Abstract
The transmembrane glycoprotein CD98 regulates multiple cellular functions, including extracellular signaling, epithelial cell adhesion/polarity, amino acid transport, and cell-cell interactions. MicroRNAs post-transcriptionally regulate gene expression, thereby functioning as modulators of numerous cellular processes, such as cell differentiation, proliferation, and apoptosis. Here, we investigated if microRNAs regulate CD98 expression during intestinal epithelial cell differentiation and inflammation. We found that microRNA-7 repressed CD98 expression in Caco2-BBE cells by directly targeting the 3′-untranslated region of human CD98 mRNA. Expression of CD98 was decreased, whereas that of microRNA-7 was increased in well-differentiated Caco2-BBE cells compared with undifferentiated cells. Undifferentiated crypt cells isolated from mouse jejunum showed higher CD98 levels and lower levels of mmu-microRNA-706, a murine original microRNA candidate for CD98, than well-differentiated villus cells. Importantly, microRNA-7 decreased Caco2-BBE cell attachment on laminin-1, and CD98 overexpression recovered this inhibition, suggesting that microRNA-7 modulates epithelial cell adhesion to extracellular matrix, which in turn could affect proliferation and differentiation during the migration of enterocytes across the crypt-villus axis, by regulating CD98 expression. In a pathological context, the pro-inflammatory cytokine interleukin 1-β increased CD98 expression in Caco2-BBE cells by decreasing microRNA-7 levels. Consistent with the in vitro findings, microRNA-7 levels were decreased in actively inflamed Crohn disease colonic tissues, where CD98 expression was up-regulated, compared with normal tissues. Together, these results reveal a novel mechanism underlying regulation of CD98 expression during patho-physiological states. This study raises microRNAs as a promising target for therapeutic modulations of CD98 expression in intestinal inflammatory disorders.
Keywords: CD98; MicroRNA; miR-7, Caco2-BBE; Intestinal Differentiation
Introduction
The cell-surface molecule CD98 is a heterodimer composed of a type II transmembrane glycosylated heavy chain and one of several non-glycosylated light chains (1, 2). CD98 was first identified as a lymphocyte activation-related antigen expressed at low levels on resting peripheral T cells and rapidly up-regulated following lectin activation (3–5). In addition to a transcriptional mechanism for regulating CD98 expression in T cells (6), a post-transcriptional mechanism has been reported to account for the strong accumulation of CD98 in activated lymphocytes (7). CD98 expression has since been reported in activated B lymphocytes (8) and some proliferating normal tissues, including cells of the blood-brain barrier (9), basal skin layer (10), kidney proximal tubes (11, 12), placenta (13, 14), testis (15), and intestinal epithelium (16, 17). CD98 expression has also been described in a wide variety of tumor cells, including hepatoma, breast cancer, and colon cancer cells (18–20). It has been shown that CD98 associates with β1 and β3 integrins and thereby regulates integrin signaling, such as cell proliferation/transformation, spreading, migration, and epithelial cell adhesion/polarity (1, 21).
The mammalian intestinal epithelium undergoes continuous and rapid renewal (22). This renewal process reflects continuous cell division of a stem cell population located in the intestinal crypt, migration of daughter cells along the villus, and extrusion of senescent cells into the intestinal lumen (22). During the migration of cells along the crypt-villus axis, extracellular matrix proteins interact continuously with enterocytes. These interactions are important in modulating several key biological activities, including cell adhesion and migration, gene expression, and cell survival (22). As enterocytes migrate toward the villus tip, they develop specialized functions (differentiation). A succession of gene expression-promoting and expression-repressing events occurs during cell migration along the crypt-villus axis; these events involve both transcriptional and post-transcriptional mechanisms.
Recently, studies have demonstrated that microRNAs (miRNAs) of ∼18–24 nucleotides negatively regulate target mRNAs by binding to their 3′-untranslated regions (UTRs)2 (23). Evidence has emerged the role of miRNAs in many cellular functions, such as cell differentiation, proliferation, and apoptosis (23, 24).
Although miRNAs are known to play a number of regulatory roles, little is known about their roles in intestinal epithelial cell (IEC) differentiation. In the present study, we investigated the role of miRNAs in the regulation of CD98 expression during IEC differentiation and under inflammatory conditions, and assessed its potential involvement in epithelial-extracellular matrix interactions.
EXPERIMENTAL PROCEDURES
Cell Culture
Caco2-BBE and J774.A1 cells were grown in Dulbecco's modified Eagle's medium and RPMI media, respectively, supplemented with 14 mm NaHCO3, 10% fetal bovine serum, and 1.5 μg/ml plasmocin (Invitrogen, Grand Island, NY). Cells were kept at 37 °C in a 5% CO2 atmosphere and 90% humidity.
Human Tissue Samples
Colonic biopsies from patients undergoing endoscopy (normal samples) or surgical specimens from patients undergoing colon resection for Crohn disease (CD) at Mount Sinai School of Medicine, New York, NY were collected and stored in liquid nitrogen until analysis.
miRNAs, Plasmid Construction, Transfection, and Luciferase Assay
Hsa-miR-7 (Pre-miR Precursor AM17100, Product ID: PM10047), anti-hsa-miR-7 (anti-miR inhibitor AM17000, Product ID: 10047), pre-miR negative control 1 (AM17110), anti-miRTM negative control 1 (AM17010), and mmu-miR-706 (Pre-miR miRNA Precursor AM17100, Product ID: PM11439) were obtained from Ambion. Caco2-BBE cells cultured on 24-well plastic plates, coverslips, or filter supports were transfected with 40 nm miRNA precursors or 40 nm miRNA antisenses using Lipofectamine 2000 (Invitrogen).
The CD98-pcDNA3.1/V5-His-TOPO was constructed as we previously described (17, 25). Caco2-BBE cells were transfected with the CD98-pcDNA3.1/V5-His-TOPO construct (Caco2-BBE/CD98) or the pcDNA3.1/V5-His-TOPO vector (Caco2-BBE/Vector) using Lipofectin (Invitrogen) and stably selected in culture medium supplemented with 1.2 mg/ml geneticin (Invitrogen). For cell attachment assay, wild-type Caco2-BBE, Caco2-BBE/CD98, and Caco2-BBE/vector cells were trypsinized, counted, and transfected with 40 nm miRNA precursors or vehicle (control) using the siPORT NeoFX Transfection Agent (Ambion).
The human (h) CD98 mRNA 3′-UTR was cloned into the SpeI/HindIII sites of the pMIR-REPORTTM Luciferase vector (Ambion) or the XhoI/HindIII sites of the pEGFP-C1 vector (BD Biosciences Clontech). For luciferase assay, Caco2-BBE cells on 24-well plastic plates were transfected with 1 μg of the hCD98 3′-UTR-luciferase construct in the presence or absence of 40 nm the indicated miRNA precursors using Lipofectamine 2000. Firefly luciferase activity was measured at 24- and 48-h post-transfection using the Dual-Luciferase Reporter Assay system (Promega) and a Luminoskan Ascent luminometer (Thermo Electron Corp., Waltham, MA). Values were normalized to lysate protein concentration. For GFP repression experiment, Caco2-BBE cells seeded on coverslips were transfected with 1 μg of the hCD98 3′-UTR-GFP construct in the presence or absence of 40 nm miRNAs using Lipofectamine 2000. After 48 h of transfection, GFP expression was assessed by Western blot analysis and fluorescent microscopy using a Zeiss Axioskop2 plus microscope.
Isolation of Epithelia from Mouse Intestinal Crypts and Villi
Isolation of epithelial cells from the intestinal crypt-villus axe was performed as previously described (26) using 6–8-week-old FVB wild-type male mice. Briefly, small intestine was reverted and cut into pieces of 1–2 cm. Pieces were washed under constant stirring in HBSS− + 0.5 mm dithiothreitol for 5 min at 4 °C (Step 1). Epithelial cells from villi were collected after an incubation of pieces at 4 °C with constant stirring for 20 min in 150 ml of chelating buffer (27 mm trisodium citrate, 5 mm Na2HPO4, 96 mm NaCl, 8 mm KH2PO4, 1.5 mm KCl, 0.5 mm dithiothreitol, 55 mm d-sorbitol, 44 mm sucrose, pH 7.3) (Step 2). Pieces were then transferred to 20 ml of fresh chelating buffer in a 50 ml tube. Tube was manually inversed 20 times, buffer was through and 20 ml of fresh buffer was added. Manual wash was repeated 10 times (Step 3). Tissues were transferred to 100 ml of fresh chelating buffer and incubated at 4 °C with constant stirring for 10 min (Step 4). Steps 3 and 4 were repeated, and buffer containing epithelial cells from crypts was collected.
Protein Extraction and Western Blot Analysis
Cells were lysed in radioimmune precipitation assay buffer (150 mm NaCl, 0.5% sodium deoxycholate, 50 mm Tris-HCl, pH 8, 0.1% SDS, 0.1% Nonidet P-40) supplemented with protease inhibitors (Roche Diagnostics) for 30 min on ice. The homogenates were centrifuged at 13,000 rpm for 20 min at 4 °C. Total cell lysates were resolved on polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). Membranes were then probed with relevant primary antibodies: goat polyclonal anti-mouse CD98 (sc-7094; Santa Cruz Biotechnology), goat polyclonal anti-human CD98 (sc-7095; Santa Cruz Biotechnology), monoclonal anti-human integrin-β1 (MAB1981, Chemicon, Temecula, CA) and monoclonal anti-GAPDH (AM4300, Ambion). After washes, membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences), and blots were detected using the Enhanced Chemiluminescence Detection kit (Amersham Biosciences).
RNA Extraction and Real-time RT-PCR
Total RNAs were extracted using TRIzol reagent (Invitrogen) or the RNeasy Mini Kit (Qiagen) and reverse transcribed using the first-strand cDNA synthesis kit (Fermentas). Real-time RT-PCR was performed using an iCycler sequence detection system (Bio-Rad). Briefly, cDNA was amplified by 40 cycles of 95 °C-15 s and 60 °C-1 min using the iQ SYBR Green Supermix (Bio-Rad) and specific primers: hCD98 sense 5′-CAGGTTCGGGACATAGAGA-3′, hCD98 antisense 5′- GAGTTAGTCCCCGCAATCAA-3′; mCD98 sense 5′-GAGGACAGGCTTTTGATTGC-3′, mCD98 antisense 5′-ATTCAGTACGCTCCCCAGTG-3′; 18S sense 5′-CCCCTCGATGACTTTAGCTGAGTGT-3′, 18S antisense 5′-CGCCGGTCCAAGAATTTCACCTCT-3′; mouse 36B4 sense 5′-TCCAGGCTTTGGGCATCA-3′, mouse 36B4 antisense 5′-CTTTATCAGCTGCACATCACTCAGA-3′. 18S and 36B4 were used as housekeeping genes, and fold-induction was calculated using the Ct method as follows: ΔΔCt = (CtTarget − Cthousekeeping) treatment − (CtTarget − Cthousekeeping) nontreatment, and the final data were derived from 2−ΔΔCT.
Quantification of Mature miRNAs
Total RNAs isolated from Caco2-BBE cells or mouse intestinal crypts and villi were polyadenylated and reverse transcribed using the NCodeTM miRNA first-strand cDNA synthesis kit (Invitrogen). Levels of mature miRNAs were quantified by real-time RT-PCR as described above using the universal reverse primer provided in the kit and the following forward primers: hsa-miR-7: 5′-ACTTCACCTGGTCCACTAGCCGT-3′; mmu-miR-706: 5′-AGAGAAACCCTGTCTCAAAAAA-3′. 18S and 36B4 were used as housekeeping genes.
Immunofluorescence Staining of hCD98 in Caco2-BBE Monolayers
Caco2-BBE cells grown on 12-filter polycarbonate Transwell Inserts (Corning) were transfected with 40 nm miR-7 precursor or vehicle (Lipofectamine 2000). Filters were washed twice with PBS supplemented with 0.1 mm CaCl2 and 1 mm MgCl2 (PBS-Ca/Mg), and fixed with 4% paraformaldehyde in PBS-Ca/Mg for 15 min at room temperature. After three washes with PBS-Ca/Mg, cells were incubated with 3% bovine serum albumin in PBS-Ca/Mg for 30 min at 37 °C, and then with mixture of goat anti-human CD98 (Santa Cruz Biotechnology) for 1 h at room temperature. Cells were washed with PBS-Ca/Mg and stained for 1 h at room temperature with Alexa Fluor 594-conjugated donkey anti-goat secondary antibodies (Molecular Probes). After washing, filters were mounted by using the Slowfade medium (Molecular Probes). Images were taken using a Zeiss epifluorescence microscope equipped with a Bio-Rad MRC600 confocal unit, computer, and laser scanning microscope image analysis software (Carl Zeiss, Jena, Germany).
Measurement of Cell Resistance and Cell Adhesion/ Spreading
Resistance of Caco2-BBE cells was monitored by using the electric cell-substrate impedance sensing (ECIS) 1600R device (Applied BioPhysics, Troy, NY). Cells were seeded in ECIS 8W1E electrodes (2 × 104 cells/400 μl/electrode), and resistance was measured in real-time at a frequency of 500 Hz and a voltage of 1 V (25, 27).
For cell attachment study, Caco2-BBE cells were seeded in electrodes precoated with 10 μg/ml of laminin-1 (Sigma) and kept at 37 °C in 5% CO2 and 90% humidity as previously described (25, 28, 29). Attachment and spreading of cells on the electrode surface change the impedance in such a way that morphological information of the attached cells can be inferred. Capacitance of Caco2-BBE cells was measured at a frequency of 40 kHz and a voltage of 1 V. Details on the operation, equivalent resistance-capacitance circuit, and modification of the ECIS system can be found in the manufacturer's instructions. The time necessary for Caco2-BBE cells to spread out on half of the available electrode (t½) and the spreading rate of cells (s) were calculated for each electrode as previously described (29). The capacitance shift (s = −ΔC/Δt) was determined between C = 1.5 nF and C = 3.5 nF by means of linear regression. We chose this capacitance range for analysis as it more or less symmetrically embraced the capacitance values at t½.
Statistical Analysis
Values were expressed as means ± S.E. Statistical analysis was performed using unpaired two-tailed Student's t test by InStat v3.06 (GraphPad) software. p < 0.05 was considered statistically significant.
RESULTS
Expression of CD98 Is Decreased during the Differentiation of Intestinal Epithelial Cells
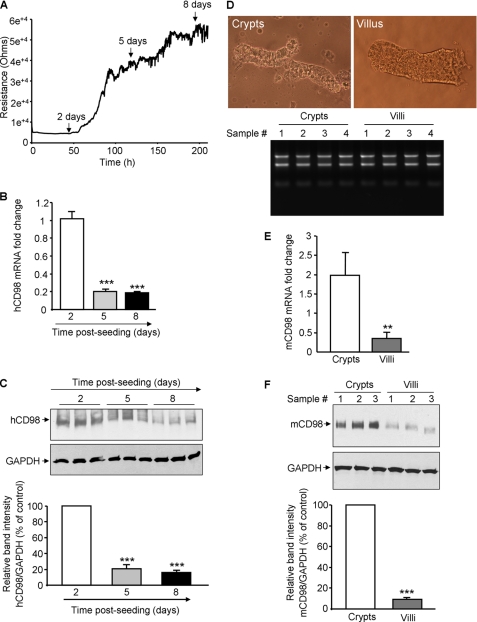
Although CD98 has been shown to be located in the basolateral membrane of well-differentiated IEC (1), its expression during IEC differentiation is not yet investigated. We first assessed the expression of human CD98 (hCD98) in intestinal epithelial Caco2-BBE cells at different time points post-plating. Caco2-BBE cell differentiation during the conventional culture period was evaluated by measuring the trans-epithelial resistance (TER) of the cells in real-time using the ECIS technique. Fig. 1A shows that Caco2-BBE cells grown as a monolayer became increasingly differentiated over time, reaching a plateau after ∼8 days of culture. Importantly, hCD98 expression at both mRNA and protein levels was decreased with increasing levels of cell differentiation: hCD98 expression levels were higher in undifferentiated Caco2-BBE cells (day 2 post-seeding) than in well-differentiated cells (day 8 post-seeding) (Fig. 1, B and C).
FIGURE 1.
Expression of CD98 is decreased during differentiation of intestinal epithelial cells. A–C, human (h) CD98 expression is decreased during Caco2-BBE cell differentiation. A, resistance of Caco2-BBE cells (2 × 104 cells/400 μl/electrode) was measured at 500 Hz, 1 V in real-time using the ECIS device. B and C, hCD98 expression levels in Caco2-BBE cells cultured on plastic plates for 2, 5, and 8 days were analyzed by real-time RT-PCR (B) and Western blot (C). Bar graphs in C show the relative intensity of blots in the upper panel. Values represent means ± S.E. of three determinations. *, p < 0.05; ***, p < 0.001; NS, not statistically significant versus day 2 (white bar). D–F, mouse (m) CD98 expression is decreased toward the crypt-villus axis of mouse intestine. D, villi and crypts were isolated from jejunum of 6–8-week-old FVB male mice. Pictures of the extracted villus and crypt fractions were taken using a Nikon Eclipse TS100 microscope at ×10 and ×40 magnifications, respectively (upper panels). Total RNAs from villi and crypts were extracted, and their integrity was shown (bottom panel). mCD98 expression levels in the villus and crypt fractions were assessed by real-time RT-PCR (E) and Western blot (F). Bar graphs in F show the relative intensity of blots in the upper panel. Values represent means ± S.E. of three determinations. **, p < 0.005; ***, p < 0.001.
To confirm the in vitro observations, we examined the expression of CD98 toward the crypt-villus axis of mouse intestine. Villi and crypts were isolated from jejunum of 6–8-week-old FVB male mice. Fig. 1D shows that the isolated villi and crypts were pure and intact (upper panel). Total RNAs from villi and crypts were extracted and their integrity was shown (Fig. 1D, bottom panel). Real-time RT-PCR and Western blot analyses showed higher levels of mouse (m) CD98 expression in undifferentiated crypt cells compared with well-differentiated villus cells (Fig. 1, E and F). Together, these data demonstrate that CD98 expression is decreased with increasing levels of IEC differentiation.
Hsa-miR-7 and mmu-miR-706 Regulate CD98 Expression during Intestinal Epithelial Cell Differentiation
Evidence has shown a role for miRNAs in cell differentiation (23, 24), we therefore hypothesized that miRNAs may be involved in the regulation of CD98 expression during IEC differentiation. To test this possibility, we first used the miRBase web site to screen the 3′-UTR sequence of hCD98 or mCD98 mRNA against the public data base for possible complementation of miRNAs. The human original hsa-miR-7 and murine original mmu-miR-706 were identified as potential miRNAs for the hCD98 and mCD98 gene, respectively (Fig. 2, A and C). Using the N-Code miRNA first-strand cDNA synthesis kit, which permits first-strand cDNA synthesis from the tailed miRNAs, and quantitative real-time RT-PCR analysis, the levels of mature forms of miR-7 in Caco2-BBE cells and of miR-706 in mouse intestinal crypt-villus axis were quantified. We found that the levels of mature miR-7 were inversely correlated with hCD98 expression levels during Caco2-BBE cell differentiation: hsa-miR-7 levels were higher in well-differentiated cells than in undifferentiated cells (Fig. 2B). In contrast, expression levels of hsa-miR-626, used as a control miRNA, in undifferentiated and well-differentiated Caco2-BBE cells were not significantly different (Fig. 2B). Consistent with this, the levels of mature mmu-miR-706 were significantly decreased in the crypt cells, where mCD98 was expressed at a high level, compared with that observed in villus cells, where mCD98 was expressed at a low level (Fig. 2D). Together, the inverse correlation between miRNA and CD98 expression levels in IECs suggests a role for miRNAs in the regulation of CD98 expression during IEC differentiation.
FIGURE 2.
Expression levels of hsa-miR-7 and mmu-miR-706 are decreased during intestinal epithelial cell differentiation. A, schematic representation of the human (h) CD98 mRNA with the 3′-UTR hsa-miR-7 binding site (upper panel) and sequence alignment of hCD98 3′-UTR miR-7 target site (bottom panel) predicted by miRBase. B, levels of mature forms of hsa-miR-7 or hsa-miR-626, used as a control miRNA, in Caco2-BBE cells cultured on plastic plates for 2, 5, and 8 days were analyzed by real-time RT-PCR. C, schematic representation of the mouse (m) CD98 mRNA with the 3′-UTR mmu-miR-706 binding site (upper panel) and sequence alignment of mCD98 3′-UTR mmu-miR-706 target site predicted by miRBase. D, levels of mature form of mmu-miR-706 in the villus and crypt fractions isolated from jejunum of 6–8-week-old FVB male mice were quantified by real-time RT-PCR. Values represent means ± S.E. of three determinations. *, p < 0.05; **, p < 0.005; NS, not statistically significant.
Hsa-miR-7 and mmu-miR-706 Inhibit CD98 Expression in Cell Culture
To directly examine the regulation of hCD98 by miR-7, Caco2-BBE cells were transiently transfected with vehicle (Lipofectamine 2000, control), miR-7 precursor (miR-7), or a negative control miRNA precursor (miR-control) containing a non-targeting sequence for the indicated time, and hCD98 expression levels were assessed by real-time RT-PCR and Western blot analyses. We found that miR-7 significantly reduced hCD98 mRNA levels in Caco2-BBE cells at 24 and 48 h post-transfection (Fig. 3A). hCD98 expression at the protein level was also decreased upon transfection of cells with miR-7 with a maximal reduction level after 48 h of transfection (Fig. 3B). In contrast, transfection of cells with miR-control or vehicle did not affect hCD98 mRNA and protein expression (Fig. 3, A and B). Furthermore, confocal microscopy showed a reduction of immunofluorescence staining for hCD98 in Caco2-BBE cells transfected with miR-7 compared with control cells (Fig. 3C), supporting the inhibition effect of miR-7 on hCD98 protein expression analyzed by Western blot.
FIGURE 3.
Hsa-miR-7 and mmu-miR-706 inhibit CD98 mRNA and protein expression in cell culture. A–C, Hsa-miR-7 inhibits human (h) CD98 expression in Caco2-BBE cells. B and C, Caco2-BBE cells were transfected with 40 nm miR-7 precursor (miR-7) or the negative control miRNA precursor (miR-control), or vehicle (Lipofectamine 2000; control). Expression of hCD98 was assessed by quantitative RT-PCR (B) and Western blot (C). Bar graphs in C show the relative intensity of blots in the upper panel. D, representative images of immunofluorescence staining for hCD98 in Caco2-BBE cells transfected with vehicle (control) or 40 nm miR-7 precursor for 48 h. Bars, 10 mm. D and E, Mmu-miR-706 inhibits mouse (m) CD98 expression in murine J774.A1 cell line. J774.A1 cells were transfected with 40 nm mmu-miR-706 precursor or vehicle (control) for the indicated times, and mCD98 expression levels were assessed by real-time RT-PCR (D) and Western blot (E). Bar graphs in E show the relative intensity of blots in the upper panel. Values represent means ± S.E. of three determinations. *, p < 0.05; **, p < 0.005; ***, p < 0.001; NS, not statistically significant versus control.
To examine if mmu-miR-706 inhibits mCD98 expression, we transfected mmu-miR-706 precursor (miR-706) into the murine macrophage cell line J774.A1. As shown in Fig. 3, D and E, miR-706 significantly down-regulated expression of mCD98 at both mRNA and protein levels after 1 and 2 days of transfection. Taken together, these results demonstrate that miRNAs down-regulate CD98 mRNA and protein expression in IECs.
MiR-7 Directly Targets the 3′- UTR of hCD98 mRNA
To examine if miR-7 directly targets the 3′-UTR of hCD98 mRNA, we cloned the hCD98 3′-UTR into the pMIR-REPORTTM Luciferase vector (hCD98 3′-UTR-luc) and measured luciferase activity in Caco2-BBE cells transiently transfected with this construct in the presence or absence of miR-7. As shown in Fig. 4A, miR-7 significantly repressed luciferase activity by up to ∼59 and 60% at 1 and 2 days post-transfection, respectively, indicating that miR-7 bound to the hCD98 3′-UTR. In contrast, miR-control had no effect on luciferase activity in cells transfected with the hCD98 3′-UTR-luc (Fig. 4A).
FIGURE 4.
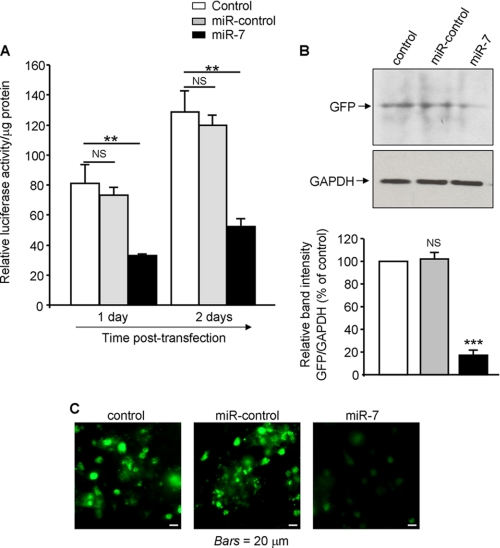
Hsa-miR-7 directly targets the hCD98 mRNA 3′-UTR. The full-length hCD98 mRNA 3′-UTR was cloned downstream a luciferase reporter gene (hCD98 3′-UTR-luc) or a GFP-coding sequence (hCD98–3′-UTR-GFP). A, Caco2-BBE cells were transfected with the hCD98 3′-UTR-luc construct in the presence or absence (control) of 40 nm miR-7 precursor or miR-control precursor. Luciferase activity was measured at 1 day and 2 days post-transfection and normalized to lysate protein concentration. B and C, Caco2-BBE cells were transfected with the hCD98 3′-UTR-GFP construct in the presence or absence of 40 nm miR-7 precursor or miR-control precursor for 48 h. GFP expression was assessed by Western blot. Bar graphs show the relative intensity of blots in the upper panel (B). Pictures of cells plated on coverslips were taken using a Zeiss Axioskop2 plus microscope. Bars, 20 μm (C). Values represent means ± S.E. of three determinations. **, p < 0.005; ***, p < 0.001; NS, not statistically significant versus control.
To confirm these results, the hCD98 3′-UTR was cloned downstream the GFP-coding sequence, and the effect of miR-7 on GFP expression in cells transfected with this construct was examined. We found consistently that miR-7 decreased the expression of the GFP as assessed by Western blot analysis (Fig. 4B). Furthermore, fluorescent microscopy showed a reduction of fluorescent intensity in Caco2-BBE cells transfected with miR-7 compared with control cells or cells transfected with the miR-control (Fig. 4C). Together, these data demonstrate that miR-7 down-regulates hCD98 expression by targeting the 3′-UTR of hCD98 mRNA.
MiR-7 Antisense Increases hCD98 Expression in Caco2-BBE Cells
To confirm the miR-7-mediated inhibition of hCD98 expression, the hCD98 3′-UTR-luc construct and an antisense of miR-7 or miR-control were co-transfected into Caco2-BBE cells. Luciferase assays revealed that miR-7 antisense increased luciferase activity in Caco2-BBE cells, whereas miR-control antisense had no significant effect (Fig. 5A). This result demonstrates that miR-7 antisense effectively bound to endogenous miR-7, thereby reducing the interaction of miR-7 with the hCD98 3′-UTR-luc inside the cells. Co-transfection of cells with the hCD98 3′-UTR-luc construct and miR-7, which was performed in parallel, decreased luciferase activity, indicating the specificity of miRNA antisense effects. The effect of miR-7 antisense on hCD98 expression in Caco2-BBE cells was further assessed by Western blot analysis. Consistent with the luciferase assay results, transfection of cells with miR-7 antisense, but not miR-control antisense, resulted in an increase in hCD98 expression (Fig. 5B). Collectively, these results strongly support the finding that miR-7 post-transcriptionally represses hCD98 expression in Caco2-BBE cells.
FIGURE 5.

MiR-7 antisense increases hCD98 expression in Caco2-BBE cells. A, Caco2-BBE cells were transfected with the hCD98 3′-UTR-luciferase reporter construct in the absence (control) or presence of 40 nm miR-7 antisense or miR-control antisense, or 40 nm miR-7 precursor. Luciferase activity was measured at 2 days post-transfection and normalized to lysate protein concentration. B, Caco2-BBE cells were transfected with vehicle (Lipofectamine 2000; control), 40 nm miR-7 antisense or miR-control antisense, or 40 nm miR-7 precursor for 2 days. hCD98 expression was assessed by Western blot. Bar graphs show the relative intensity of blots in the upper panel. *, p < 0.05; **, p < 0.005; ***, p < 0.001; NS, not statistically significant versus control.
MiR-7 Inhibits β1-Integrin Activation in Caco2-BBE Cells by Down-regulating hCD98 Expression
As CD98 has been shown to interact with and thereby function as a regulator of β1-integrin (1), we next investigated if miR-7 affects β1-integrin activity by regulating hCD98 expression. As shown in Fig. 6A, transfection of Caco2-BBE cells with either miR-7 or miR-control did not affect β1-integrin expression. By using the ECIS technique, we then examined the effect of miR-7 on the attachment and spreading of Caco2-BBE cells to laminin-1, which was previously demonstrated to be completely dependent on β1-integrin (28). Capacitance of Caco2-BBE cells was monitored quantitatively and in real-time on ECIS electrodes coated with 10 μg/ml of laminin-1. The time necessary for cells to spread out on half of the available electrode (t½) and the spreading rate of cells (s) were accordingly determined. Fig. 6, B and C show that Caco2-BBE cells transfected with miR-7 attached and spread on laminin-1 more slowly compared with control Caco2-BBE cells ([control: black solid line, t½ = 2.93 ± 0.46 h, s = 0.29 ± 0.04 nF/h, n = 4] versus [+miR-7: gray solid line, t½ = 7.3 ± 0.88 h, s = 0.21 ± 0.01 nF/h, n = 4]). These results demonstrate that miR-7 decreases β1-integrin activation but not β1-integrin expression, leading to a reduction in Caco2-BBE cell attachment to laminin-1. To examine if hCD98 was involved in the miR-7-mediated decrease of β1-integrin activation, the effects of miR-7 on attachment and spreading of Caco2-BBE cells stably transfected with the hCD98-pcDNA3.1/V5-His-TOPO construct (CD98) or the empty vector (vector) were determined. The hCD98-pcDNA3.1/V5-His-TOPO construct, which we previously generated, lacks of the 3′-UTR of hCD98 mRNA (17, 25) and therefore is not targeted by miR-7. Fig. 6B shows that overexpression of hCD98 significantly recovered the miR-7-mediated decrease in Caco2-BBE cell attachment ([+miR-7: gray solid line, t½ = 7.3 ± 0.88 h, s = 0.21 ± 0.01 nF/h, n = 4] versus [CD98+miR-7: black dotted line, t½ = 2.38 ± 0.18 h, s = 0.47 ± 0.04 nF/h, n = 4]). MiR-7-transfected Caco2-BBE/CD98 cells attached and spread on laminin-1 almost as fast as control cells ([control: black solid line, t½ = 2.93 ± 0.46 h, s = 0.29 ± 0.04 nF/h, n = 4] versus [CD98+miR-7: black dotted line, t½ = 2.38 ± 0.18 h, s = 0.47 ± 0.04 nF/h, n = 4]). However, transfection of cells with the empty vector failed to suppress the inhibition effect of miR-7 on cell attachment ([+miR-7: gray solid line, t½ = 7.3 ± 0.88 h, s = 0.21 ± 0.01 nF/h, n = 4] versus [vector+miR-7: gray dotted line, t½ = 6.83 ± 0.49 h, s = 0.25 ± 0.02 nF/h, n = 4]). Consistent with these data, microscopic images taken at 10 h post-seeding showed that the control cells were totally confluent, whereas the miR-7-transfected cells were not (Fig. 6D). Attachment and spreading of Caco2-BBE/CD98 cells were not affected by miR-7, therefore cells reached confluence at 10 h post-seeding as control cells did. However, miR-7-transfected Caco2-BBE/Vector cells were not confluent at 10-h post-seeding (Fig. 6D). Together, these results demonstrate that miR-7 inhibits β1-integrin activation in Caco2-BBE cells by directly down-regulating hCD98 expression.
FIGURE 6.
Hsa-miR-7 decreases β1-integrin activation in Caco2-BBE cells by down-regulating hCD98 expression. A, Caco2-BBE cells cultured on 12-well plastic plates were transfected without (control) or with 40 nm miR-7 precursor or miR-control precursor using Lipofectamine 2000, and β1-integrin expression was assessed by Western blot. B, Caco2-BBE cells or Caco2-BBE cells stably transfected with the CD98- pcDNA3.1/V5-His-TOPO construct (Caco2-BBE/CD98) or the empty vector (Caco2-BBE/Vector) were trypsinized, counted, and transfected without (control) or with 40 nm miR-7 precursor using the siPORT NeoFX transfection agent. Cells were then seeded on electrodes coated with 10 μg/ml of laminin-1 (105 cells/400 μl/electrode). Capacitance was measured at 40 kHz and 1 V in real-time using the ECIS device. C, half-time (t½) and spreading rate (s) of the cells were determined for each electrode. Data are means ± S.E. of two determinations. *, p < 0.05; **, p < 0.005; ***, p < 0.001. D, images were taken at 0 h and 10 h post-seeding using a Nikon Eclipse TS100 microscope at ×20 magnification. Each image is representative of quadruplicate electrodes.
MiR-7 Regulates hCD98 Expression during Intestinal Inflammation
We first assessed the effect of the pro-inflammatory cytokine IL1-β on hCD98 expression in Caco2-BBE cells. Quantitative real-time RT-PCR and Western blot analyses revealed that stimulation of Caco2-BBE cells with 2 ng/ml of IL1-β for 24 h significantly increased hCD98 mRNA and protein expression levels, respectively (Fig. 7, A and B). Interestingly, the levels of mature miR-7 assessed by quantitative RT-PCR were decreased upon stimulation of cells with IL1-β (Fig. 7C). To test whether miR-7 was directly involved in the IL1-β-mediated hCD98 up-regulation, Caco2-BBE cells were transfected with miR-7 and subsequently stimulated with IL1-β. Quantitative RT-PCR analysis revealed that IL1-β increased hCD98 mRNA levels by ∼1.5-fold ([control: 1.02 ± 0.15-fold] versus [+IL1-β: 1.5 ± 0.08-fold]; n = 5; *, p = 0.01), and this effect was repressed when cells were pretransfected with miR-7 ([+IL1-β: 1.5 ± 0.08-fold] versus [+miR-7+IL1-β: 0.92 ± 0.05-fold]; n = 5; ***, p = 0.0004) (Fig. 7D). These results suggest that miR-7 is involved in the IL1-β-mediated up-regulation of hCD98. Furthermore, miR-7 antisense increased hCD98 mRNA levels by ∼1.4-fold ([control: 1.02 ± 0.15-fold] versus [+miR-7 antisense: 1.44 ± 0.08-fold]; n = 5; *, p = 0.04), and IL1-β failed to significantly increase hCD98 mRNA levels in cells pretransfected with miR-7 antisense ([+miR-7 antisense: 1.44 ± 0.08-fold] versus [+miR-7 antisense+IL1-β: 1.63 ± 0.14-fold]; n = 5; p = 0.18, non-significant). This overlapping in the effects of miR-7 antisense and of IL-1β on hCD98 expression indicates that both miR-7 antisense and IL-1β up-regulate hCD98 expression by reducing the levels of miR-7.
FIGURE 7.
MiR-7 is involved in IL1-β-mediated hCD98 up-regulation in Caco2-BBE cells. A–C, IL1-β increases hCD98 expression and decreases miR-7 levels in Caco2-BBE cells. Cells were cultured on 12-well plastic plates for 5 days, serum starved for 24 h and stimulated or not with 2 ng/ml of IL1-β in serum-free Dulbecco's modified Eagle's medium for 24 h. hCD98 expression in untreated (control) and IL1-β-treated cells were analyzed by real-time RT-PCR (A) and Western blot (B). Bar graphs in B show the relative intensity of blots in the upper panel. Levels of mature miR-7 were quantified by real-time RT-PCR (C). D, MiR-7 is involved in IL1-β-induced hCD98 up-regulation in Cacao2-BBE cells. Cells were cultured on 24-well plastic plates and transfected with 40 nm miR-7 precursor or 40 nm miR-7 antisense or vehicle (Lipofectamine 2000; control) for 24 h. Cells were serum starved and treated or not with 2 ng/ml of IL1-β for 24 h. hCD98 expression was analyzed by real-time RT-PCR. Values represent means ± S.E. of two determinations. *, p < 0.05; ***, p < 0.001; NS, not statistically significant.
In an effort to verify the in vitro findings, we assessed hCD98 expression levels in actively inflamed colonic tissues from Crohn disease (CD) patients and the possible involvement of miR-7 in regulation of hCD98 expression during intestinal inflammation. Quantitative RT-PCR analysis showed that hCD98 mRNA expression was significantly increased in CD group compared with normal group ([CD group: 1.84 ± 0.29-fold, n = 8] versus [normal group: 1.19 ± 0.23-fold, n = 6]; *, p = 0.048). Importantly, the levels of mature miR-7 in CD group were markedly decreased compared with normal group ([CD group: 0.44 ± 0.09-fold, n = 8] versus [normal group: 1.06 ± 0.16-fold, n = 6]; **, p = 0.002) (Fig. 8). These results, which are consistent with in vitro observations, demonstrate that down-regulation of miR-7 is one of the mechanisms underlying up-regulation of hCD98 during intestinal inflammation.
FIGURE 8.

MiR-7 is involved in hCD98 up-regulation in Crohn disease. Total RNAs were extracted from normal (n = 6) and actively inflamed Crohn disease (CD, n = 8) colonic tissues using the RNAeasy mini kit and reverse transcribed using the NCodeTM miRNA first-strand cDNA synthesis kit. Expression levels of hCD98 mRNA (A) and mature miR-7 (B) were quantified by real-time RT-PCR.
DISCUSSION
In this study, we demonstrate, for the first time, that the expression of CD98 in IECs is post-transcriptionally down-regulated by miRNAs. MiRNAs have recently been shown to have a critical role in important biological processes including development, differentiation, proliferation, and apoptosis (23, 24). Using a candidate miRNA approach and the miRBase website we demonstrate that hsa-miR-7 inhibits intestinal epithelial hCD98 expression in Caco2-BBE cells by directly targeting the 3′-UTR sequence of hCD98 mRNA. We also identified mmu-miR-706 as a murine miRNA for mCD98 and demonstrated that this miRNA down-regulates mCD98 expression in the murine J744.A1 cell line.
Importantly, we show that CD98 expression is inversely related to IEC differentiation as shown by higher levels of CD98 expression in undifferentiated intestinal epithelial Caco2-BBE cells compared with well-differentiated cells, as well as in mouse intestinal crypt cells compared with villus cells. It is known that mitotically active, undifferentiated cells located at the crypts give rise to differentiated non-proliferative cells, which migrate and reach the villi (22). Our results suggest that CD98 may have different functional roles in proliferative and non-proliferative cells. Indeed, it has been shown that CD98 has a role in lymphocyte activation, cell proliferation, and malignant transformation (21). In agreement with this view, Kelly et al. (30) showed that the expression of CD98 is rate-limiting for pre-T-cell growth. In this context, it is plausible that the CD98 expression signature may have a defining role in IEC differentiation and proliferation.
Interestingly, the expression levels of mature miR-7 were higher in well-differentiated than in undifferentiated Caco2-BBE cells. Similarly, mmu-miR-706 levels were significantly increased in villus cells compared with crypt cells. These findings strongly suggest that miRNAs could regulate CD98 expression during the differentiation of IECs. It has been shown that miR-194 is up-regulated during IEC differentiation (31), miR-273 is implicated in the neuronal differentiation of Caenorhabditis elegans (32), miR-181 is involved in human hematopoietic cell differentiation (33), miR-375 is implicated in the development of pancreatic inlets (34), miR-1 and miR-133 in skeletal muscle differentiation (35), and miR-143 is thought to play a role in adipocyte differentiation (36). Recently, screening the conserved 3′-UTR sequences from the Drosophila melanogaster genome identified potential targets for miR-7 including Notch genes, which are known to be important in cell proliferation and differentiation (37). Together with these studies, our study supports the role of miRNAs in IEC differentiation via regulating expression of target genes.
Furthermore, we show that miR-7 decreased attachment and spreading of Caco2-BBE cells on laminin-1, and that overexpression of CD98 supressed the inhibition effect of miR-7 on cell attachment. Given that Caco2-BBE cell attachment is dependent on the interactions of β1-integrin with laminin-1 (28), and because miR-7 did not affect β1-integrin expression, this reduction of cell attachment and spreading behavior could reflect a decrease in the interactions between β1-integrin and laminin-1. Because CD98 has been shown to act as a mediator of β1-integrin signaling (1, 21), these data suggest that miR-7 modulates β1-integrin-laminin-1 interactions by regulating CD98 expression, which in turn could affect proliferation and differentiation during the migration of enterocytes across the crypt-villus axis. In a physiological context, we propose that well-differentiated IECs, such as villus cells, express high levels of miR-7, which represses CD98 expression. A low level of CD98 expression would decrease β1-integrin-dependent attachment of villus cells to the extracellular matrix, and eventually results in cell exfoliation. In contrast, undifferentiated IECs, such as crypt cells, have low levels of miR-7 and high levels of CD98, which would increase the attachment of crypt cells to the extracellular matrix via β1-integrin and lead to an increase in cell migration along the crypt-villus axis. Therefore, the regulation of CD98 expression by miRNAs may have an important role in maintaining cellular homeostasis along the crypt-villus axis.
Importantly, we demonstrate that miR-7 is involved in up-regulation of hCD98 during intestinal inflammation. Exposure of human intestinal epithelial Caco2-BBE cells to the pro-inflammatory cytokine IL1-β increased hCD98 expression and decreased mature miR-7 levels. Furthermore, miR-7 is directly involved in the IL1-β-mediated hCD98 up-regulation since transfection of cells with miR-7 suppressed the IL1-β-induced hCD98 expression levels. Consistent with the in vitro findings, the levels of mature miR-7 were markedly decreased in actively inflamed Crohn disease colonic tissues, where hCD98 expression was up-regulated, compared with normal tissues. A role for CD98 in the etiology of inflammatory disorders has been suggested (8, 38–40). Pro-inflammatory cytokines have been shown to up-regulate CD98 expression in IECs (39, 40). Increased levels of lymphocyte-activation antigens, including CD98, have been found at the cell surface of intestinal B cells, CD4+ T cells and CD8+ T cells isolated from patients with inflammatory bowel disease (8). We have recently shown that CD98 expression is highly up-regulated in colonic tissues from mice with active colitis, and that activation of epithelial CD98 aggravates intestinal inflammation (38). To our knowledge, our study is the first supported case showing the involvement of miRNAs in the up-regulation of CD98 during intestinal inflammation. In a pathological context, such as in inflammatory bowel disease, we propose that an abnormal expression of miR-7 could affect CD98 expression and subsequently IEC differentiation along the crypt-villus axis, which can alter the crypt-villus architecture. Although the role of miRNAs in immune system development and response has recently become evident, the mechanisms underlying involvement of miRNAs in immunity, such as their role in regulation of cytokine responses, are poorly understood (41). Therefore, it would be of interest to further study the mechanisms by which the pro-inflammatory cytokines regulate miR-7 expression and the role of miRNAs in the regulation of epithelial inflammatory responses in general.
In conclusion, our data show that 1) miR-7 down-regulates hCD98 expression by directly targeting the 3′-UTR of hCD98 mRNA, 2) miR-7 acts via regulating hCD98 expression to modulate β1-integrin-laminin-1 interactions, which in turn could affect proliferation and differentiation of enterocytes during the migration across the crypt-villus axis, and 3) miR-7 regulates hCD98 expression during intestinal inflammation. Together, our study reveals a novel mechanism underlying the regulation of CD98 during patho-physiological states, and raises miRNAs as a promising target for therapeutic modulations of the multiple functions of CD98 in intestinal inflammatory disorders.
This work was supported, in whole or in part, by National Institutes of Health Grants R24-DK-064399 center grant, R01-DK-071594 (to D. M.), and R01-DK55850 (to S. V. S.) from the NIDDK. This work was also supported by the Crohn and Colitis Foundation of America (a research fellowship award (to G. D.) and a research career development award (to S. D.)).
- UTR
- untranslated region
- GFP
- green fluorescent protein
- PBS
- phosphate-buffered saline
- TER
- trans-epithelial resistance
- ECIS
- electric cell-substrate impedance sensing
- IL
- interleukin
- IEC
- intestinal epithelial cell.
REFERENCES
- 1.Yan Y., Vasudevan S., Nguyen H. T., Merlin D. (2008) Biochim. Biophys. Acta 1780, 1087–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devés R., Boyd C. A. (2000) J. Membr. Biol. 173, 165–177 [DOI] [PubMed] [Google Scholar]
- 3.Lindsten T., June C. H., Thompson C. B., Leiden J. M. (1988) Mol. Cell Biol. 8, 3820–3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottesdiener K. M., Karpinski B. A., Lindsten T., Strominger J. L., Jones N. H., Thompson C. B., Leiden J. M. (1988) Mol. Cell Biol. 8, 3809–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotner T., Williams J. M., Christenson L., Shapiro H. M., Strom T. B., Strominger J. (1983) J. Exp. Med. 157, 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leidesn J. M., Yang L. H., Morle G. D., June C. H., Lindsten T., Thompson C. B., Karpinski B. (1989) J. Autoimmun 2, (Suppl.), 67–79 [DOI] [PubMed] [Google Scholar]
- 7.Teixeira S., Kühn L. C. (1991) Eur. J. Biochem. 202, 819–826 [DOI] [PubMed] [Google Scholar]
- 8.Schreiber S., MacDermott R. P., Raedler A., Pinnau R., Bertovich M. J., Nash G. S. (1991) Gastroenterology 101, 1020–1030 [DOI] [PubMed] [Google Scholar]
- 9.Kageyama T., Nakamura M., Matsuo A., Yamasaki Y., Takakura Y., Hashida M., Kanai Y., Naito M., Tsuruo T., Minato N., Shimohama S. (2000) Brain Res. 879, 115–121 [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Herrera J., Sánchez-Madrid F., Díez A. G. (1989) J. Invest. Dermatol. 92, 247–250 [DOI] [PubMed] [Google Scholar]
- 11.Quackenbush E. J., Gougos A., Baumal R., Letarte M. (1986) J. Immunol. 136, 118–124 [PubMed] [Google Scholar]
- 12.Rossier G., Meier C., Bauch C., Summa V., Sordat B., Verrey F., Kühn L. C. (1999) J. Biol. Chem. 274, 34948–34954 [DOI] [PubMed] [Google Scholar]
- 13.Novak D. A., Matthews J. C., Beveridge M. J., Yao S. Y., Young J., Kilberg M. S. (1997) Placenta 18, 643–648 [DOI] [PubMed] [Google Scholar]
- 14.Okamoto Y., Sakata M., Ogura K., Yamamoto T., Yamaguchi M., Tasaka K., Kurachi H., Tsurudome M., Murata Y. (2002) Am. J. Physiol. Cell Physiol. 282, C196–C204 [DOI] [PubMed] [Google Scholar]
- 15.Parmacek M. S., Karpinski B. A., Gottesdiener K. M., Thompson C. B., Leiden J. M. (1989) Nucleic Acids Res. 17, 1915–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X., Charrier L., Gewirtz A., Sitaraman S., Merlin D. (2003) J. Biol. Chem. 278, 23672–23677 [DOI] [PubMed] [Google Scholar]
- 17.Merlin D., Sitaraman S., Liu X., Eastburn K., Sun J., Kucharzik T., Lewis B., Madara J. L. (2001) J. Biol. Chem. 276, 39282–39289 [DOI] [PubMed] [Google Scholar]
- 18.Ohkame H., Masuda H., Ishii Y., Kanai Y. (2001) J. Surg. Oncol. 78, 265–271 [DOI] [PubMed] [Google Scholar]
- 19.Quackenbush E., Clabby M., Gottesdiener K. M., Barbosa J., Jones N. H., Strominger J. L., Speck S., Leiden J. M. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6526–6530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shennan D. B., Thomson J., Gow I. F., Travers M. T., Barber M. C. (2004) Biochim. Biophys. Acta 1664, 206–216 [DOI] [PubMed] [Google Scholar]
- 21.Cantor J. M., Ginsberg M. H., Rose D. M. (2008) Immunol. Rev. 223, 236–251 [DOI] [PubMed] [Google Scholar]
- 22.Gordon J. I. (1989) J. Cell Biol. 108, 1187–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 24.Ambros V. (2004) Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 25.Nguyen H. T., Dalmasso G., Yan Y., Obertone T. S., Sitaraman S. V., Merlin D. (2008) PLoS One 3, e3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flint N., Cove F. L., Evans G. S. (1991) Biochem. J. 280 (Pt 2), 331–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charrier L., Yan Y., Driss A., Laboisse C. L., Sitaraman S. V., Merlin D. (2005) Am. J. Physiol. Gastrointest. Liver Physiol. 288, G346–G353 [DOI] [PubMed] [Google Scholar]
- 28.Driss A., Charrier L., Yan Y., Nduati V., Sitaraman S., Merlin D. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 290, G1228–G1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegener J., Keese C. R., Giaever I. (2000) Exp. Cell Res. 259, 158–166 [DOI] [PubMed] [Google Scholar]
- 30.Kelly A. P., Finlay D. K., Hinton H. J., Clarke R. G., Fiorini E., Radtke F., Cantrell D. A. (2007) EMBO J. 26, 3441–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hino K., Tsuchiya K., Fukao T., Kiga K., Okamoto R., Kanai T., Watanabe M. (2008) Rna 14, 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang S., Johnston R. J., Jr., Frøkjaer-Jensen C., Lockery S., Hobert O. (2004) Nature 430, 785–789 [DOI] [PubMed] [Google Scholar]
- 33.Chen C. Z., Li L., Lodish H. F., Bartel D. P. (2004) Science 303, 83–86 [DOI] [PubMed] [Google Scholar]
- 34.Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P. E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. (2004) Nature 432, 226–230 [DOI] [PubMed] [Google Scholar]
- 35.Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esau C., Kang X., Peralta E., Hanson E., Marcusson E. G., Ravichandran L. V., Sun Y., Koo S., Perera R. J., Jain R., Dean N. M., Freier S. M., Bennett C. F., Lollo B., Griffey R. (2004) J. Biol. Chem. 279, 52361–52365 [DOI] [PubMed] [Google Scholar]
- 37.Stark A., Brennecke J., Russell R. B., Cohen S. M. (2003) PLoS Biol. 1, E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kucharzik T., Lugering A., Yan Y., Driss A., Charrier L., Sitaraman S., Merlin D. (2005) Lab. Invest. 85, 932–941 [DOI] [PubMed] [Google Scholar]
- 39.Fais S., Pallone F. (1989) Gastroenterology 97, 1435–1441 [DOI] [PubMed] [Google Scholar]
- 40.Yan Y., Dalmasso G., Sitaraman S., Merlin D. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G535–G545 [DOI] [PubMed] [Google Scholar]
- 41.Pedersen I., David M. (2008) Cytokine 43, 391–394 [DOI] [PMC free article] [PubMed] [Google Scholar]