sequencing with an Illumina Genome Analyzer. Two independent ChIP replicates were performed using K562 cells grown on separate days, and two independent libraries were prepared and sequenced. After mapping to the University of California, Santa Cruz human HG18 assembly, 10,322,952 and 11,102,465 reads that uniquely map to the human genome were obtained for ZNF263 replicate A and B, respectively. To cluster these mapped reads into peaks, we used a bin-based strategy to distribute each read into a particular bin on a particular chromosome. In other words, each human chromosome was first divided into a window-size bin from the chromosome start to end, and reads were then located into a corresponding bin interval based on their genomic coordinates. This analysis provides a landscape signal map for each sample that also generates a quantitative visualization of the enrichment of each binding site. As shown in Fig. 2A, the binding patterns of the two replicate samples are very similar. We then applied our genome-wide peak calling program, BELT (see “Experimental Procedures” and supplemental Methods), to identify the binding sites for ZNF263 in K562 cells. Briefly, our BELT program uses a percentile scoring method to determine the enrichment threshold values for each level of a set of top percentiles from the entire genome followed by identifying the number of the binding sites at each level, then uses randomly simulated reads as a background to estimate the FDR that measures the significance of each percentile. For example, 9,523 and 12,763 binding sites for ZNF263 Replicate A and B in K562 cells are detected at the Top 1% level with a FDR of less than 0.01, respectively (Table 1), whereas only 1838 and 1753 binding sites were detected at the Top 0.1% level. Binding sites that are identified in two independent experiments are more likely to be bona fide targets. Therefore, as a final step we compared the sets of targets identified in the two replicates at the top 0.1, 0.5, 1, 5, and 10% levels. We found that the sets of binding sites identified in the Top 0.5% level of replicate A and B overlapped by 76% (Fig. 2B) and provided a large number of common binding sites (5273). Therefore, this set of 5273 binding sites was used for further analyses (see below). The quantitative PCR analysis of several targets identified by ChIP-seq is shown in supplemental Fig. S2; the sequence of all primers used in this study can be found in supplemental Table S1.
TABLE 1.
A summary of binding sites of ZNF263 in K526 cells identified by ChIP-seq
Peaks (binding sites) were identified by the BELT program at the Top 0.1% with a FDR less than 0.001.
| ZNF263 | Reads | Mapped reads | Unique reads | Peaks |
||||
|---|---|---|---|---|---|---|---|---|
| Top 0.1% | Top 0.5% | Top 1% | Top 5% | Top 10% | ||||
| A | 27,069,576 | 15,052,915 | 10,322,952 | 1,838 | 6,763 | 9,523 | 30,548 | 67,008 |
| B | 25,451,310 | 15,895,922 | 11,102,465 | 1,753 | 7,070 | 12,763 | 33,155 | 81,788 |
| Overlap | 1,473 | 5,273 | 7,282 | 21,123 | 47,183 | |||
Location Analysis of ZNF263 Binding Sites
Previous studies of site-specific transcription factors have shown that some factors (e.g. E2F1) have a strong preference for binding near core promoters (18), whereas other factors (19, 20) show a more promiscuous binding pattern with respect to the start site of transcription. To determine the location preference for ZNF263 binding, we matched each binding site to the nearest gene and then divided the sites into different categories (Fig. 3, A and B). We found that two categories comprised the majority of the binding sites; these were regions spanning −2 to +2 kb relative to the start site of transcription (the promoter region) and intragenic sites (other than within 2 kb downstream of a start site). Lists of the 1473 and the 5273 binding sites identified in the Top 0.1% and Top 0.5% sets, respectively, are shown in supplemental Tables S2 and S3; information regarding the peak location with respect to the nearest gene is also provided in these tables. To determine whether the intragenic sites were scattered throughout the gene or if they localized in a more restricted manner, we performed additional analyses. First, the distance of each binding site from the start site of transcription was plotted. The sharp peak at the start site with a downstream shoulder show that a large percentage of the intragenic sites are within 10 kb of the start site (Fig. 3C). Second, the location with respect to exons and introns was determined (Fig. 3C, inset). For this analysis the sites within 2 kb downstream of the start site were included in the intragenic sites (unlike for the analysis shown in Fig. 3B). We found that 20% of all sites are within 2 kb upstream of the start of transcription, 40% are intergenic but not within 2 kb of the start site, and 40% are intragenic; of the intragenic sites, 76% are located in introns.
FIGURE 3.
Location analysis of ZNF263 binding sites. A, the location, relative to the transcription start site (5′TSS) and termination codon (3′TSS), is diagrammatically illustrated for the different categories of binding sites shown in panel B. B, the percentage of ZNF263 binding sites residing in the different location categories is shown for the peaks called at the Top 0.5% level for each replicate and for the overlap set of 5273 sites. C, the 5273 ZNF263 binding sites are plotted with respect to the distance from the nearest transcription start site. The inset indicates the percentage of sites in the 2-kb upstream core promoter regions, all other intergenic sites, and intragenic sites; the percentage of intragenic sites located in introns versus exons is also shown.
Derivation of a Binding Motif for ZNF263
We used the set of 1473 ZNF263 binding sites that we identified in common in the two ChIP-seq experiments at the top 0.1% level to derive an in vivo binding motif for ZNF263. As noted above, it was possible that ZNF263 could bind to a large motif (using most of its multiple adjacent fingers) or it could bind to several different smaller motifs (using different subsets of the nine fingers). Using a de novo motif discovery approach ChIPMotifs developed in our previous study (11), we identified a 24-nt motif. It is important to note that this motif (Fig. 4A) was identified using only the top 1473 binding sites as the training set. To determine whether this motif was specific only to the sites showing the highest enrichment or if it was present in other regions bound by ZNF263, we determined its prevalence in the entire set of 5273 sites. We found that 75% of the 5273 sites contained a good match (Core/position weight matrix 0.80/0.75) to this motif. We next examined the distribution of this motif in the two largest categories of ZNF263 binding site locations, promoters, and introns. We found that 86% of the 5′ transcription start site category and 73% of the intragenic category contained this site. Therefore, it seems that ZNF263 is recruited to the intragenic sites using the same motif as used in the core promoter regions.
Recent in vitro studies (21) have shown that approximately half of a set of 104 mouse DNA-binding proteins recognized multiple different sequence motifs. Therefore, we performed additional analyses to determine whether the ZNF263 binding sites that lacked a good match to the 24-nt consensus instead recruited ZNF263 via a different motif. The 1297 binding sites that were present in the top 5273 binding but did not contain a match to the 24-nt motif were reanalyzed as a separate subset of ZNF263 sites. A motif (GAGCAC) resembling a half-site for the androgen receptor was identified (supplemental Figs. S5 and S6) that showed high enrichment (p = 1.24e-46 for ZNF263W1) in this subset of ZNF263 binding sites as compared with a set of negative control data (∼22,000 randomly selected sequences with a length of ∼500 bp; each sequence was from human promoter regions). Of 1297 binding sites, 1196 (92%) have at least one copy of the ZNF263W1 motif. The secondary 6-nt ZNF263W1 motif could be specific to the binding sites that lacked the primary 24-nt motif or could also be enriched in the binding sites that have the primary 24-nt motif. To distinguish these possibilities, we searched the top 1473 ZNF263 binding sites (86% of which contain the 24-nt motif) for the presence of the 6-nt secondary motif. We found that 57% of these sites also contained the secondary 6-nt motif. As a test to determine whether the 6-nt motif was commonly found in other regulatory regions, we examined the top 1500 binding sites derived from genome-wide ChIP-seq data for the transcription factor AP2γ.4 We found that only 31% of the AP2γ binding sites contained the ZNF263W1 motif. Thus, the 6-nt motif is greatly enriched in ZNF263 binding sites that lack the 24-nt motif (92%) and is more enriched in the set of all top ZNF263 binding sites (57%) than in the set of top AP2γ binding sites (31%). Future studies are required to determine whether these motifs directly recruit ZNF263 or if ZNF263 is recruited via protein-protein interactions with another factor.
As indicated above, a zinc finger code has been developed that can be used to predict binding motifs for ZNFs. It would be very useful if this code could help to identify promoters that are bound by a particular ZNF. However, very few comparisons between predicted motifs and experimental motifs (derived from in vivo binding data) have been performed. Therefore, our next step was to define the predicted binding site for ZNF263 and determine whether (a) it is similar to the experimentally derived motif and (b) if it could have been used to identify ZNF263 target genes. The arrangement of the finger domains in ZNF263 suggests that finger 1, which is isolated from the other fingers (see Fig. 1), may not contribute to binding specificity. In support of this hypothesis, we used a prediction program (5) to predict motifs for ZNF263, and only fingers 2–5 and 6–9 were predicted to be involved in DNA binding. The predicted motifs for fingers 2–5 and 6–9 were merged (see supplemental Fig. S3) and compared with our experimentally derived motif in Fig. 4. Although the experimentally identified motif and the predicted motif are both GA-rich, the predicted motif differs from the experimentally derived motif in several positions (6, 18, and 21); the predicted motif is less specific, having many N residues that were not specified as a preferred nucleotide by the zinc finger code.
ChIP-seq experiments can provide very high resolution mapping of transcription factor binding sites. For example, three-quarters of all ChIP-seq peak positions for the DNA binding proteins CCCTC binding factor, neuron-restrictive silencer factor, and signal transducer and activator of transcription 1 are within 18, 27, and 51 bp, respectively, of the nearest motif for that factor (22). Such studies suggest that a motif should fall near the center of the binding site if in fact that motif specifies binding. We, therefore, examined the location of the experimentally derived and the predicted motifs in relation to the center of the ZNF263 binding regions (Fig. 5). We found that the experimentally derived motif was fairly well centered within the binding regions. There were too few examples of the entire 24-nt predicted motif for this analysis. However, it is possible that only one of the clusters of zinc fingers is involved in DNA binding. Therefore, we examined the location of the left and right halves of the predicted motif (specified by fingers 6–9 and 2–5, respectively). In general these motifs seemed to be less localized to the center of the binding region than the experimentally derived motif, suggesting that some of the predicted sites may not be related to recruitment of ZNF263 but instead are identified due to the GA content of the entire binding region (see Table 2 for 20 examples each of matches to the experimentally derived motif and to the left and right halves of the predicted motif). To more closely examine the predicted motifs near the center of the binding regions, we identified 656 and 495 regions that had a match to the left and right halves, respectively, of the predicted motif with the additional requirement that the match fell within 200 bp of the center of the binding region (see supplemental Table S7). We then used these matches to develop a position weight matrix for the left and right half motifs (predicted binding by fingers 6–9 and 2–5, respectively); see supplemental Fig. S7. We found that positions 6, 18, and 21 are highly specified. Using matches to the predicted motif, position 6 is 81% G, 16% A, and <4% C plus T, position 18 is 77% G, 16% A, and <7% C plus T, and position 21 is 67% G, 23% A, and <9% C plus T. These results are similar to the nucleotide distribution at the matches to the experimental motif; position 6 is 81% G, 13% A, and <7% C plus T, position 18 is 81% G, 16% A, and 47% C plus T, and position 21 is 78% G, 17% A, and <5% C plus T. Thus, a C or T in these three positions seems to be incompatible with binding.
FIGURE 5.
Location of experimental and predicted motifs in the ZNF263 ChIP-seq peaks. The locations of the 24-nt experimentally derived motif (Fig. 4A) and the left half (specified by fingers 6–9) and the right half (specified by fingers 2–5) of the predicted motif (Fig. 4B) were analyzed in the set of 5273 top ranked ZNF263 binding sites. The distance of each motif relative to the center of the binding region is shown.
TABLE 2.
Examples of matches to the predicted and experimentally derived ZNF263 motifs
| Position̂a | Left halfb | Position̂ | Right halfc | Position̂ | De novod |
|---|---|---|---|---|---|
| −4 | AGGGAGGAAGGA | −4 | GGGGAAGAGGGA | −3 | GAAGAGGAGGAGGAGGGGGAGGAG |
| −4 | GGGGAAGAGGGA | −4 | GGGGATGAGGGA | −3 | GAGGAGGAAAGGGAGAGGGAAAAG |
| −3 | GAGGAGGAGGGA | −4 | AGGGAGGAAGGA | −3 | AAGAAGGAGGAGAGGGAGAAAGAG |
| −3 | AGGGAGGAGGGA | −4 | GGGGAAGAGGGA | −3 | GAGAAGAAAGAGAAAAGAGAGGAG |
| −2 | GGGGAGGAGGGA | −3 | AGGGAGGAGGGA | −2 | AGAGAGGGGGGGGAGGAGGGGGAG |
| −1 | GAGGAGGAGGGA | −3 | AGGGAGAAGGGA | −2 | AAGGGAAAAGGGAAAGAAAAGGAG |
| −1 | AGGGAGGAGGGA | −2 | GGGGAGGAGGGA | −1 | AGGGAGGAGGAGAGGGAGGAGGAG |
| −1 | AGGGAGGAGGGA | −1 | AGGGAGGAGGGA | −1 | GGGGAGGAAGGAGGAAGGGAGGAG |
| 0 | GAGGAGGAAGGA | −1 | AGGGAGGAGGGA | −1 | GGAGAGGGAAGAGAGGAGGGAGAG |
| 0 | CAGGAGGAAGGA | 0 | CGGGAGAAGGGA | 0 | GGAGAGAGGGGGAAAGGGGAGGAA |
| 1 | AGGGAAGAAGGA | 1 | AGGGAAGAAGGA | 0 | GAGGAGGAGGGAGGGAAGAGGGAG |
| 1 | GAGGAGGAGGGA | 1 | AGGGAGGAGGGA | 0 | AGGGAGGGGAAAAGAGGGAAGAGG |
| 2 | CGGGATGACGGA | 2 | CGGGATGACGGA | 1 | GAAGAGGAGGAGGAAGGGGAGGAG |
| 2 | GAGGACGATGGA | 2 | AGGGAAAAGGGA | 1 | GGAGAGGGGGAGGGAGGAGAGGGG |
| 2 | GCGGAGGACGGA | 3 | GGGGAACAGGGA | 1 | GGAGAGGGAGGGAGGGAGGAGGGA |
| 3 | GTGGAGGAGGGA | 3 | GGGGAGTAGGGA | 1 | AGGGAGAGGAGGGAGGAGGAGGAG |
| 4 | GAGGAGGATGGA | 3 | AGGGACAATGGA | 1 | GAGGAGGAGGAGGAAGAGGAGGAG |
| 4 | GAGGAGGAGGGA | 3 | GGGGAGAAGGGA | 1 | GAGAGGAAAGAGGGAGGAAAGGAA |
| 4 | GAGGAGGAGGGA | 5 | GGGGAGGAGGGA | 1 | GGAGGAAGAGGGGAGGAGAGGGAG |
| 5 | TAGGAGGAGGGA | 6 | GGGGAGGAGGGA | 2 | GGAGAGGGAAGGAGGGAAGAGGGA |
a Relative to the center of the peak.
b Matches to left half of predicted motif.
c Matches to right half of predicted motif.
d Matches to the 24-nt de novo ChIPs motif.
Having defined both a predicted and an experimental motif for ZNF263, we could now compare the prevalence of these two motifs in the set of promoters bound by ZNF263 versus in the set of all human core promoters (Table 3). We found that 29% of all human promoters contain a good match to the 24-nt experimentally defined ZNF263 motif, whereas 86% of the promoters bound by ZNF263 contain this motif. Clearly, the experimentally derived motif is significantly more enriched in target promoters than in the set of all promoters. To determine whether the motif predicted to be recognized by ZNF263 by the zinc finger code would have allowed a bioinformatically based identification of ZNF263 target promoters, we examined the prevalence of the predicted motif in the set of all human core promoters versus in the ZNF263 target promoters. We found that a very low percentage of promoters bound by ZNF263 contained the motif predicted for fingers 2–9. However, it is important to point out that the analysis of the 24-nt experimental motif was performed using a position weight matrix, but the analysis of the 24-nt predicted motif was performed using a single motif having both specified and unspecified positions (see the Table 3 and Fig. 4 legends). We also analyzed the promoters bound by ZNF263 for a match to the two half-sites. We found that the two 12-nt motifs were present in the set of promoters bound by ZNF263 at a lower percentage than in the set of all promoters. Thus, neither the 24-nt site nor the 12-nt half-sites predicted by the zinc finger code could predict ZNF263 recruitment to target promoters. However, we note that fingers 4 and 3 are predicted to bind GANNAN (see supplemental Fig. S3), which resembles the ZNF263W1 motif. Therefore, it is possible that these two fingers mediate some of the binding specificity of ZNF263.
TABLE 3.
Summary of predictive abilities of experimental versus predicted motif
| Type of motif | ZNF263 target promoters (1496)a | All promoters (24,872)b |
|---|---|---|
| ChIPMotifs_Identifiedc | 1290 (86%) | 7286 (29%) |
| ZIFIBI_predicted: for fingers 2–9d | 3 (0.2%) | 8 (0.03%) |
| ZIFIBI_predicted: for fingers 2–5e | 336 (22%) | 7681 (31%) |
| ZIFIBI_predicted: for fingers 6–9f | 441 (29%) | 8324 (34%) |
a1496 of the 5273 ZNF263 binding sites fall within a core promoter, as defined as the region +/− 2 kb around a transcription start site.
b All (24,872) human core promoters from the USCS HG18 RefSeq dataset were analyzed using a length of 4 kb for each promoter (from −2 to +2 kb around the transcription start site).
c The PWM derived from the experimentally identified ZNF263 sites was used, with the criteria of Core/PWM 0.80/0.75.
d The sequence NNGGANGANGGANGGGANNANGGA was used as the motif bound by fingers 2–9; supplemental Fig. 5.
e The sequence NGGGANNANGGA was used as the motif bound by fingers 2–5.
f The sequence NNGGANGANGGA was used as the motif bound by fingers 6–9.
As noted above, there have been no studies of the human ZNF263 binding specificity. However, one previous study did analyze the mouse homolog of ZNF263, called Zfp263 ((also called NT2 (23)). This protein was shown to bind in vitro to a 24-bp sequence in the Col11a2 promoter. Interestingly, the core binding sequence identified via gel shift competition experiments for the Col11a2 promoter was also GA-rich (GAGGAGGGAG). Sequence comparison of the human and mouse ZNF263 homologs show an overall amino acid identity of ∼85%, with a 97.4% amino acid identity in the zinc finger motifs. This high degree of amino acid conservation and the similarity of the Col11a2 binding site to the experimentally derived site suggest that human ZNF263 might bind to the human Col11a2 promoter. However, there is no signal detected in the ChIP-seq data near the Col11a2 promoter. To more specifically examine the binding to Col11a2, we made primers to the promoter region and performed ChIP-PCR. We did observe very low levels of binding of ZNF263 to the Col11a promoter in K562 cells (supplemental Fig. S4), suggesting that the low levels of binding were simply not detected in the ChIP-seq library.
Is ZNF263 a Transcriptional Repressor?
As described above, approximately one-third of the set of C2H2 ZNFs also contain a KRAB domain. KRAB domains have been shown to cause transcriptional repression when artificially recruited to DNA through a GAL4 DNA binding domain. The mechanism of repression is postulated to be due to the KRAB domain recruiting TRIM28 (KAP1), which in turn recruits the histone methyltransferase SETDB1. SETDB1 can mediate trimethylation of lysine 9 of histone H3, which then results in the formation of repressed chromatin (24). Most of the previous repression studies of KRAB domains have been performed by artificially tethering an isolated KRAB domain (or TRIM28 (KAP1) itself) to the chromatin using transiently introduced or stably integrated artificial reporter constructs (25). However, several studies have used cotransfection of a KRAB-ZNF with a reporter construct to study repression. In particular, cotransfection of mouse Zpf263 (NT2) with the Col11a2 promoter into RCS cells revealed that Zpf263 (NT2) could repress transcription but only if the KRAB domain was present in the protein construct; the SCAN domain was not required for this repressive activity (23). Taken together, the previous work suggests that perhaps human ZFN263 is a repressor that functions via recruitment of TRIM28 (KAP1) and SETDB1, resulting in the formation of heterochromatin.
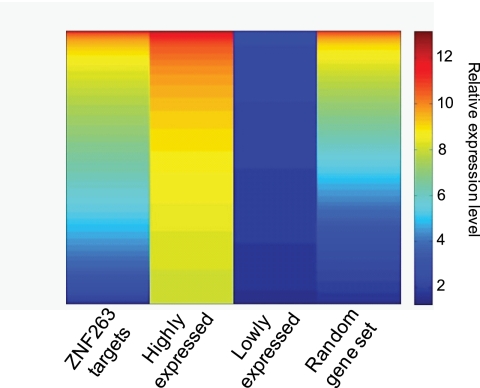
As a first step to determine whether ZNF263 functions as a repressor, we examined the expression level of ZNF263 target genes (supplemental Table S4). We found that most ZNF263 target genes were modestly expressed, with the set of targets having a similar overall expression profile as a randomly chosen set of genes (Fig. 6). It was possible that ZNF263 was more likely to function as a repressor for the set of 1496 genes that have ZNF263 bound in the promoter region. Therefore, we examined the expression of only that subset of ZFN263 target genes. We found that the range of expression levels of ZNF263 promoter-localized target genes was very similar to the range of expression levels of the entire set of ZNF263 target genes (data not shown). Thus, restricting the analysis to genes having a bound ZNF263 near the promoter did not provide stronger evidence in support of the repression model. However, it remains possible that ZNF263 could serve as a repressor for a subset (∼15–20%) of its target genes.
FIGURE 6.
Heatmap of expression data for ZNF263 targets. The expression levels of the subset of ZNF263 target genes (identified as the nearest gene to each binding site in the 5273 set of peaks) present on the Affymetrix Human Exon 1.0 ST (HuEx-1_0-st-v2) arrays is shown compared with the expression levels of the same number of genes that are expressed at the highest and lowest levels in K562 cells. The expression profile of a set of randomly chosen genes is also shown.
As a next step in the analysis of ZNF263 target genes, we analyzed RNA expression levels before and after reduction of ZNF263 by siRNA treatment using Illumina expression arrays. We tried several times to knock down ZNF263 in K562 cells, but regardless of which method we used the knockdowns were never very efficient. However, we could obtain robust knockdowns of ZNF263 in HeLa cells. As shown in Fig. 7, ZNF263 RNA (Fig. 7A) and protein (Fig. 7B) levels were greatly reduced upon siRNA treatment of HeLa cells. Upon reduction of ZNF263 levels, we identified 195 genes whose expression was increased, 61 of which had also been identified as ZNF263 target genes by ChIP-seq in K562 cells, and 118 genes whose expression was decreased, 37 of which had also been identified as ZNF263 target genes by ChIP-seq in K562 cells (supplemental Table S5). To confirm the array results, we tested by quantitative real-time PCR several genes identified as up-regulated in the absence of ZNF263 (i.e. they are normally repressed by ZNF263) and several genes identified as down-regulated in the absence of ZNF263 (i.e. they are normally activated by ZNF263). As shown in Fig. 7, C and D, the expression array results for three up-regulated and three down-regulated genes were confirmed by quantitative real-time PCR analysis. We note that a simple comparison of the list of K562 ChIP-seq targets and the genes deregulated in HeLa cells upon loss of ZNF263 will not necessarily identify the set of genes bound by and regulated by ZNF263 because different cell lines were used for the two experimental approaches. As indicated above, we could not achieve knockdown of ZNF263 in K562 cells, and multiple attempts to perform ZNF263 ChIP-seq in HeLa cells resulted in very high background. Therefore, to confirm that we had identified genes that were both bound by and regulated by ZNF263, we performed ChIP assays in HeLa cells and tested binding using PCR at several genes that were responsive to loss of ZNF263 in HeLa cells. Binding of ZNF263 in HeLa cells to six deregulated genes was confirmed by PCR analysis of ChIP samples (Fig. 7E). Thus, binding of ZNF263 to a regulatory region of a target gene can either positively or negatively influence transcriptional regulation, depending on the target gene. To gain insight into the types of genes deregulated upon loss of ZNF263, we performed a gene ontology analysis (supplemental Table S6) using DAVID (david.abcc.ncifcrf.gov/). We found that one of the largest categories of genes whose expression decreased upon loss of ZNF263 (i.e. ZNF263 normally has a positive effect on their transcription) was “cellular component organization and biogenesis”; many of these genes are involved in cytoskeletal formation. In contrast, the largest categories of genes whose expression increased upon loss of ZNF263 (i.e. ZNF263 normally has a repressive effect on their transcription) were “negative regulation of biological process and negative regulation of cellular process.” Thus, ZNF263 may play a critical role in maintaining cell structure (by up-regulating components of the cytoskeleton) and proliferation (by down-regulating negative regulators of proliferation).
FIGURE 7.
ZNF263 can have both positive and negative effects on transcription. A–C, HeLa cells were transfected with control siRNA (siCONTROL) or with siRNAs against ZNF263 (siZNF263). After 72 h cells were harvested, and total RNA or protein extracts were prepared. A, reverse transcription-quantitative real-time PCR was performed for detection of transcript levels of ZNF263. Results were normalized using glyceraldehyde-3-phosphate dehydrogenase mRNA as a reference and reported as a percentage of the transcript levels in control siRNA-treated cells. B, protein levels of ZNF263 and actin were analyzed by Western blot from 50 μg of protein extract. C and D, transcript levels of EBI3, GPER, KIAA1324, FOXA1, ITPKA, and RGS10 in cells treated with siZNF263 are expressed as either an increase (C) or decrease (D) relative to transcript levels in control siRNA cells. The values shown (with S.D.) are the averages from three different siRNA knockdown experiments (all of which are different from the preparations of RNA used for the arrays); RNA levels were normalized using glyceraldehyde-3-phosphate dehydrogenase. E, chromatin immunoprecipitation was performed with antibodies against ZNF263 and used in quantitative real-time PCR assays. ChIP samples were analyzed by real-time PCR using primers specific for the binding sites within the ZNF263 target genes and compared with total (input) DNA; three negative control regions in which ZNF263 does not bind (CCNA1, ZNF333, glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) were also analyzed. Data were normalized using a negative control region CDH10; shown are the mean of three independent replicates with S.D.
DISCUSSION
There are hundreds of ZNF proteins encoded in the human genome whose DNA binding specificity has not been studied. The zinc finger family has arisen through duplication and divergence, producing subsets of highly related factors that are predicted to bind to distinct sets of target genes. Expanding the collection of ChIP-seq experiments of this set of factors would provide a rich source of transcriptional regulatory and DNA-protein interaction data. However, it is very helpful to have a positive control binding site to ensure that the antibody for the factor being studied is working well in the ChIP experiment. Using a motif predicted from the zinc finger code to identify a set of putative target promoters to test in ChIP assays could possibly provide the needed control. To determine whether this approach is valid, we used the zinc finger code to develop a predicted motif for ZNF263 and then determined the percentage of all human promoters and the percentage of promoters experimentally determined to be bound by ZNF263 that contain a good match to the predicted motif. Unfortunately, we found that the motif predicted for ZNF263 by the zinc finger code is not useful for identifying an in vivo ZNF263 binding site. For example, less than 1% of the promoters bound by ZNF263 contain a match to the 24-nt site predicted if both clusters of 4 fingers recognize DNA. Also, the two “half-sites” predicted for fingers 2–5 and fingers 6–9 are found at a lower frequency in the set of promoters bound by ZNF263 than in the full set of all human promoters. In contrast, 86% of the promoters bound by ZNF263 contain a good match to the 24-nt experimentally derived motif (which was identified using only the top 1473 binding sites, not the set of all ZNF263 target promoters). Thus, we suggest that the zinc finger code is not sufficient for determining the in vivo binding pattern of a zinc finger protein. To our knowledge, this is the first time in which a predicted motif for a particular ZNF has been compared with an experimentally derived motif derived from a genome-wide ChIP-seq analysis. These results taken along with our finding that only 25% of all ZNF263 binding sites fall within core promoter regions suggest that an unbiased, comprehensive, genome-wide experimental analysis using ChIP-seq is required to identify the complete set of binding sites for a particular ZNF. However, we conclude that the inability to use the zinc finger code to predict target sites is due to the lack of specificity (characterized by N residues in positions 6, 18, and 21) of the predicted motif, not to an “inaccuracy” of the prediction. Perhaps our in vivo binding data for ZNF263 will allow the zinc finger code to be modified such that more specificity can be incorporated into future predictions of other ZNF binding motifs.
In summary, we have performed ChIP-seq for ZNF263, a C2H2 ZNF that contains nine finger domains and a KRAB domain. Before this work no in vivo binding sites for ZNF263 had been identified, no target genes were known, and it was not known if this transcription factor functioned as an activator or a repressor. Our studies have provided a resource of >5000 binding sites for this zinc finger factor, have shown that ZNF263 binding sites are primarily located near start sites or within introns, and have identified a 24-nt motif that is highly enriched in both the promoter and intron subsets of binding sites. We found that in general ZNF263 target genes are expressed over a large range and that reduction of ZNF263 can lead to an increase in one set of targets and a decrease in another. Thus, our studies suggest that this KRAB-ZNF can have both positive and negative effects on transcription.
Supplementary Material
Acknowledgments
We thank the members of the Farnham laboratory for helpful discussions and Sushma Iyengar for assistance with the preliminary ZNF263 experiments. We also thank the ENCODE Consortium, in particular the groups headed by Mike Snyder (Yale), Brad Bernstein (Broad), Greg Crawford (Duke), John Stamatoyannopoulos (University of Washington), Rick Myers (Stanford), Scott Tenenbaum (SUNY Albany), and Tom Gingeras (Cold Spring Harbor Laboratory) for allowing the use of the unpublished K562 RNA expression data.
This work was supported, in whole or in part, by National Institutes of Health Grants CA45250 and 1U54HG004558 (United States Public Health Service).
The ChIP-seq data and the RNA expression array data of this protein can be accessed through the NCBI Gene Expression Omnibus under NCBI accession numbers GSE19235 and GSE19146.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods,” Figs. S1–S7, and Tables S1–S7.
A. R. Cao and P. J. Farnham, unpublished data.
- ZNF
- zinc finger factor
- siRNA
- small interfering RNA
- ChIP
- chromatin immunoprecipitation
- BELT
- bin-based enrichment threshold level
- FDR
- false discovery rate
- nt
- nucleotide(s)
- EGR
- early growth response.
REFERENCES
- 1.Tupler R., Perini G., Green M. R. (2001) Nature 409, 832–833 [DOI] [PubMed] [Google Scholar]
- 2.Kadonaga J. T., Tjian R. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 5889–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iuchi S. (2001) Cell. Mol. Life Sci. 58, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J., Stormo G. D. (2008) Bioinformatics 24, 1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho S. Y., Chung M., Park M., Park S., Lee Y. S. (2008) BBRC 369, 845–848 [DOI] [PubMed] [Google Scholar]
- 6.Segal D. J., Barbas C. F., 3rd (2000) Curr. Opin. Chem. Biol. 4, 34–39 [DOI] [PubMed] [Google Scholar]
- 7.Yokoyama M., Nakamura M., Okubo K., Matsubara K., Nishi Y., Matsumoto T., Fukushima A. (1997) BBA 1353, 13–17 [DOI] [PubMed] [Google Scholar]
- 8.Williams A. J., Blacklow S. C., Collins T. (1999) Mol. Cell. Biol. 19, 8526–8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huntley S., Baggott D. M., Hamilton A. T., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L. (2006) Genome Res. 16, 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., Thiessen N., Griffith O. L., He A., Marra M., Snyder M., Jones S. (2007) Nat. Methods 4, 1–2 [DOI] [PubMed] [Google Scholar]
- 11.Jin V. X., O'Geen H., Iyengar S., Green R., Farnham P. J. (2007) Genome Res. 17, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavesi G., Mereghetti P., Mauri G., Pesole G. (2004) Nucleic Acids Res. 32, W199–W203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hon L. S., Jain A. N. (2006) Bioinformatics 22, 1047–1054 [DOI] [PubMed] [Google Scholar]
- 14.Bailey T. L., Gribskov M. (1997) J. Comput. Biol. 4, 45–59 [DOI] [PubMed] [Google Scholar]
- 15.Ho-Sui S. J., Mortimer J. R., Arenillas D. J., Brumm J., Walsh C. J., Kennedy B. P., Wasserman W. W. (2005) Nucleic Acids Res. 33, 3154–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Prüss M., Reuter I., Schacherer F. (2000) Nucleic Acids Res. 28, 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahony S., Benos P. V. (2007) Nucleic Acids Res. 35, W253–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bieda M., Xu X., Singer M. A., Green R., Farnham P. J. (2006) Genome Res. 16, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 20.Cawley S., Bekiranov S., Ng H. H., Kapranov P., Sekinger E. A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A. J., Wheeler R., Wong B., Drenkow J., Yamanaka M., Patel S., Brubaker S., Tammana H., Helt G., Struhl K., Gingeras T. R. (2004) Cell 116, 499–509 [DOI] [PubMed] [Google Scholar]
- 21.Badis G., Berger M. F., Philippakis A. A., Talukder S., Gehrke A. R., Jaeger S. A., Chan E. T., Metzler G., Vedenko A., Chen X., Kuznetsov H., Wang C. F., Coburn D., Newburger D. E., Morris Q., Hughes T. R., Bulyk M. L. (2009) Science 324, 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jothi R., Cuddapah S., Barski A., Cui K., Zhao K. (2008) Nucleic Acids Res. 36, 5221–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka K., Tsumaki N., Kozak C. A., Matsumoto Y., Nakatani F., Iwamoto Y., Yamada Y. (2002) Mol. Cell. Biol. 22, 4256–4267 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 25.Sripathy S. P., Stevens J., Schultz D. C. (2006) Mol. Cell. Biol. 26, 8623–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.