Abstract
DNA double strand break (DSB) repair by non-homologous end joining (NHEJ) is initiated by DSB detection by Ku70/80 (Ku) and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) recruitment, which promotes pathway progression through poorly defined mechanisms. Here, Ku and DNA-PKcs solution structures alone and in complex with DNA, defined by x-ray scattering, reveal major structural reorganizations that choreograph NHEJ initiation. The Ku80 C-terminal region forms a flexible arm that extends from the DNA-binding core to recruit and retain DNA-PKcs at DSBs. Furthermore, Ku- and DNA-promoted assembly of a DNA-PKcs dimer facilitates trans-autophosphorylation at the DSB. The resulting site-specific autophosphorylation induces a large conformational change that opens DNA-PKcs and promotes its release from DNA ends. These results show how protein and DNA interactions initiate large Ku and DNA-PKcs rearrangements to control DNA-PK biological functions as a macromolecular machine orchestrating assembly and disassembly of the initial NHEJ complex on DNA.
Introduction
DNA double strand breaks (DSBs)4 are among the most cytotoxic forms of DNA damage (1). In non-replicating mammalian cells, the major pathway for repair of DSBs, such as those induced by ionizing radiation, is non-homologous end joining (NHEJ) (2–4). Initial committed steps in NHEJ are DSB detection by the Ku70/80 heterodimer, followed by recruitment of the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) to form the active DNA-dependent protein kinase holoenzyme (DNA-PK) (reviewed in Refs. 5–7). DNA-PK complex assembly at the DSB results in trans-autophosphorylation of two DNA-PKcs molecules across the DSB (8, 9), which, in vitro, leads to disruption of the DNA-PK complex (10) (reviewed in Ref. 5). Multiple DNA-PKcs autophosphorylation sites have been identified, including serine 2056 (11, 12), a cluster of sites between residues 2609–2647 (referred to as the ABCDE or Thr-2609 cluster) (13, 14), and threonine 3950 (15). Although DNA-PKcs in which the ABCDE sites have been mutated to alanine has normal protein kinase activity, its ability to dissociate from the Ku·DNA complex is reduced, suggesting that phosphorylation of the ABCDE sites plays a major role in regulating disassembly of the initial DNA-PK complex (15, 16). DNA-PKcs is phosphorylated at multiple sites in vivo in response to DNA damage, including serine 2056, the ABCDE cluster, and threonine 3950 (9, 12, 14, 15); cells expressing DNA-PKcs that is unable to undergo autophosphorylation at the ABCDE/Thr-2609 cluster display extreme radiation sensitivity and multiple DSB repair defects (14, 17). Moreover, autophosphorylation-defective DNA-PKcs in which the ABCDE cluster plus serine 2056 have been mutated to alanine as well as kinase-dead DNA-PKcs are retained longer at in vivo sites of DNA damage than is wild type DNA-PKcs (18). Together, these data support a model in which the Ku heterodimer first binds the DSB, followed by recruitment of DNA-PKcs which leads to stimulation of DNA-PKcs protein kinase activity, autophosphorylation, and dissociation of DNA-PKcs. Assembly and disassembly of the initial NHEJ complex thus regulates accessibility of the DSB to other repair factors as well as pathway progression (reviewed in Refs. 5–7), but the structural and mechanistic regulation of this critical process has been poorly understood.
Here, we determined the overall structures and structural rearrangements of DNA-PKcs and Ku with DNA in solution under conditions mimicking DSBs and DNA-PKcs autophosphorylation. Understanding the molecular basis of how DNA-PKcs, Ku, and DNA interact and how post-translational modifications control NHEJ presents a significant challenge. Although such dynamic events are not amenable to x-ray crystallography, they can be accurately defined by solution methods, such as small angle x-ray scattering (SAXS) (19). Here, we combined SAXS with live cell imaging and biochemical approaches to characterize the regulation of DNA-PK complex assembly and disassembly. These results reveal a dramatically opened conformation of trans-autophosphorylated DNA-PKcs. Our combined in vitro and in vivo results reveal that structural plasticity involving the flexible Ku80 C-terminal domain, DNA-PKcs dimers, and phosphorylation-induced conformational changes regulate the interaction of DNA-PKcs with its partners Ku and DNA to regulate the initial stages of NHEJ.
EXPERIMENTAL PROCEDURES
Data Collection and Evaluation
SAXS data were collected at the ALS beamline 12.3.1 (Lawrence Berkeley National Laboratory, Berkeley, CA) (20). Tunable wavelength λ 1.0–1.5 Å and the sample-to-detector distances were set to 1.5 m, resulting in scattering vectors, q, ranging from 0.007 to 0.31 Å−1. The scattering vector is defined as q = 4π sinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20 °C, and data were processed as described (20). Briefly, the data acquired at short and long time exposure (6 and 60 s) were merged for calculations using the entire scattering spectrum. The experimental SAXS data for different protein concentrations were investigated for aggregation using Guinier plots (21). The radius of gyration (RG) was derived by the Guinier approximation I(q) = I(0) exp(−q2RG2/3) with the limits qRG < 1.6. The program GNOM (22) was used to compute the pair distance distribution functions, P(r). This approach also provided the maximum dimension of the macromolecule, Dmax. The overall shapes were restored from the experimental data using the program DAMMIN (23) or GASBOR (24), respectively. In our rigid body modeling strategy, BILBOMD, molecular dynamics (MD) simulations were used to explore conformational space adopted by the Ku80 C-terminal region (Ku80CTR) and the SAP (SAF-A/B, Acinus, and PIAS) domain. A minimal ensemble search (MES) was used to identify the minimal ensemble required to best fit the experimental data (25). For more details about data evaluation, see the supplemental material.
Other Methods
Experimental procedures for protein purification, cell culture and transfection, live cell imaging, biotin pull-down assays, and limited proteolysis assays are provided in the supplemental material.
RESULTS
Ku80 C-terminal Region Forms a Flexible Arm for DNA-PKcs Recruitment
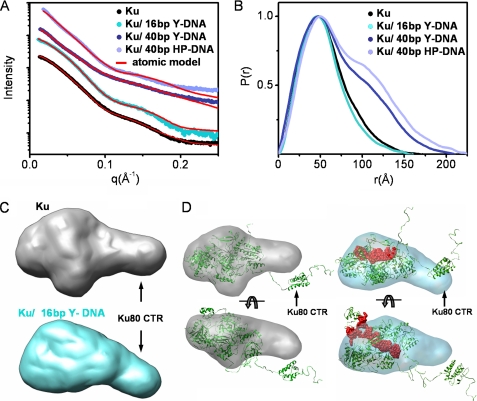
To define the dynamic nature of the Ku heterodimer alone and in combination with DNA, we employed SAXS (19), which allows analyses in solution, in combination with information from the crystallography (26–28) and biochemistry of the protein and DNA components (29, 30). The SAXS profile and linear radius of gyration (RG) of the free Ku heterodimer over a concentration range of 1–10 mg/ml indicated that the protein is well behaved over a wide concentration range and adopts an aggregation-free state in solution (Fig. 1A and supplemental Fig. S1A). The maximal dimension (Dmax) of the heterodimer was 165 Å (Fig. 1B and Table 1). To better define the geometric shape of the Ku heterodimer, we calculated the SAXS envelopes. Calculation of independent models gave confidence in the consistency of the SAXS architectures because objective algorithms showed small variations between independent runs (supplemental Fig. S1C). Together, the data reveal that Ku displays a globular structure with an elongated arm on one side of the molecule (Fig. 1C, top, gray). The SAXS envelope differs from the three-dimensional reconstruction obtained by EM (31), particularly with respect to the presence of the extended region. Based on the manual superimposition of the Ku crystal structure on the SAXS envelopes, we conclude that the extended region observed in the SAXS envelopes corresponds to the Ku80CTR, which was not present in the original crystal structure of the core DNA binding domain.
FIGURE 1.
Solution structures of Ku and Ku-DNA complexes. A, experimental scattering profiles of Ku alone (black), Ku with 16-bp Y-DNA (cyan), 40-bp Y-DNA (dark blue), or 40-bp HP-DNA (blue) (see also supplemental Fig. S1). The theoretical scattering from the final MES models for Ku (χ2 = 2.8) and multicomponent model for Ku·DNA assemblies (χ2 = 3.0 for Ku·16-bp Y-DNA; χ2 = 4.5 for Ku·40-bp Y-DNA; χ2 = 7.0 for Ku·40-bp HP-DNA) are shown by the red lines. B, distance distribution functions P(r) of the Ku assemblies computed from experimental SAXS data shown in the same colors as in A. The P(r) functions are normalized to unity at their maxima. C, the average SAXS envelopes of the Ku assemblies calculated with DAMMIN, as displayed in volumetric representation and colored as in A. D, the best fit atomic model of Ku (shown in Fig. 2B) and Ku/16-bp Y-DNA (Ku (green) and DNA (red)) were superimposed with the average SAXS envelopes, displayed in a transparent, volumetric representation.
TABLE 1.
Global SAXS parameters
| Assembly | RGa | Dmaxb | Molecular mass (SAXS)c |
|---|---|---|---|
| Å | Å | kDa | |
| Ku | 42.0 ± 0.1 | ∼165 | ∼140 |
| Ku·16-bp Y-DNA | 40.6 ± 0.1 | ∼155 | ∼130 |
| Ku·40-bp Y-DNA | 51.9 ± 0.2 | ∼210 | ∼220 |
| Ku·40-bp HP-DNA | 60.0 ± 0.4 | ∼220 | ∼240 |
| DNA-PKcs | 55.0 ± 0.1 | ∼155 | ∼405 |
| DNA-PKcs (dimer) | 91.6 ± 0.6 | ∼300 | ∼800 |
| DNA-PKcs·40-bp Y-DNA | 91.1 ± 0.7 | ∼330 | ∼900 |
| DNA-PKcs·40-bp Y-DNA·Ku | 105 ± 1.0 | ∼330 | ∼1000 |
| DNA-PKcs·40-bp HP-DNA | 85.0 ± 1.0 | ∼300 | ∼800 |
| DNA-PKcs/40bp HP-DNA/Ku | 85.0 ± 1.0 | ∼300 | ∼900 |
| Phospho-DNA-PKcs | 58.5 ± 0.2 | ∼180 | ∼425 |
| Phospho-DNA-PKcs (dimer) | 90.4 ± 1.4 | ∼300 | ∼900 |
The Ku80CTR high resolution structure shows that the globular domain most probably interacts with the core via a disordered, flexible linker (27). The C-terminal SAP domain of Ku70 was visualized together with the original crystal structure of the core DNA binding domain (26, 32); however, the electron density of the linker region could not be defined, presumably due to disorder (32). To examine these extended regions and the flexible character of Ku, we used rigid body modeling by BILBOMD (25). In general, rigid body modeling involves preparing many possible atomic models and comparing predicted models to experimental data. These models, which can either be directly refined against the experimental data (33) or independently, provide a set from which the best model structure for the experimental data is selected (25, 34, 35). MD simulations on the Ku80CTR and the Ku70 SAP domains were performed at high temperature, where the additional kinetic energy prevents the molecule from becoming trapped in a local minimum. The MD simulations provided an ensemble of Ku models from which SAXS curves were calculated and compared with the experimental curve (Fig. 2A). From the entire pool of modeled conformers, the best fit structure (χ2 = 4.0) shows that the Ku80CTR domain exists a distance from the core DNA binding domain (Fig. 2B), which is consistent with the extension observed in the SAXS envelope (Fig. 1D). Attempting to reconstruct a single best model for flexible proteins can be misleading and at best provides an average of the conformations (19). In the minimal ensemble search (MES) applied in this study, a genetic algorithm identified the minimal ensemble required to best fit the experimental data (25, 36). MES-selected models represent probable components of the population of many conformations that may better describe flexible proteins than a single model directly refined against experimental data (37–39). Considering the conformational disorder of the C-terminal domains, the coexistence of different protein conformations in solution that could contribute to the experimental scattering needed to be taken into account. Assembly of five selected conformers improved the fit to the experimental data (χ2 = 2.8) and supports large conformational variability of the Ku80CTR domain (Fig. 2, A and C). The extreme C terminus of the Ku80CTR contains a conserved region that is important for interaction with DNA-PKcs (29, 30); thus, we propose that the observed flexibility of the Ku80CTR is probably important for recruitment and targeting of DNA-PKcs to DSBs.
FIGURE 2.
Atomic models of Ku. A, graph showing a comparison of RG values for all 10,000 models obtained in MD conformational sampling with their maximal dimensions. The values for the best fit model (red dots) and MES ensemble (cyan) with five conformers are indicated. The distribution of RG values (40–54 Å) for MES conformers indicates flexibility of the Ku80CTR. The MES fit to the experimental data is shown in Fig. 1A. B, the best fit model of Ku (χ2 = 4.0) for Ku70 (blue) and Ku80 (cyan) shown schematically. C, five MES-selected conformers (χ2 = 2.8), representing the probable conformational space adopted by the Ku80CTR domain. The positions of the Ku80CTR shown schematically are highlighted (red stars). MES conformers are superimposed on the Ku crystal structure (Protein Data Bank code 1jeq) shown in a surface representation.
To examine the flexibility of the Ku heterodimer experimentally, we compared the structure of the Ku heterodimer in the free state to that of complexes composed of Ku and DNA. We used either 16-bp double-stranded DNA (dsDNA) with a Y-shaped structure at one end (16-bp Y-DNA), similar to that utilized in the crystal structure of the DNA binding core of Ku (26), a 40-bp duplex with a Y-shaped structure at one end (40-bp Y-DNA) similar to that used for structural studies on the DNA-PKcs·Ku complex (40), or a 40-bp duplex with a hairpin at one end (40-bp HP-DNA) (supplemental Fig. S2). Ku bound to the 16-bp Y-DNA adopts a similar elongated conformation to that described for Ku alone, with a maximal dimension of 155 Å (Table 1). However, narrowing of the pair distribution function P(r) (Fig. 1B) supports DNA binding within the Ku ring. Interaction with the 16-bp Y-DNA did not induce a major conformational change, and the extended conformation of the Ku80CTR persisted in the DNA-bound state (Fig. 1, C and D), consistent with the structure of truncated Ku in crystals (26). Rigid body modeling yielded representative Ku/16-bp Y-DNA conformations showing an elongated arrangement of the Ku80CTR that matched well with the experimental data (χ2 = 4.5). Due to the possible presence of free Ku, we matched the experimental data to theoretical scattering profiles of uncomplexed and complexed Ku and found that allowing for ∼50% free Ku in the sample improved the fit to the data (χ2 = 3.0) (Fig. 1A). For additional data analysis, see supplemental material.
To further examine Ku interactions with DNA, we analyzed Ku assemblies with longer DNA molecules (40-bp Y-DNA and 40-bp HP-DNA). The results show that two Ku molecules can be accommodated on the longer DNA. When an excess of 40-bp DNA was present, a shoulder in the P(r) function was observed (Fig. 1B), suggesting the formation of Ku dimers. P(r) functions at different molar ratios show similar shapes, indicating that the complex is in equilibrium with free DNA. These data explain electrophoretic mobility shift assay observations that show two bands implying 1 or 2 Ku proteins per DNA molecule (supplemental Fig. S1B) and indicate that the association of Ku with DNA is dynamic.
The Dynamic Nature of DNA-PKcs Alone and in Complex with DNA
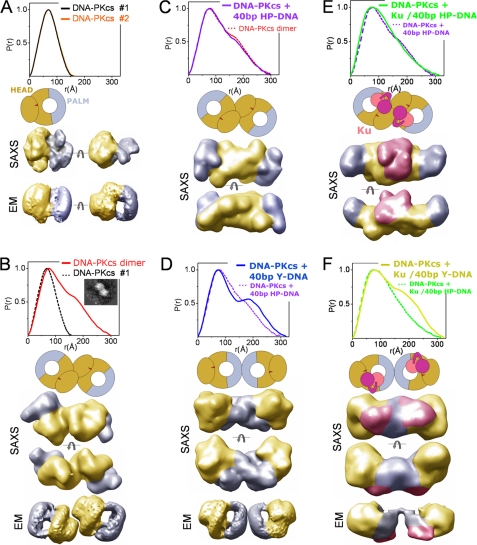
DNA-PKcs is a protein of over 4100 amino acids that is composed of a large N-terminal helical region and a C-terminal kinase domain (reviewed in Ref. 5). Cryo-electron EM structures reveal that DNA-PKcs consists of a globular “head domain” (Fig. 3A, yellow) connected to a “palm” region (Fig. 3A, blue) by a flexible arm (41, 42). To examine the DNA-PKcs structure in solution, we generated SAXS profiles over a protein concentration range of 1.5–15 mg/ml (supplemental Fig. S3C). The SAXS curves displayed a concentration dependence arising from the tendency of DNA-PKcs to undergo self-association at high protein concentrations (supplemental Fig. S3C). Interference-free SAXS profiles were estimated by extrapolating the measured scattering curves to infinite dilution. Analysis of the interference-free SAXS profile, which represents the monomeric state of DNA-PKcs, shows a typical globular particle with a Dmax of ∼155 Å (Fig. 3A and Table 1). Although variation exists between the individual SAXS envelopes, the position of the head and palm regions previously described in cryo-EM envelopes (41, 42) could be visualized (Fig. 3A and supplemental Fig. S4A). Similar to the three-dimensional EM reconstructions, the SAXS envelopes show distinct hollow features located inside the head region (supplemental Fig. S4A). Furthermore, SAXS envelopes show flattening of the shape in one direction, which is consistent with the structure determined earlier by electron crystallography (43) (Fig. 3A and supplemental Fig. S4A). To test the accuracy of the DNA-PKcs shape, we measured a second independently prepared sample. The P(r) functions matched well (Fig. 3A), and the reconstructed average shapes show identical structural features in both possible hands as expected (supplemental Fig. S5). Although the missing handedness precluded superposition onto EM reconstructions, the relative positions of the palm and head regions were evident.
FIGURE 3.
DNA-PKcs solution assemblies with and without DNA. A, top, two P(r) functions for DNA-PKcs alone calculated for two independent data sets shown in supplemental Fig. S3A. Middle, schematic view of DNA-PKcs adapted from cryo-EM reconstructions (41) with colored head (yellow) and palm (blue) regions. SAXS, two views of representative SAXS envelope rotated by 90° with head (yellow) and palm (blue) regions highlighted, as displayed in a volumetric representation. EM, cryo-EM envelopes (rotated by 90°) obtained for DNA-PKcs (41) and colored as above. See supplemental Fig. S4 for added structural analyses. B, DNA-PKcs dimers, showing the P(r) function, a schematic model, two views of representative SAXS envelope, and cryo-EM envelopes as in A. The inset shows the DNA-PKcs dimer described by cryo-EM (44). C, DNA-PKcs plus 40-bp HP-DNA, showing the P(r) function, a schematic model, and two views of representative SAXS envelope, as in A. D, DNA-PKcs plus 40-bp Y-DNA, showing the P(r) function, a schematic model, two views of representative SAXS envelope, and a cryo-EM three-dimensional reconstruction as in A. E, DNA-PKcs·Ku·40-bp HP-DNA, showing the P(r) function, a schematic model, and two views of representative SAXS envelope as in C. Ku is shown in pink/purple in the schematic and the SAXS envelopes. F, DNA-PKcs·Ku·40-bp Y-DNA, showing the P(r) function, a schematic model, two views of representative SAXS envelope, and a reconstructed cryo-EM envelope, obtained for DNA-PKcs·Ku·DNA (40).
Scattering profiles observed for conditions of high DNA-PKcs concentration (15 mg/ml) represent the protein in a dimeric state. However, inspection of the Guinier region (supplemental Fig. S3B) indicates that the sample is aggregation-free. Analysis of size exclusion chromatography equipped with a multiangle light scattering detector (SEC-MALS) (supplemental Fig. S6A), estimations of molecular weight (Table 1), and the shape of the pair distribution function P(r) (Fig. 3B) all reveal the formation of DNA-PKcs dimers. The P(r) suggests a state containing two globular particles with a Dmax of ∼300 Å. The reconstructed envelopes of this concentration-induced, self-assembly DNA-PKcs dimer indicate close contact between the DNA-PKcs molecules, resembling those described but not interpreted in prior EM studies (Fig. 3B, inset) (44). A thorough inspection of the commonalities among individual reconstructions of SAXS envelopes highlights head-to-head interactions with the palm regions pointing outwards (Fig. 3B and supplemental Fig. S4B).
We next examined the interaction of DNA-PKcs with DNA. dsDNA is predicted to bind to a central region of DNA-PKcs, inducing the head and palm regions to come together (41, 42). Alone, DNA-PKcs binds about 10 bp of DNA at the extreme termini of the DSB (45), whereas the DNA-PKcs·Ku·DNA complex requires about 30 bp of duplex DNA, and Ku is repositioned such that it binds internally with DNA-PKcs occupying the extreme termini of the DSB (46). To test the effect of DNA binding on DNA-PKcs, we examined scattering results with the short (16-bp Y-DNA) and long oligonucleotides (40-bp Y-DNA and 40-bp HP-DNA) described above. As expected, the 40-bp DNA molecules interacted with DNA-PKcs by electrophoretic mobility shift assay and supported the kinase activity of DNA-PK in vitro (supplemental Fig. S7).
No observed effects on the DNA-PKcs structure were seen in the presence of short 16-bp Y-DNA (data not shown). On the other hand, DNA-PKcs complexes in the presence of longer DNA constructs formed a complex with similar dimensions to the concentration-induced, self-assembly dimers described above. In these experiments, DNA was added to monomeric DNA-PKcs (i.e. protein concentration 1 mg/ml) at different molar ratios. In the experiment with hairpin DNA, the largest proportion of dimers was formed at a DNA-PKcs·40-bp HP-DNA ratio of 1:0.5 (supplemental Fig. S3E), a result that was confirmed by SEC-MALS experiments (supplemental Fig. S6B). Interestingly, SEC-MALS analysis of samples containing limiting amounts of DNA (molar ratio 1:0.3) showed a broad signal between 500 and 1000 kDa, suggesting sample polydispersity and indicating that the DNA-PKcs·HP-DNA complex is dynamic in nature with a high dissociation rate (supplemental Fig. S6B). The calculated P(r) function resembled the function determined for the concentration-induced DNA-PKcs dimer (Fig. 3C). In addition, the SAXS envelopes also show similar structural features to the concentration-induced DNA-PKcs dimer, adopting a head-to-head interface with the palm regions pointing outward (Fig. 3, B and C, and supplemental Fig. S4, B and C). Thus, in the presence of the hairpin DNA, two DNA-PKcs molecules adopt a surprisingly similar arrangement to that of the concentration-induced, self-assembly dimer.
In contrast, the binding of 40-bp Y-DNA resulted in a different protein-DNA complex. The larger RG, Dmax (Table 1) and shift in the P(r) shoulder all indicate a larger mass of the protein at the extremity of the DNA-PKcs complex (Fig. 3D). This is interpreted in the SAXS envelopes as a palm-palm interaction, with the bulky head regions pointing outward (Fig. 3D and supplemental Fig. S4D). The distinct P(r) shoulder is also consistent with the larger head regions being located at the dimer extremities. The reconstructed SAXS envelopes of the DNA-PKcs·40-bp Y-DNA complex resemble the cryo-EM envelopes reconstructed for the DNA-PKcs· DNA·Ku complex (40) (Fig. 3E). This may indicate that one molecule of DNA-PKcs is bound to the free dsDNA termini of the duplex and the other to the Y end. Increasing the ratio of 40-bp Y-DNA (1:1.2 and 1:2, DNA-PKcs·40-bp Y-DNA) caused the gradual disappearance of the dimeric state from solution (supplemental Fig. S3D), suggesting the favorable association of DNA-PKcs with the free duplex end before the Y-ended DNA end. In contrast, the DNA-PKcs dimers formed at high protein concentrations resemble the DNA-PKcs assembly formed in the presence of HP-DNA. We therefore propose that this head-to-head interface may represent a protein arrangement of the synaptic complex involved in the initial NHEJ step.
The DNA-PKcs Self-assembly Dimer Mimics the DNA-PKcs·Ku· DNA Synaptic Complex
To determine the structure of the DNA-PK complex (i.e. DNA-PKcs plus Ku assembled on DNA), we incubated DNA-PKcs with Ku and different molar ratios of either 40-bp Y-DNA or 40-bp HP-DNA. The structural features of the DNA-PKcs·Ku assembly in the presence of 40-bp HP-DNA resembled those observed in the absence of Ku (Fig. 3E and supplemental S4E). Broadening of the P(r) maxima is consistent with Ku being located in the central region of the DNA-PKcs dimer (Fig. 3E). The addition of DNA to give higher molar ratios did not disengage the DNA-PKcs dimers, as was seen in the absence of Ku (supplemental Fig. S3F), indicating stabilization of the DNA-PKcs·DNA assembly in the presence of Ku. In the absence of DNA, DNA-PKcs plus Ku did not support formation of DNA-PKcs dimers, because the P(r) function was similar to that obtained for DNA-PKcs by itself (supplemental Fig. S3F). Narrowing of the P(r) peak also indicates the absence of DNA-PKcs·Ku complexes (supplemental Fig. S3F).
The structural features of the DNA-PKcs·Ku assembly in the presence of 40-bp Y-DNA were surprisingly different from those observed in the presence of 40-bp HP-DNA. Similar to the arrangement observed in the absence of Ku, the DNA-PKcs·Ku·40-bp Y-DNA assembly shows DNA-PKcs dimers with a palm-palm interface, where Ku is most probably located between the palm and head regions (Fig. 3F and supplemental Fig. S4F). This assembly displays a striking similarity to the complex described by cryo-EM (40) (Fig. 3F and supplemental Fig. S4F), where the three-dimensional reconstruction was interpreted as a synaptic complex with two DNA-PKcs molecules tethered together by two fragments of DNA, such as those generated at a break.
Based on our results, we propose that only one Y-DNA duplex is located between the two DNA-PKcs molecules. Furthermore, the significant differences in the interaction of DNA-PKcs with HP-DNA compared with Y-DNA and its identical arrangement within the self-assembly dimer all suggest that the DNA-PKcs self-assembly dimer mimics the synaptic complex. The high dissociation rate of DNA-PKcs from HP-DNA (supplemental Figs. S3E and S6B) and the complex stabilization in the presence of Ku (supplemental Fig. S3F) support the high plasticity in the formation of the synaptic complex in vitro.
Autophosphorylation of DNA-PKcs Causes Release from DSBs in Vitro and in Vivo
We next examined the effects of autophosphorylation on DNA-PKcs function in vivo. We recently showed that DNA-PKcs in which seven in vitro/in vivo autophosphorylation sites (the ABCDE/Thr-2609 cluster plus serine 2056) had been converted to alanine (7A mutant) was retained at in vivo DSBs longer than WT DNA-PKcs (18), suggesting that autophosphorylation stimulates release of DNA-PKcs from DSBs in vivo. To extend these studies, we created a 7D mutant, in which the seven autophosphorylation sites were replaced by the phosphomimic, aspartic acid. Chinese hamster V3 cells, which are deficient for DNA-PKcs, were complemented with YFP-tagged WT, 7A, or 7D forms of human DNA-PKcs, and stable clones were isolated. DSBs were induced using a 365-nm UV laser, and the kinetics of accumulation of YFP-proteins at sites of DNA damage were examined using live cell imaging (see supplemental material). Both wild-type and mutant DNA-PKcs localized at sites of laser-induced DSBs, indicating that phosphorylation at these sites is not required for recruitment of DNA-PKcs to DSBs in vivo (supplemental Fig. S8A), supporting and extending earlier data (18). After initial localization, 7D DNA-PKcs dissociated from DSBs faster than WT DNA-PKcs, whereas release of the 7A mutant was considerably slower (supplemental Fig. S8A). Combined, these results support a model in which DNA-PKcs autophosphorylation facilitates its dissociation from DSB ends (reviewed in Ref. 5).
We next employed fluorescence recovery after photobleaching (FRAP) assays to examine the stability and dynamics of the DNA-PKcs-DNA interaction at DSBs. V3 cells expressing WT, 7A, or 7D forms of DNA-PKcs were microirradiated and incubated to allow DNA-PKcs accumulation at DSBs to reach the maximum (∼10 min after irradiation), and then the area of accumulated DNA-PKcs protein was photobleached. The recovery of fluorescence signals at various time points was monitored. As shown in supplemental Fig. S8B, recovery of FRAP occurred at much faster rates in 7D and WT cells than in 7A mutant cells. The recovery rate constants between WT, 7A, and 7D at the DSB sites were determined by fitting the raw data (supplemental Fig. S8C) to a reaction dominant model where the diffusion occurs much faster than binding and FRAP measurement (47). To test that the data fitting was appropriate, we compared the data (supplemental Fig. S8C) with the full reaction model and found an excellent fit of our experimental data to the reaction dominant model (supplemental Fig. S9). In this model, recovery represents the exchange of DNA-bound protein with free protein, and the recovery rate constant of the FRAP curve is identical to the dissociation rate constant, koff, of DNA-PKcs (supplemental Fig. S8C). The rate of recovery reflects the strength of complex binding and indicates that DNA-PKcs 7A has stronger (tighter) binding to DSBs than either WT DNA-PKcs or the 7D mutant. These results show that the exchange of DNA-bound protein with free protein is much faster in 7D or WT cells than in DNA-PKcs 7A, and therefore this exchange is influenced by the phosphorylation status of DNA-PKcs. Together, these experiments provide in vivo evidence that the phosphorylation status of DNA-PKcs modulates its dynamics and stability at DSBs.
To determine the mechanistic basis for the release of phosphorylated DNA-PKcs from DSBs, we examined the interaction of DNA-PKcs and Ku with DNA in vitro using biotinylated DNA pull-down assays. dsDNA oligonucleotides that were biotinylated at either one or both ends were generated and incubated with purified DNA-PKcs and Ku (see the supplemental material). As expected, neither protein bound to the doubly biotinylated species, whereas Ku bound to the singly biotinylated dsDNA, and DNA-PKcs bound only in the presence of Ku (supplemental Fig. S10A). When DNA-PKcs was preincubated under conditions that support autophosphorylation, Ku remained bound to the DNA, whereas the amount of retained DNA-PKcs was significantly reduced (supplemental Fig. S10B). Moreover, preincubation with the DNA-PK inhibitor wortmannin restored binding of DNA-PKcs to the DNA beads (supplemental Fig. S10C). Similar results were seen with DNA with long 5′- or 3′-overhangs (data not shown). Together, these data show that autophosphorylation promotes release of DNA-PKcs from DNA ends and the Ku-DNA complex.
Autophosphorylation of DNA-PKcs Induces a Conformational Change That Enlarges the Cleft between the Head and Palm Regions
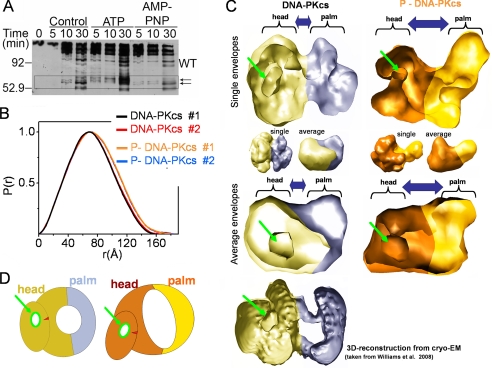
We hypothesized that autophosphorylation-induced dissociation of DNA-PKcs might result from a conformational change that reduces its ability to interact with Ku and/or DNA ends. We therefore first used limited proteolysis assays to probe for autophosphorylation-induced conformational changes in DNA-PKcs. Purified DNA-PKcs and Ku were incubated in the presence or absence of ATP or the non-hydrolyzable analogue AMP-PNP then digested with chymotrypsin and reaction products analyzed by SDS-PAGE and immunoblot (Fig. 4A). The presence of a unique series of polypeptides in the samples incubated with ATP but not AMP-PNP suggests that autophosphorylation induces a conformational change in DNA-PKcs (Fig. 4A, highlighted by arrows). To analyze the effects of autophosphorylation on the solution structure of DNA-PKcs, in vitro autophosphorylated and mock-phosphorylated forms of purified DNA-PKcs were isolated (Fig. S11, A and B) and analyzed using SAXS. Comparison of the SAXS data from phosphorylated (phospho-DNA-PKcs) and unphosphorylated DNA-PKcs revealed different scattering profiles over the entire scattering range (supplemental Fig. S11C): broadening in the P(r) maxima (Fig. 4B), an increase in particle dimension (from 155 to 180 Å), and increased RG values (from 55 to 57 Å) (Table 1). These data all indicate opening of the DNA-PKcs structure and show that a large conformational change has occurred over the entire internal structure, probably involving relocation of DNA-PKcs domains rather than extension of a single domain. Comparison of the reconstructed envelopes for the phosphorylated and unphosphorylated forms similarly reveals a large opening of the entire DNA-PKcs molecule as well as conformational changes in the head region (Fig. 4C). Both averaged and individual SAXS models reveal the presence of the palm and head regions and the cavity in the head region, as seen in EM reconstructions (41–43) (Fig. 4C). Comparison of the position of the enclosed cavity in the SAXS and EM envelopes (Fig. 4C, highlighted with a green arrow) suggests that autophosphorylation leads to enlargement of the cleft between the head and palm regions. Notably, the concentration-induced dimer remains present under autophosphorylation conditions and therefore does not dissociate upon autophosphorylation (supplemental Fig. S11E). Considering that DNA-PKcs autophosphorylation facilitates the release of DNA ends (supplemental Fig. S10) and that DNA is bound in the cleft between the head and palm regions (41, 44), we propose that this conformational change releases the interaction of DNA-PKcs with DNA ends.
FIGURE 4.
Autophosphorylation of DNA-PKcs causes a large conformational change and opening of the structure. A, purified DNA-PKcs and Ku were preincubated in the absence of ATP (control) or in the presence of either ATP or the non-hydrolyzable ATP analogue, AMP-PNP. Reactions were analyzed by SDS-PAGE and Western blot using an antibody to amino acids 2016–2136 of DNA-PKcs (see the supplemental material for details). The box highlights the region of the gel corresponding to the putative conformational change. B, P(r) functions calculated for the two data sets measured for DNA-PKcs (red and black) and two data sets measured for phospho-DNA-PKcs (P-DNA-PKcs) (orange and blue). Corresponding SAXS profiles are shown in supplemental Fig. S11. C, single (top) and average (bottom) SAXS envelope for DNA-PKcs (yellow, blue) and phospho-DNA-PKcs (orange, gold) are displayed in a clipped volumetric representation. Head and palm regions are highlighted. The enclosed cavity in the head region, which can be localized in the SAXS and EM envelopes (see Ref. 41), is highlighted by the green arrow. Unclipped envelopes are shown in the middle panel. D, schematic model describing proposed conformational changes during autophosphorylation of DNA-PKcs.
DISCUSSION
The dynamic structure of DNA-PKcs and its interactions with Ku and DNA in solution provide insights into the dynamic architectural changes whereby these complexes orchestrate NHEJ initiation. The Ku70/80 heterodimer solution structure reveals that Ku80CTR forms a long flexible arm in solution, which extends from the truncated DNA binding core determined by x-ray crystallography and cryo-EM reconstruction. These results are consistent with earlier studies showing proteolytic sensitivity of the Ku80CTR (48, 49) as well as subsequent structural studies revealing a disordered linker region (27). Because the extreme C terminus of the Ku80CTR contains a region important for interaction with DNA-PKcs (29, 30), the flexibility of this region is probably functional and appears suitable to recruit and stabilize DNA-PKcs at DSBs. Notably, the Ku80CTR flexibility and dimensions observed by SAXS are sufficient to allow interactions not only with a DNA-PKcs molecule bound to the same DSB end but also with the DNA-PKcs molecule bound on the opposing side of the DSB. Thus, the Ku80CTR extended structure may recruit and retain DNA-PKcs molecules on both sides of the DSB, promoting trans-autophosphorylation.
We furthermore show that DNA-PKcs in solution can form dimers in which the head regions of the molecule are in direct contact. We estimate that the concentration of DNA-PKcs in the nucleus of a human cell is ∼1 μm (equivalent to ∼5 mg/ml).5 We therefore suggest that the increased local concentration of DNA-PKcs at sites of damage may promote dimerization. Our results, which show that DNA-PKcs interacts with DNA in the absence of Ku, support and extend previous studies (46, 50–52). Interestingly, DNA-PKcs·DNA complexes formed with Y-DNA and HP-DNA had inverse arrangements, forming head-to-head dimers on HP-DNA but palm-to-palm dimers on Y-DNA. Similarly, DNA-PKcs molecules in DNA-PKcs·Ku complexes formed on HP-DNA adopted a head-to head arrangement, whereas dimers in DNA-PKcs·Ku complexes formed on Y-DNA were in a palm-to-palm arrangement. Interestingly, several studies have suggested that interaction of DNA-PKcs with the termini of a DSB results in melting or opening of the DNA ends within the DNA-PKcs molecule (8, 53, 54). These observations are supported by cryo-EM structures of DNA-PKcs that reveal potential sites of binding for both single-stranded DNA and dsDNA (41, 43). Thus, we propose that the different DNA-PKcs·DNA-PK complexes formed on Y-DNA and HP-DNA may reflect the ability of DNA-PKcs to open the Y-end of the Y-DNA structure, thus providing opening of both ends of the DNA, whereas the hairpin DNA would present only one open DNA end. However, we caution that the ability of DNA-PKcs and the DNA-PK complex to assemble on both ends of the Y-DNA structure as observed here and in previous studies (40) may generate a structure that is not physiologically relevant, because in vivo DNA damage-induced DSBs would each have only one exposed DNA end. In contrast, the DNA-PKcs assembly formed in the presence of hairpin DNA, which resembles the self-association in the DNA-PKcs dimer, provides an appropriate protein arrangement for the initial NHEJ step. Although high resolution structures of DNA-PKcs·DNA-PK in complex with DNA will be needed to determine precisely how DNA-PKcs and the DNA-PK complex interacts with DNA ends, these SAXS results reveal the dynamic DNA-PKcs assembly states and architectures.
The studies presented here indicate that autophosphorylation causes a dramatic conformational change in vitro that results in expansion of the region between the head and palm domains of DNA-PKcs. Although these observations provide a explanation for the observed effects of autophosphorylation on DNA-PKcs dissociation from DNA ends in vitro (10, 51) and in vivo (18), the regulation of DNA-PKcs by phosphorylation may be complex both in vitro and in vivo. To date, 16 in vitro autophosphorylation sites have been identified in DNA-PKcs: serines 2023, 2029, 2041, and 2053 (11); serine 2056 (11, 12); threonine 2609 (13, 14); serine 2612, threonine 2620, serine 2624, threonines 2638 and 2647, and serine 3205 (13); threonine 3950 (15); and serines 3821 and 4026 and threonine 4102 (55). In fact, unpublished data from our laboratory5 suggest that the total number of in vitro autophosphorylation sites may exceed 30 (6). The precise phosphorylation stoichiometry of these sites within the DNA-PKcs population analyzed by SAXS is unknown, and in addition, the autophosphorylated protein analyzed here may represent a mixture of phosphorylated forms.
Yet, several lines of evidence suggest that the in vitro phosphorylation sites, in particular the ABCDE cluster, are important for DNA-PKcs function in vivo. Serine 2056 (12, 15) and threonines 2609, 2638, 2647, and 3950 (9, 12, 14, 15) are phosphorylated in vivo in response to DNA damage and are required for DSB repair (9, 11, 12, 14, 15, 17, 56). Moreover, mutation of the ABCDE cluster to alanine reduces the ability of DNA-PKcs to dissociate from the Ku-DNA complex in vitro (this study) (15) and in vivo (this study) (18). Together, these studies suggest that phosphorylation of the ABCDE cluster plays an important role in the conformational changes observed here. However, phosphorylation of the ABCDE and PQR clusters have opposing effects on NHEJ in vivo (11); thus, different phosphorylation events may have different effects on DNA-PKcs structure. Therefore, further studies may help determine the effects of individual phosphorylation events on DNA-PKcs structure and function both in vitro and in vivo.
Despite uncertainties regarding the precise kinase domain location (41), placing the kinase domain in the DNA-PKcs head region (42) reveals that the head-to-head interaction visualized in our putative synaptic complex could facilitate trans-autophosphorylation by the kinase domains. In contrast, the palm-to-palm arrangement seen with Y-DNA would require kinase domain placement in the palm for trans-autophosphorylation, but this appears less physiologically relevant. Precise placement of the DNA-PKcs kinase and other functional domains within the DNA complex will require high resolution structures; however, our combined SAXS, biochemical, and biological data favor the physiological relevance of the head-to-head interaction and support the placement of the kinase in the head domain. Notably, we show that autophosphorylation of DNA-PKcs results in opening of the central DNA binding region located between the head and palm domains. We propose that this large conformational change facilitates dissociation of DNA-PKcs from DNA ends and DNA-bound Ku in vitro and in vivo. The experimentally defined dynamic plasticity of the initial NHEJ complex reported here supports and extends the novel two-phase model for the dynamic assembly of NHEJ factors to DSBs in vivo (57). In this model, recruited factors assemble into a large complex, which is stabilized by multiple protein interactions. In addition, the observed autophosphorylation-induced shape change also has the potential to affect the interaction of DNA-PKcs with accessory proteins, which may further regulate NHEJ in vivo.
Our results show that efficient association and dissociation of the DNA-PKcs at DSBs is regulated by Ku and autophosphorylation of DNA-PKcs. This exchange of DNA-PKcs on DSBs provides a probable determinant to facilitate transition and progression of NHEJ reactions for proper timing of end joining. Furthermore, the dramatically opened conformation of trans-autophosphorylated DNA-PKcs, which alters the relationship of the kinase domain to its regulatory domains, provides a probable molecular mechanism to alter DNA-PKcs interactions with downstream targets for signaling. These results thereby extend our previous structural understanding of DSB repair (59, 60) and provide an overall view of DNA-PK activation and flexibility for comparison with the recently defined Ctp1-Nbs1 complex acting in DSB repair by homologous recombination with its flexible linkage to Mre11-Rad50 and its ATM binding motif (61). Specifically, these combined SAXS, biochemical, and mutational results provide dynamic models that suggest how Ku and DNA-PKcs combine and assemble enzymatic and structural activities for NHEJ initiation and the choreography of DSB pathway progression.
Supplementary Material
Acknowledgments
We thank the Berkeley Laboratory Advanced Light Source and SIBYLS beamline staff at 12.3.1 for aiding solution scattering data collection and D. W. Chan for preliminary studies.
This work was supported, in whole or in part, by National Institutes of Health Structural Cell Biology of DNA Repair Machines P01 Grant CA92584 (to J. A. T., G. L. H., and S. P. L.-M.). This work was also supported by Canadian Institutes of Health Research Grant 13639 (to S. P. L.-M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S11.
Experimental and theoretical scattering profiles, P(r) functions, SAXS envelopes, and atomic models have been deposited in the BIOISIS data base, available on the World Wide Web.
Y. Yu and S. P. Lees-Miller, unpublished data.
- DSB
- double strand break
- NHEJ
- non-homologous end joining
- Ku
- Ku70/80
- DNA-PK
- DNA-dependent protein kinase
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- SAXS
- small angle x-ray scattering
- MES
- minimal ensemble search
- dsDNA
- double-stranded DNA
- SEC-MALS
- size exclusion chromatography equipped with a multiangle light scattering detector
- FRAP
- fluorescence recovery after photobleaching
- MD
- molecular dynamics
- EM
- electron microscopy
- Ku80CTR
- Ku80 C-terminal region
- Y-DNA
- dsDNA with a Y-shaped structure at one end
- HP-DNA
- duplex with a hairpin at one end
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate.
REFERENCES
- 1.Hall E. J., Giaccia A. J. (2006) Radiobiology for the Radiobiologist, 6th Ed., pp. 16–46, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 2.Helleday T., Lo J., van Gent D. C., Engelward B. P. (2007) DNA Repair 6, 923–935 [DOI] [PubMed] [Google Scholar]
- 3.Löbrich M., Jeggo P. A. (2007) Nat. Rev. Cancer 7, 861–869 [DOI] [PubMed] [Google Scholar]
- 4.O'Driscoll M., Jeggo P. A. (2006) Nat. Rev. Genet 7, 45–54 [DOI] [PubMed] [Google Scholar]
- 5.Mahaney B. L., Meek K., Lees-Miller S. P. (2009) Biochem. J. 417, 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meek K., Dang V., Lees-Miller S. P. (2008) Adv. Immunol. 99, 33–58 [DOI] [PubMed] [Google Scholar]
- 7.Weterings E., Chen D. J. (2008) Cell Res. 18, 114–124 [DOI] [PubMed] [Google Scholar]
- 8.DeFazio L. G., Stansel R. M., Griffith J. D., Chu G. (2002) EMBO J. 21, 3192–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meek K., Douglas P., Cui X., Ding Q., Lees-Miller S. P. (2007) Mol. Cell. Biol. 27, 3881–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan D. W., Lees-Miller S. P. (1996) J. Biol. Chem. 271, 8936–8941 [DOI] [PubMed] [Google Scholar]
- 11.Cui X., Yu Y., Gupta S., Cho Y. M., Lees-Miller S. P., Meek K. (2005) Mol. Cell. Biol. 25, 10842–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B. P., Chan D. W., Kobayashi J., Burma S., Asaithamby A., Morotomi-Yano K., Botvinick E., Qin J., Chen D. J. (2005) J. Biol. Chem. 280, 14709–14715 [DOI] [PubMed] [Google Scholar]
- 13.Douglas P., Sapkota G. P., Morrice N., Yu Y., Goodarzi A. A., Merkle D., Meek K., Alessi D. R., Lees-Miller S. P. (2002) Biochem. J. 368, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan D. W., Chen B. P., Prithivirajsingh S., Kurimasa A., Story M. D., Qin J., Chen D. J. (2002) Genes Dev. 16, 2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglas P., Cui X., Block W. D., Yu Y., Gupta S., Ding Q., Ye R., Morrice N., Lees-Miller S. P., Meek K. (2007) Mol. Cell. Biol. 27, 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Block W. D., Yu Y., Merkle D., Gifford J. L., Ding Q., Meek K., Lees-Miller S. P. (2004) Nucleic Acids Res. 32, 4351–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding Q., Reddy Y. V., Wang W., Woods T., Douglas P., Ramsden D. A., Lees-Miller S. P., Meek K. (2003) Mol. Cell. Biol. 23, 5836–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uematsu N., Weterings E., Yano K., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P. O., van Gent D. C., Chen B. P., Chen D. J. (2007) J. Cell Biol. 177, 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putnam C. D., Hammel M., Hura G. L., Tainer J. A. (2007) Q. Rev. Biophys. 40, 191–285 [DOI] [PubMed] [Google Scholar]
- 20.Hura G. L., Menon A. L., Hammel M., Rambo R. P., Poole F. L., 2nd, Tsutakawa S. E., Jenney F. E., Jr., Classen S., Frankel K. A., Hopkins R. C., Yang S. J., Scott J. W., Dillard B. D., Adams M. W., Tainer J. A. (2009) Nat. Methods 6, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guinier A., Fournet F. (1955) Small Angle Scattering of X-rays, Wiley Interscience, New York [Google Scholar]
- 22.Svergun D. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 23.Svergun D. I. (1999) Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelikan M., Hura G. L., Hammel M. (2009) Gen. Physiol. Biophys. 28, 174–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker J. R., Corpina R. A., Goldberg J. (2001) Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 27.Harris R., Esposito D., Sankar A., Maman J. D., Hinks J. A., Pearl L. H., Driscoll P. C. (2004) J. Mol. Biol. 335, 573–582 [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z., Hu W., Cano L., Lee T. D., Chen D. J., Chen Y. (2004) Structure 12, 495–502 [DOI] [PubMed] [Google Scholar]
- 29.Falck J., Coates J., Jackson S. P. (2005) Nature 434, 605–611 [DOI] [PubMed] [Google Scholar]
- 30.Gell D., Jackson S. P. (1999) Nucleic Acids Res. 27, 3494–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivera-Calzada A., Spagnolo L., Pearl L. H., Llorca O. (2007) EMBO Rep. 8, 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z., Zhu L., Lin D., Chen F., Chen D. J., Chen Y. (2001) J. Biol. Chem. 276, 38231–38236 [DOI] [PubMed] [Google Scholar]
- 33.Petoukhov M. V., Svergun D. I. (2005) Biophys. J. 89, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boehm M. K., Woof J. M., Kerr M. A., Perkins S. J. (1999) J. Mol. Biol. 286, 1421–1447 [DOI] [PubMed] [Google Scholar]
- 35.Förster F., Webb B., Krukenberg K. A., Tsuruta H., Agard D. A., Sali A. (2008) J. Mol. Biol. 382, 1089–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., Svergun D. I. (2007) J. Am. Chem. Soc. 129, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 37.Zhuang M., Calabrese M. F., Liu J., Waddell M. B., Nourse A., Hammel M., Miller D. J., Walden H., Duda D. M., Seyedin S. N., Hoggard T., Harper J. W., White K. P., Schulman B. A. (2009) Mol. Cell 36, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta A. B., Hura G. L., Wolberger C. (2009) J. Mol. Biol. 392, 1117–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernstein N. K., Hammel M., Mani R. S., Weinfeld M., Pelikan M., Tainer J. A., Glover J. N. (2009) Nucleic Acids Res. 37, 6161–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spagnolo L., Rivera-Calzada A., Pearl L. H., Llorca O. (2006) Mol. Cell 22, 511–519 [DOI] [PubMed] [Google Scholar]
- 41.Williams D. R., Lee K. J., Shi J., Chen D. J., Stewart P. L. (2008) Structure 16, 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivera-Calzada A., Maman J. P., Spagnolo L., Pearl L. H., Llorca O. (2005) Structure 13, 243–255 [DOI] [PubMed] [Google Scholar]
- 43.Leuther K. K., Hammarsten O., Kornberg R. D., Chu G. (1999) EMBO J. 18, 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boskovic J., Rivera-Calzada A., Maman J. D., Chacón P., Willison K. R., Pearl L. H., Llorca O. (2003) EMBO J. 22, 5875–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoo S., Dynan W. S. (1999) Nucleic Acids Res. 27, 4679–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West R. B., Yaneva M., Lieber M. R. (1998) Mol. Cell. Biol. 18, 5908–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sprague B. L., Pego R. L., Stavreva D. A., McNally J. G. (2004) Biophys. J. 86, 3473–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paillard S., Strauss F. (1993) Proteins 15, 330–337 [DOI] [PubMed] [Google Scholar]
- 49.Mo X., Dynan W. S. (2002) Mol. Cell. Biol. 22, 8088–8099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiu C. Y., Cary R. B., Chen D. J., Peterson S. R., Stewart P. L. (1998) J. Mol. Biol. 284, 1075–1081 [DOI] [PubMed] [Google Scholar]
- 51.Merkle D., Douglas P., Moorhead G. B., Leonenko Z., Yu Y., Cramb D., Bazett-Jones D. P., Lees-Miller S. P. (2002) Biochemistry 41, 12706–12714 [DOI] [PubMed] [Google Scholar]
- 52.Yaneva M., Kowalewski T., Lieber M. R. (1997) EMBO J. 16, 5098–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pawelczak K. S., Turchi J. J. (2008) Nucleic Acids Res. 36, 4022–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jovanovic M., Dynan W. S. (2006) Nucleic Acids Res. 34, 1112–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma Y., Pannicke U., Lu H., Niewolik D., Schwarz K., Lieber M. R. (2005) J. Biol. Chem. 280, 33839–33846 [DOI] [PubMed] [Google Scholar]
- 56.Convery E., Shin E. K., Ding Q., Wang W., Douglas P., Davis L. S., Nickoloff J. A., Lees-Miller S. P., Meek K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yano K., Morotomi-Yano K., Wang S. Y., Uematsu N., Lee K. J., Asaithamby A., Weterings E., Chen D. J. (2008) EMBO Rep. 9, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porod G. (1982) in Small Angle X-ray Scattering (Glatter O., Kratky O. eds) pp. 17–51, Academic Press, London [Google Scholar]
- 59.Shin D. S., Chahwan C., Huffman J. L., Tainer J. A. (2004) DNA Repair 3, 863–873 [DOI] [PubMed] [Google Scholar]
- 60.Hopfner K. P., Putnam C. D., Tainer J. A. (2002) Curr. Opin. Struct. Biol. 12, 115–122 [DOI] [PubMed] [Google Scholar]
- 61.Williams R. S., Dodson G. E., Limbo O., Yamada Y., Williams J. S., Guenther G., Classen S., Glover J. N., Iwasaki H., Russell P., Tainer J. A. (2009) Cell 139, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.