Abstract
When glucose is added to yeast cells that are starved for 3 days, fructose-1,6-bisphosphatase (FBPase) and malate dehydrogenase 2 are degraded in the vacuole via the vacuole import and degradation (Vid) pathway. In this study, we examined the distribution of FBPase at the ultrastructural level. FBPase was observed in areas close to the plasma membrane and in cytoplasmic structures that are heterogeneous in size and density. We have isolated these intracellular structures that contain FBPase, the Vid vesicle marker Vid24p, and the endosomal marker Pep12p. They appeared irregular in size and shape. In yeast, actin polymerization plays an important role in early steps of endocytosis. Mutants that affect actin polymerization inhibited FBPase degradation, suggesting that actin polymerization is important for FBPase degradation. Both FBPase and malate dehydrogenase 2 were associated with actin patches. Vid vesicle proteins such as Vid24p or Sec28p were also at actin patches, although they dissociated from these structures at later time points. We propose that Vid24p and Sec28p are present at actin patches during glucose starvation. Cargo proteins arrive at these sites following the addition of glucose, and the endocytic vesicles then pinch off from the plasma membrane. Following the fusion of endosomes with the vacuole, cargo proteins are then degraded in the vacuole.
Keywords: Cell/Endocytosis, Organisms/Yeast, Protein/Turnover, Subcellular Organelles/Cytoskeleton, Subcellular Organelles/Lysosomes, Subcellular Organelles/Vesicles, FBPase, Vid Vesicles
Introduction
Protein synthesis and degradation play important roles in determining the levels of proteins in cells. For protein degradation, two major pathways have been characterized (1–6). These are the ubiquitin/proteasome pathway and the lysosomal targeting pathway (1–6). Lysosomes degrade intracellular proteins as well as proteins that are internalized from the plasma membrane. The lysosomal pathway can be regulated by nutrients or by hormones in the media (4–7). During starvation, a macroautophagy pathway is induced that degrades proteins or organelles non-selectively (8–10). Another chaperone-mediated autophagy pathway degrades proteins based on the presence of a particular KFERQ sequence (11–14).
The yeast vacuole is homologous to mammalian lysosomes (15–17). A number of protein trafficking pathways lead to the yeast vacuole. For example, the Vps pathway targets carboxypeptidase Y from the endoplasmic reticulum to the Golgi, and then to the vacuole for maturation (18, 19). The Cvt pathway targets aminopeptidase I from the cytosol to the vacuole for processing (20, 21). During nitrogen starvation, a non-selective autophagy pathway is induced, and ATG genes play important roles in this pathway (20–23). Interestingly, many of these ATG genes are utilized by the Cvt pathway (20–23). ATG genes are also needed for micro- or macro-pexophagy pathways that degrade peroxisomes when yeast cells are transferred from methanol or ethanol to glucose (24, 25). The macroautophagy pathway is inhibited by Tor1 and is stimulated by rapamycin. The induction of autophagy by rapamycin has been used to clear large aggregates that cannot be cleared by the proteasome pathway (26, 27). Abnormal autophagy may have a causative effect for diseases such as cancer, neurodegeneration, or aging (28–30).
Our laboratory studies a selective degradation pathway in Saccharomyces cerevisiae (31–39). The key gluconeogenic enzyme fructose-1,6-bisphosphatase (FBPase)2 is induced when cells are starved of glucose. However, when cells are replenished with glucose, this enzyme, as well as malate dehydrogenase 2 (MDH2) are targeted for degradation via the Vid pathway (35). Degradation of these gluconeogenic enzymes occurs either in the proteasome or in the vacuole, depending on the duration of starvation (40). For example, when cells are starved for 1 day, FBPase is degraded in the proteasome. However, when cells are starved for 3 days, FBPase is degraded in the vacuole. The vacuole-dependent pathway consists of several steps. First, FBPase is associated with Vid (vacuole import and degradation) vesicles (41). These Vid vesicles subsequently merge with the endocytic pathway (42), although the exact point of this merger is not clear. Finally, FBPase is delivered to the vacuole and degraded in the lumen of this organelle.
Although we have purified Vid vesicles to homogeneity (41), we do not know their site of biogenesis, or whether they are present prior to a glucose shift. To address these questions, we screened for the presence of Vid vesicle protein markers. Coat protein 1 (COPI) coatomer proteins were identified as components of Vid vesicles. Of these, we have extensively characterized the localization and function of Sec28p, the ϵ subunit of the coatomer complex (42). We propose that Sec28p is on Vid vesicles and then becomes associated with endosomes following the addition of glucose. After the fusion of endosomes with vacuoles, Sec28p is distributed on the vacuole membrane and then in retrograde vesicles that form from the vacuole membrane.
The presence of Sec28p in retrograde vesicles raises a number of questions. For example, how are these vesicles formed? Are Vid vesicles derived from these retrograde vesicles? Alternatively, do retrograde vesicles travel to the plasma membrane and then become Vid vesicles? If the plasma membrane is the site of Vid vesicle formation and/or function, might FBPase be targeted to this location? Along these lines, early steps of endocytosis occur at the plasma membrane, and these are tightly linked to actin polymerization in yeast (43–48). Protein molecules involved in the polymerization of actin cytoskeleton are recruited to actin patches in a stepwise manner. For example, Sla1p and Las17p are localized to this area early in the process. This is followed by recruitment of actin and various actin-binding proteins. Finally, proteins involved in the scission of the endocytic vesicles such as Rvs161p and Rvs167p are recruited toward the end of the process (43–48). Therefore, these proteins may play an important role in FBPase degradation, if the Vid pathway utilizes early steps of endocytosis to deliver FBPase to the vacuole.
To address the above questions, we first examined the distribution of FBPase at the ultrastructural level. Following the addition of glucose to wild-type and Δpep4 cells that were glucose-starved, a portion of FBPase was found in regions underneath the plasma membrane. FBPase was also seen in abundant intracellular structures that were heterogeneous in size and in density. These FBPase-containing structures were purified; they contained the endosomal marker Pep12p, the cargo protein FBPase, and the Vid vesicle marker Vid24p.
A number of actin polymerization mutants blocked FBPase degradation. Furthermore, FBPase and MDH2 were targeted to actin patches following the addition of glucose to starved cells. Likewise, the Vid vesicle proteins Sec28p and Vid24p were associated with actin patches. Taken together, our results suggest that yeast cells utilize early steps of the endocytic pathway and actin polymerization to deliver FBPase and MDH2 to the vacuole for degradation.
EXPERIMENTAL PROCEDURES
Strains, Media, and Antibodies
Strains used in this study are listed in Table 1. Deletion strains were purchased from Euroscarf, and these deletions were derived from the parental BY4741 strain. The Abp1p-GFP strain was purchased from Invitrogen. For most experiments, cells were grown in YPKG (1% yeast extract, 2% peptone, 1% potassium acetate, and 0.5% glucose) for 3 days at 30 °C in an environmental shaker (300 rpm). Cells were transferred to YPD (1% yeast extract, 2% peptone, and 2% glucose) for the indicated times. Yeast transformations were carried out using the lithium acetate/Tris EDTA protocol. Western blotting was performed with anti-FBPase antibodies at 1:10,000 dilution and detected with ECL kits. For EM studies, anti-FBPase antibodies were affinity-purified over an FBPase affinity column followed by incubation with fixed cells that lacked the FBP1 gene. Antibodies reacted with FBPase as a single protein band that migrated at 38 kDa. This protein was absent in cells lacking the FBP1 gene. Purified anti-FBPase antibodies were incubated at 1:5 dilution with thin sections of embedded cells for 4 h, followed by a 2-h incubation with secondary antibodies conjugated with 10 nm colloid gold particles. Mouse anti-HA antibodies were purchased from Roche Applied Science. FM4-64 dye was purchased from Invitrogen, whereas rhodamine-conjugated phalloidin was purchased from Sigma.
TABLE 1.
Strain used in this studyα Δ
| Strain | Genotype |
|---|---|
| W303 | MATα ura3 leu2 his3 trp1 lys2 ade2 |
| Δpep4 | MATα ura3 leu2 his3 trp1 lys2 ade2 pep4::URA3 |
| HLY228 | MATα ura3 leu2 his3 trp1 lys2 ade2 VID24-HA::HIS3 |
| HLY635 | MATα ura3–52 LEU2 trp1Δ63 his3Δ200 GAL2 |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| Δvph1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vph1::kanMX4 |
| Δubc1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ubc1::kanMX4 |
| Δend3 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 end3::kanMX4 |
| Δsla1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sla1::kanMX4 |
| Δmyo3 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 myo3::kanMX4 |
| Δrvs161 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 rvs161::kanMX4 |
| Δrvs167 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 rvs167::kanMX4 |
| Δsac6 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 sac6::kanMX4 |
| Δvrp1 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vrp1::kanMX4 |
| HLY1747 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vph1::kanMX4 FBPase-GFP::HIS3 |
| HLY1160 | MATαleu2Δ0 lys2Δ0 ura3Δ0 sla1::kanMX4 FBPase-GFP::HIS3 |
| HLY1159 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 rvs167::kanMX4 FBPase-GFP::HIS3 |
| Abp1p-GFP | MATα leu2Δ0 lys2Δ0 ura3Δ0 Abp1-GFP::HIS3 |
| HLY1418 | MATα leu2Δ0 lys2Δ0 ura3Δ0 FBPase-GFP::HIS3 |
| HLY2251 | MATα leu2Δ0 lys2Δ0 ura3Δ0 MDH2-GFP::HIS3 |
| HLY2139 | MATα leu2Δ0 lys2Δ0 ura3Δ0 myo3::kanMX4 MDH2-GFP::HIS3 |
| HLY2123 | MATα his3Δ leu2Δ0 lys2Δ0 ura3Δ0 sla1::kanMX4 Sec28p-GFP::HIS3 |
| HLY2132 | MATα his3Δ1leu2Δ0 lys2Δ0 ura3Δ0 rvs167::kanMX4 Sec28p-GFP::HIS3 |
| HLY1419 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Sec28p-GFP::HIS3 |
| HLY2586 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 GFP-Vid24p::URA3 |
| HLY2153 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 rvs167::kanMX4 GFP-Vid24p::URA3 |
Sephacryl-1000 Chromatography
Cells were grown in 500 ml of YPKG in an environmental shaker. Cells were pelleted and re-suspended in 500 ml of YPD. At t = 15 min following a glucose shift, 10 mm sodium azide was added to cells. Cells were collected using low speed spins and spheroplasted. Spheroplasts were re-suspended in 10 ml of TEA buffer (10 mm triethanolamine, pH 7.5, 100 mg/ml phenylmethylsulfonyl fluoride, 10 mM NaF, 10 mm NaN3, 1 mm EDTA, and 0.8 m sorbitol). Spheroplasts were homogenized on ice with 20 strokes of a Dounce homogenizer. Lysates were centrifuged at 15,000 × g for 20 min. The resulting supernatant was further centrifuged at 100,000 × g for 2 h. Pellets were re-suspended in 5 ml of TEA and applied to a pre-packed 90- × 1.5-cm Sephacryl S-1000 column (Sigma). Material was eluted with 500 ml of TEA with a flow rate of 8 ml/h. Samples (4 ml) were collected from the column and precipitated with 10% trichloroacetic acid (final concentration). Trichloroacetic acid precipitates were washed, solubilized in 200 μl of SDS-PAGE buffer, and subjected to SDS-PAGE electrophoresis. The distribution of FBPase was examined by Western blotting of samples collected from fractions 11–30.
Sucrose Density Gradients
Cells were grown in 50 ml of YPKG and transferred to 50 ml of YPD for 15 min. Cells were spheroplasted and homogenized. Lysates were centrifuged at 15,000 × g for 20 min, and the resulting supernatant was centrifuged at 100,000 × g for 2 h. The 100,000 × g pellets were re-suspended in 1 ml of TEA buffer and loaded onto a 20–70% sucrose density gradient. These gradients were prepared by adding 2 ml each of 70, 50, 40, 30, and 20% sucrose sequentially. Gradients were then centrifuged at 100,000 × g for 16 h at 4 °C using a Sorvall TH-641 rotor. Following centrifugation, samples (0.7 ml) were collected from the top, and each fraction was precipitated by using 10% trichloroacetic acid. Trichloroacetic acid precipitants were washed and re-suspended in 100 μl of SDS-PAGE buffer followed by SDS-PAGE electrophoresis. The distribution of FBPase, HA-Vid24p, and Pep12p on sucrose gradients was examined by Western blotting with anti-FBPase, anti-HA, and anti-Pep12p antibodies.
Differential Centrifugation
Cells were grown in 10 ml of YPKG and then were transferred to 10 ml of YPD for 15 min. Cells were re-suspended in 0.8 ml of FBPase buffer containing 0.8 m sorbitol and lysed by vortexing vigorously with glass beading on ice. Lysates were subjected to differential centrifugation at 13,000 × g for 20 min. The resulting supernatants were centrifuged at 100,000 × g for 2 h, followed by centrifugation at 200,000 × g for 2 h. The 200,000 × g pellets were re-suspended in 75 μl of SDS-PAGE buffer, whereas the 200,000 × g supernatant was precipitated with 15% trichloroacetic acid, washed, and re-suspended in 200 μl of SDS-PAGE buffer. Vid vesicles were enriched in the 200,000 × g pellets, whereas cytosol was enriched in the 200,000 × g supernatant. The distribution of FBPase was examined by Western blotting with anti-FBPase antibodies.
Isolation of FBPase-containing Structures
Wild-type cells were grown in 500 ml of YPKG and transferred to 500 ml of YPD for 15 min. Cells were spheroplasted and homogenized by Dounce homogenization. Lysates were centrifuged at 13,000 × g for 20 min, followed by centrifugation at 100,000 × g for 1 h. Pellets were re-suspended in 10 ml of TEA and subjected to S-1000 column chromatography. The FBPase-containing peaks were examined by Western blotting with anti-FBPase antibodies. Fractions 19–22 were pooled and centrifuged at 100,000 × g for 2 h. Pellets were re-suspended in 1 ml of TEA and further separated using 20–70% sucrose density gradients. The distribution of FBPase was examined by Western blotting of each fraction from the sucrose density gradient. Fraction 12 from the first sucrose density gradient was collected, pelleted, and centrifuged on the second 20–70% sucrose density gradient. Fraction 12 from the second sucrose density gradient was collected for further EM analysis. Samples (5 μl) were adsorbed to a carbon-coated grid. After a brief wash with water to remove sucrose, samples were stained with 1% uranyl acetate for 1 min. Grids were examined using a 1200 EX transmission electron microscope (JOEL). The same experiments were conducted to isolate endosomes from fractions 15–18 from the S-1000 column. These fractions were pooled and centrifuged at 100,000 × g for 2 h. Pellets were re-suspended in 1 ml of TEA and applied to 20–70% sucrose density gradients and centrifugation at 100,000 × g for 16 h. Fraction 12 from the first sucrose density gradient was collected, pelleted, and centrifuged on a second 20–70% sucrose density gradient. Fraction 12 from the second sucrose density gradient was collected and examined by negative staining and electron microscopy. For FBPase and Vid24p staining, these FBPase-containing organelles were purified, fixed, and embedded in LR White. Thin sections were then incubated with affinity-purified FBPase antibodies followed by goat anti-rabbit antibodies conjugated with 10 nm gold particles. For Vid24p staining, thin sections were incubated with HA antibodies followed by goat anti-mouse antibodies conjugated with 15 nm gold particles. For surface labeling of Vid24p, purified FBPase-containing structures were fixed and incubated with or without HA antibodies followed by goat anti-mouse antibodies conjugated with 15 nm gold particles.
Fluorescence Microscopy
GFP-tagged proteins were amplified using PCR-based reactions. Primers used for PCR amplification are listed in Table 2. PCR products were purified and concentrated using Qiagen PCR Cleanup kit (Qiagen). PCR products were transformed into cells, and genomic integration was confirmed by PCR. Cells expressing FBPase-GFP, MDH2-GFP, Sec28p-GFP, or GFP-Vid24p were grown in YPKG and then transferred to YPD media. In some experiments, cells were transferred to glucose in the presence of FM. In other experiments, FM was added 16 h prior to a glucose shift. Cells were then transferred to YPD in the absence of FM for the indicated time points. For the staining of actin patches, cells expressing various tagged GFP fusion proteins were grown in 3 ml of YPKG for 3 days and transferred to 1 ml of YPD. Actin staining was performed as described previously (49, 50), with modifications. At various time points, 300 μl of cells was collected and formaldehyde (22 μl) was added for 5 min at 2.5% (final concentration). Cells were centrifuged and washed in 400 μl of phosphate-buffered saline. Cells were spun down and fixed with 300 μl of ice-cold acetone while vortexing. Cells were incubated at −20 °C for 5 min. Fixed cells were centrifuged and washed with phosphate-buffered saline and re-suspended in 80 μl of phosphate-buffered saline. Rhodamine-conjugated phalloidin (1 μl) was added, and the mixture was incubated in the dark for 30 min. Cells were immobilized on 1% agar and visualized with fluorescence microscopy. GFP, FM, and rhodamine-phalloidin were then visualized using a Zeiss fluorescence microscope and Axiovision software.
TABLE 2.
Primers used in this study
| FBPase-GFP | |
| F | ATTTGGTTGGGTTCTTCAGGTGAAATTGACAAATTTTTAGACCATATTGGCAAGTCACAGCGGATCCCCGGGTTAATTAA |
| R | CCATCCCATTCCATTCGCTACTTCCTTTCTCTTTTCCTAAGAATTTTCATTATTAGAAGGGAATTCGAGCTCGTTTAAAC |
| MDH2-GFP | |
| F | TAAGGGCTTGGAATTCGTTGCATCGAGATCTGCATCATCTCGGATCCCCGGGTTAATTAA |
| R | TGTATGTAGGGATGGTGCGCGTTTGTTGTGTGACATTTGCGAATTCGAGCTCGTTTAAAC |
| Sec28p-GFP | |
| F | CACCAAGAAATTGACGCAAAATTCGATGAATTAGTGAGGAAATATGATACGTCCAACCGGATCCCCGGGTTAATTAA |
| R | ATG AAATATTTTTTTCTTTTTCTAAAAAACCTACATGTTTAATGTGAGATATTACGTAAAGAATTCGAGCTCGTTTAAAC |
| Vid24p-TOPO | |
| 155F | AGGCCCTCGGTTTTGGCGCGTTTGGCCACCGGCCGCC |
| 152R | GTTAATTAAAGCAAACTCAAAAGAACAATCACTGGACTCG |
| Vid24p-TOPO | |
| 177F | CATTTGTGGCGTCTTGTGCATGACACCAACACATATCGCAAGCTTGAGTCTTAATAACAGTAAAGGAGAAGAA |
| 178R | GTCACAGCTTTGGGTTTCTCCGCTACACTGTCTACCTTAGGATTATTGATCATCTCGAGTTTGTATAGTTCAT |
GFP-Vid24p Construct
Vid24p was amplified by PCR reactions using the forward and reverse primers (P152 and P155, see Table 2). The resulting PCR product contained a 558-bp promoter region and the open reading frame ending with an engineered Pac1 site. DNA was gel-purified and cloned into the pYES2.1 vector (Invitrogen) according to the manufacturer's protocol. DNA was isolated from bacteria containing Vid24p-TOPO (B434) and was further digested with MscI and SnaB1 to remove the Bsu361 site. Following digestion, the plasmid was re-ligated to produce B461. GFP was amplified with P177 and P178 (see Table 2) using the pFA6a-GFP-S65T-HIS3MX6 plasmid as the template. Amplified DNA was cloned into a TOPO vector to produce B483. This plasmid was digested with HindIII and Bsu361 and ligated into plasmid B461 that was also digested with HindIII and Bsu361. The final plasmid contained 558 bp of the Vid24p promoter region, and GFP was fused in-frame with Vid24p at the N terminus (B498). This plasmid was linearized with SacII and integrated into wild-type cells at the VID24 promoter region, thereby allowing for expression under the control of the VID24 gene.
RESULTS
FBPase Is in Areas Near the Plasma Membrane
Previously, we demonstrated that the Vid pathway utilizes the endocytic pathway (42). However, the exact point of merger is not clear. Vid vesicles may fuse with endosomes that are released into the cytoplasm. If this is the case, FBPase may be targeted to these structures in the cytoplasm. Alternatively, Vid vesicles may merge with endocytic vesicles that are forming on the plasma membrane. Under these circumstances, FBPase may be targeted to the plasma membrane.
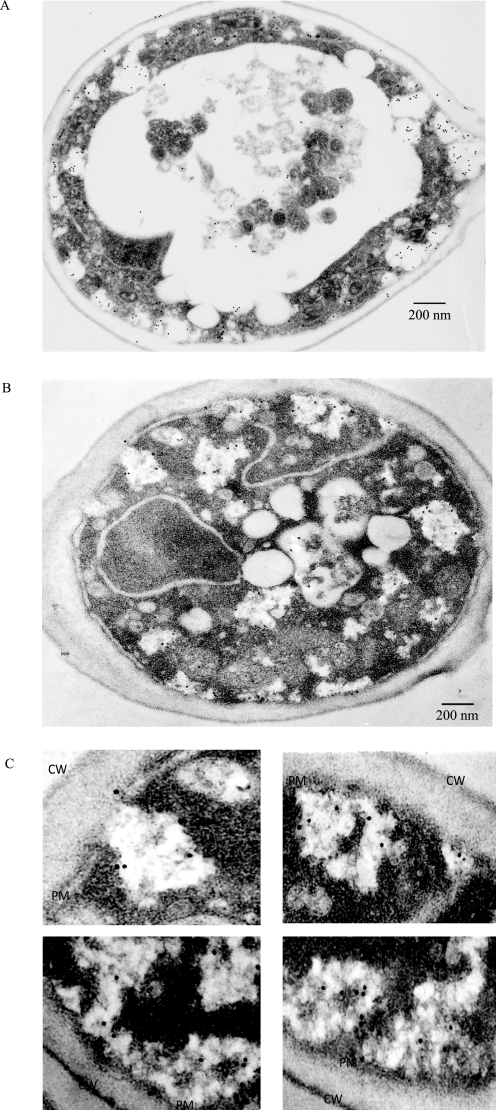
To distinguish between the above possibilities, we first examined the distribution of FBPase at the ultrastructural level. Wild-type (Fig. 1A) and Δpep4 (Fig. 1B) cells were starved of glucose and transferred to media containing fresh glucose. FBPase distribution was examined by immunoelectron microscopy using affinity-purified anti-FBPase antibodies followed by secondary antibodies conjugated with 10 nm colloid gold particles. We found that FBPase was in multiple locations. However, in this study, we focused on the portion of FBPase that appeared to be associated with the endocytic pathway. Along these lines, a high percentage of FBPase was localized to abundant intracellular structures that had a lower electron density than the cytoplasm (Fig. 1, A and B). These FBPase-positive structures appeared to exhibit irregular shapes, and they were of different sizes. Interestingly, high levels of FBPase were inside these structures.
FIGURE 1.
Immunoelectron microscopy studies of FBPase distribution in wild-type and Δpep4 cells following the addition of glucose. A, wild-type cells were glucose-starved and transferred to media containing fresh glucose for 15 min. Cells were fixed and embedded. Immuno-EM was performed on thin sections of cells using affinity-purified FBPase antibodies followed by goat anti-rabbit antibodies conjugated with 10 nm colloid gold. B, Δpep4 cells were glucose-starved and transferred to media containing glucose for 15 min. Cells were fixed, and immuno-EM was performed on thin sections of cells. C, enlarged views of FBPase distribution in areas near the plasma membrane. CW, cell wall; PM, plasma membrane.
A significant amount of FBPase was also found closely associated with the plasma membrane (Fig. 1, A–C). Fig. 1C shows enlarged views of FBPase-positive structures in regions near the plasma membrane. In some cases, FBPase was found in areas that appeared to be connected directly to the periplasm (Fig. 1C, upper left panel). These FBPase-associated structures had irregular shapes, and they were not homogeneous in density. Inside these compartments, small dark structures appeared to intermix with light structures. Based on FBPase distribution at the ultrastructural level, we suspected that at least a percentage of FBPase is targeted to areas near the plasma membrane or in regions that form endocytic vesicles. These studies also suggest that the Vid pathway utilizes early steps of the endocytic pathway to deliver FBPase to the vacuole for degradation.
Isolation of Intracellular Structures that Contain FBPase, Vid24p, and Pep12p
We next set out to purify these FBPase-containing structures. Previously we demonstrated that FBPase is resolved in four distinct peaks, when Δpep4 cells were transferred from low to high glucose media for 15 min (42). The first peak (fractions 11–14) co-localized with the plasma membrane marker Pma1p (35). The second (fractions 15–18) and the third (fractions 19–22) peaks co-localized with the endosomal marker Pep12p (42). The last peak (fractions 23–30) contained small vesicles of 30–50 nm that we termed Vid (vacuole import and degradation) vesicles. These Vid vesicles were subsequently purified from wild-type cells (41) and appeared as small round structures with defined membranes.
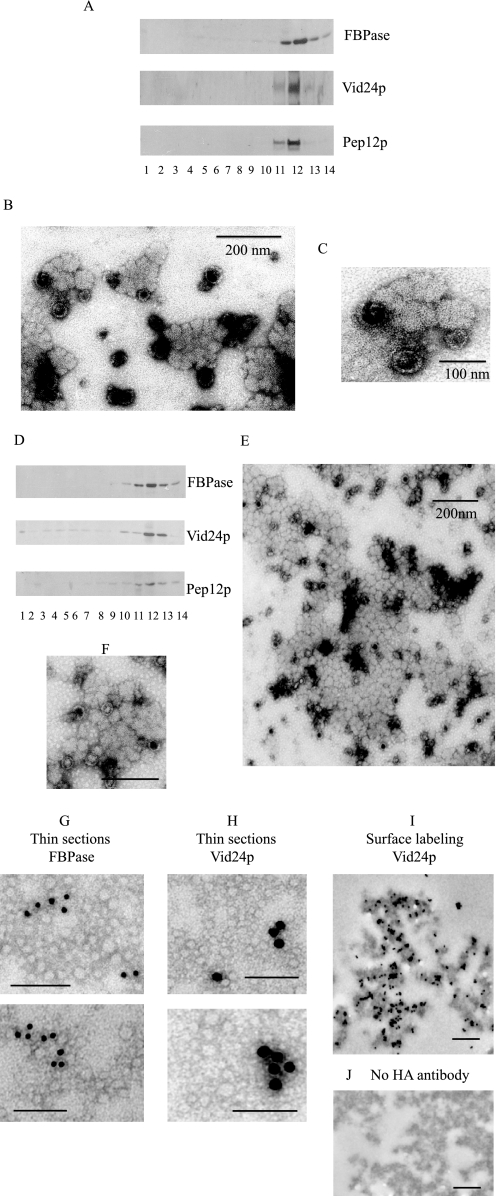
In the present study, we attempted to purify intracellular compartments that contained both FBPase and Pep12p. Wild-type cells expressing HA-Vid24p were transferred to media containing fresh glucose for 15 min. The high speed pellets (100,000 × g pellet) were re-suspended and passed over an S-1000 column. The FBPase- and Pep12p-containing fractions 19–22 were pooled and centrifuged at 100,000 × g. The high speed pellets were re-suspended and subjected to 20–70% sucrose gradient purification. Fraction 12 contained most of the FBPase and was pooled from the first sucrose density gradient, re-centrifuged, and separated using a second sucrose density gradient. On the second gradient, the majority of FBPase was distributed at the 50–70% sucrose density interface (fraction 12, Fig. 2A). These fractions also contained Vid24p and Pep12p. When FBPase-enriched structures (fraction 12) from the second sucrose gradients were examined by negative staining under EM, they were irregular in shape and they varied in size (Fig. 2, B and C).
FIGURE 2.
Isolation of intracellular structures that contain FBPase, Vid24p, and Pep12p. A, wild-type cells expressing HA-Vid24p were transferred to media containing glucose for 15 min. Cells were spheroplasted and subjected to differential centrifugation. The 100,000 × g pellets were re-suspended and further fractionated over an S-1000 column. Fractions 19–22 were pooled and centrifuged at 100,000 × g. Pellets were re-suspended and subjected to two sequential 20–70% sucrose density gradients. On the second gradient, the distribution of FBPase, HA-Vid24p, and Pep12p was determined by Western blotting with anti-FBPase, anti-HA, and anti-Pep12p antibodies. B, fraction 12 from the second sucrose gradient was collected and examined by negative staining and visualized by electron microscopy. C, enlarged views of organelles isolated from fraction 12. D, fractions 15–18 were collected from the S-1000 column and subjected to two sequential sucrose density gradients. The distribution of FBPase, HA-Vid24p, and Pep12p was examined by Western blotting. E, fraction 12 was collected and examined by electron microscopy using negative staining. F, enlarged views of FBPase-containing structures purified from fraction 12. Bars equal 200 nm. G, FBPase-containing organelles were purified, fixed, and embedded in LR White. Thin sections were incubated with anti-FBPase antibodies followed by goat anti-rabbit secondary antibodies conjugated with 10 nm gold particles. H, thin sections were incubated with anti-HA antibodies followed by goat anti-mouse secondary antibodies conjugated with 15 nm gold particles. I, FBPase-containing organelles were purified, fixed, and incubated with anti-HA antibodies followed by anti-mouse antibodies conjugated with 10 nm gold particles. J, control samples were processed the same way as in I but in the absence of anti-HA antibodies. Bars equal 200 nm.
Because materials in fractions 15–18 and fractions 19–22 migrated at different positions on the S-1000 column, they might represent different types of organelles. Alternatively, they may be similar types of structures that vary in size. To address this issue, fractions15–18 were pooled from the S-1000 column and purified over two sequential sucrose gradients using the same protocol mentioned above. The majority of FBPase, Vid24p, and Pep12p were distributed at the 50–70% sucrose density interface (Fig. 2D). At the ultrastructural level, they appeared very similar to those isolated from fractions 15–18 (Fig. 2, B and C). They had irregular shapes, and they contained both light and dark structures that appeared to be mixed together (Fig. 2, E and F).
If Vid vesicles are present at the periphery of the FBPase-containing organelles, then Vid24p should be detected at the periphery of these structures. To test this, wild-type cells expressing HA-Vid24p were starved of glucose and refed with glucose for 15 min. FBPase-containing structures were purified and fixed. Samples were processed and embedded. Immunogold labeling was carried out on thin sections of embedded FBPase-containing structures. For FBPase staining, affinity-purified FBPase antibodies were used, followed by goat anti-rabbit antibodies conjugated with 10 nm gold particles. Vid24p was stained with anti-HA antibodies followed by goat anti-mouse antibodies conjugated with 15 nm gold particles. Although the overall morphology was somewhat compromised following the processing and embedding, both FBPase (Fig. 2G) and Vid24p (Fig. 2H) were found inside these FBPase-containing structures.
We next performed surface labeling of Vid24p on purified FBPase-containing organelles. Samples were purified from the sucrose density gradient and fixed. They were then incubated with or without anti-HA antibodies followed by secondary antibodies conjugated with 10 nm gold particles. Again, the overall morphology was compromised following processing and incubations. However, HA staining appeared in multiple areas on the surfaces of these FBPase-containing structures (Fig. 2I). There was minimal HA staining in control samples without the HA antibodies incubation (Fig. 2J).
FBPase Levels in Vid Vesicle Fractions Are Reduced in Mutants That Block Early Steps of Endocytosis
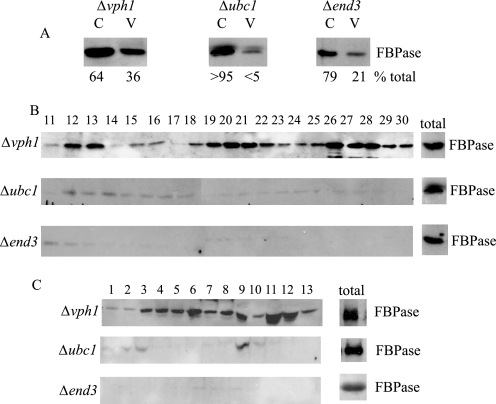
Because FBPase was observed near the plasma membrane, we next asked whether early steps of endocytosis were needed for the Vid pathway. For these experiments, FBPase distribution was examined in the Δend3 mutant. End3p interacts with Sla1p and the actin nucleation factor Pan1p to promote actin assembly at actin patches (42). Accordingly, the deletion of this gene blocks early steps of endocytosis. The Δvph1 mutant was used as a positive control. This mutant blocks FBPase trafficking at a late step, thereby accumulating FBPase in intermediate compartments following a glucose addition (42). By contrast, cells lacking the UBC1 (ubiquitin conjugating enzyme) gene were used as negative controls, because this mutation reduces the level of FBPase with the Vid vesicle fraction (38). Cells were transferred to media containing fresh glucose, and lysates were subjected to differential centrifugation. Using this method, Vid vesicles are enriched in the 200,000 × g pellet (V), whereas cytosol is enriched in the 200,000 × g supernatant (C). In the Δvph1 mutant, a portion of FBPase was in the Vid vesicle fraction (Fig. 3A). By contrast, FBPase level in the Vid vesicle fraction was reduced in the Δubc1 mutant (Fig. 3A). In the Δend3 mutant, FBPase level in Vid vesicle fraction was also low (Fig. 3A). Note that, in the Δubc1 and Δend3 cells, most of the FBPase was in the cytosolic fractions. Therefore, these results suggest that early steps of endocytosis are needed for FBPase association with Vid vesicles.
FIGURE 3.
FBPase levels in the Vid vesicle fraction are reduced in Δend3 mutants that block early steps of endocytosis. A, cells lacking VPH1, UBC1, and END3 were shifted to glucose for 15 min, and cell lysates were subjected to differential centrifugation. FBPase distribution in the Vid vesicle-enriched fraction (V, 200,000 × g pellet) and cytosolic fraction (C, 200,000 × g supernatant) was examined by Western blotting with anti-FBPase antibodies. Levels of FBPase in C and V fractions were quantitated by NIH ImageJ software. B, the same cells were transferred to media containing fresh glucose for 15 min and subjected to differential centrifugation. The 100,000 × g pellets were re-suspended and further separated by S-1000 columns. Fractions were collected, and the distribution of FBPase was examined. C, the same cells were glucose-starved and re-fed with fresh glucose for 15 min. Cell lysates were centrifuged by differential centrifugation. The 100,000 × g pellets were re-suspended and loaded onto 20–70% sucrose density gradients. Fractions were precipitated by trichloroacetic acid, and the distribution of FBPase was examined.
Because FBPase distribution in the plasma membrane, endosome, and Vid vesicle fractions can be resolved on the S-1000 column, we used this method to examine FBPase association with these compartments. If Vid vesicle function is linked to early steps of endocytosis, then the Δend3 mutant should reduce FBPase levels in Vid vesicle fraction. Furthermore, FBPase levels in the putative endosomal fraction should also be low in the mutant. By contrast, if Vid vesicle function is not linked to plasma membrane internalization, then the amounts of FBPase in the Vid vesicle fraction should not be affected in the Δend3 mutant. However, FBPase levels in putative endosomal fractions should be inhibited in this strain. In the Δvph1 mutant, a significant amount of FBPase was in plasma membrane, endosomes, and Vid vesicle fractions (Fig. 3B). The Δvph1 mutant blocks a late step of the endocytic pathway, perhaps resulting in the accumulation of endosomes with different sizes. Therefore, they cannot be separated clearly into two distinct peaks. The Δubc1 mutant had decreased amounts of FBPase in Vid vesicle fractions. FBPase levels in the plasma membrane fraction and endosomal fraction were also reduced in this mutant. Likewise, FBPase levels in the plasma membrane, endosomes, and Vid vesicles fractions were reduced in the Δend3 mutant (Fig. 3B).
The above materials were also examined using sucrose density gradient centrifugation (Fig. 3C). For the Δvph1 mutant, a number of FBPase-containing peaks were observed on the sucrose density gradient, indicating that FBPase is associated with multiple compartments. By contrast, FBPase levels were low in all fractions on the sucrose density gradients in the Δubc1 mutant and the Δend3 mutant (Fig. 3C). Taken together, these results showed that FBPase levels in the Vid vesicle fractions were affected when early steps of endocytosis were blocked. Thus, we suggest that these early steps are necessary for FBPase association with Vid vesicles.
FBPase Degradation Is Inhibited in Endocytosis Mutants That Affect Actin Polymerization
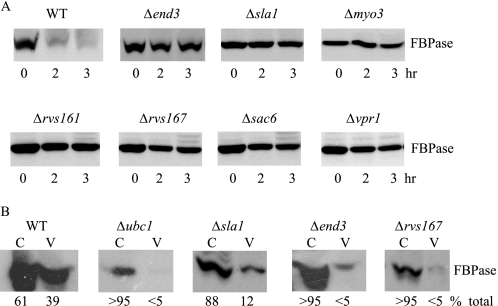
In yeast, endocytic vesicles are formed on the plasma membrane. These vesicles utilize actin polymerization to pinch off from the plasma membrane. Actin polymerization can be divided into multiple steps (48, 51, 52). Molecules such as Las17p and Sla1p are recruited early on in the process. Myo3p and Myo5p are recruited at actin patches at a later step (53, 54). Fission of endocytic vesicles also requires two amphiphysins Rvs161p and Rvs167p (End6p) (55). Assembled F-actin is then cross-linked by Sac6p (fimbrin) (56). When select mutants that affect actin polymerization at different steps were examined, they all inhibited FBPase degradation (Fig. 4A). Thus, this implies a role for actin polymerization in the FBPase degradation pathway.
FIGURE 4.
FBPase degradation is defective in mutants that block early steps of endocytosis. A, wild-type and various mutant cells that block early steps of endocytosis were grown in low glucose media. Cells were transferred to media containing fresh glucose for 0, 2, and 3 h, and FBPase degradation was examined. B, wild-type and endocytosis mutants were transferred from low to high glucose for 15 min. Cell lysates were subjected to differential centrifugation and FBPase distribution in the cytosolic (C) and Vid vesicle (V)-enriched fractions was examined. FBPase levels in these fractions were quantitated using ImageJ software (National Institutes of Health).
We next examined which step of the FBPase targeting pathway was affected by these endocytosis mutants. If FBPase association with Vid vesicles is blocked, then most of FBPase should remain in the cytosol in these mutants. However, if FBPase association with Vid vesicles is not affected, then FBPase should be in the Vid vesicle-enriched fractions. Wild-type and mutant cells were transferred to media containing fresh glucose for 15 min, and lysates were subjected to differential centrifugation. In wild-type cells, FBPase was in Vid vesicle-enriched fraction and also in the cytosolic fraction (Fig. 4B). In contrast, the majority of FBPase was in the cytosolic fraction in the Δsla1 and Δend3 mutants that block actin polymerization at an early step as well as in the Δrvs167 mutant that blocks fission of endocytic vesicles at a later step. Thus, these results suggest that FBPase association with Vid vesicles was inhibited when actin polymerization was affected in these mutants.
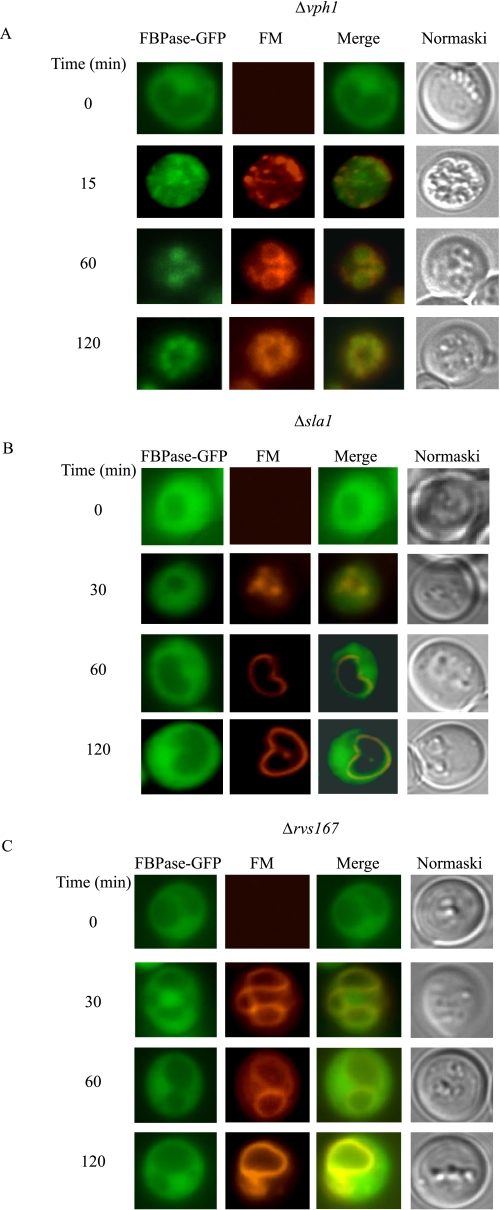
To confirm the above results, we examined the distribution of a GFP-tagged form of FBPase expressed in Δsla1, Δrvs167, and Δvph1 cells and expressed under its own promoter. Cells were transferred from low glucose to high glucose media in the presence of the FM4-64 dye. This dye is taken up from the plasma membrane, internalized into endosomes, and delivered to the vacuole membrane (57). As expected, glucose starvation induced a high level of FBPase-GFP expression in the cytosol in Δvph1 cells. However, following the addition of glucose, a high percentage of FBPase was in endosomes that were also labeled with FM dye (Fig. 5A). In the Δsla1 (Fig. 5B) and Δrvs167 (Fig. 5C) mutants, FBPase was in the cytosol before and after the addition of glucose. Thus, FBPase association with Vid vesicles appeared to be impaired when actin polymerization was blocked.
FIGURE 5.
FBPase remains in the cytosol when early steps of endocytosis are blocked. The Δvph1 cell expressing FBPase-GFP (A), Δsla1 cells expressing FBPase-GFP (B), and Δrvs167 cells expressing FBPase-GFP (C) were grown in low glucose media. Cells were transferred to media containing fresh glucose in the presence of FM for the indicated times. FBPase-GFP, FM, and cells were visualized using fluorescence microscopy.
FBPase and MDH2 Are Targeted to Actin Patches
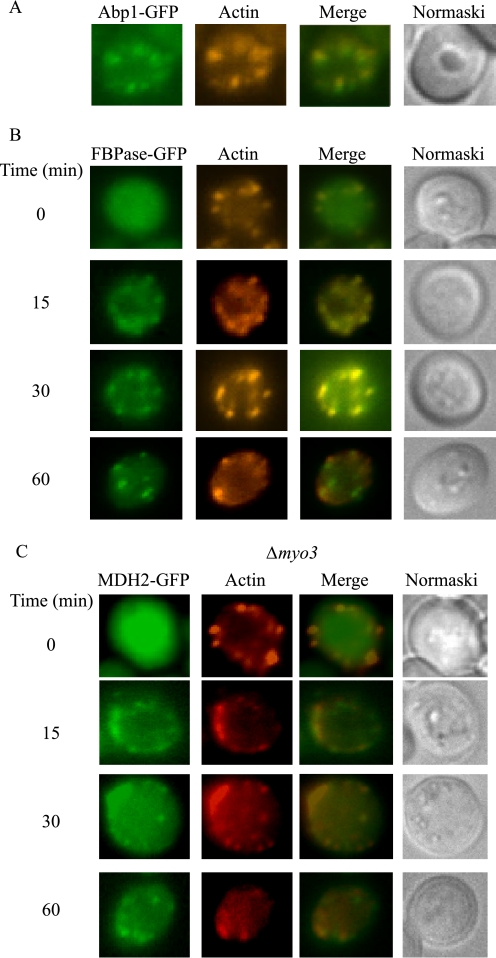
Our above studies indicated a role for actin polymerization in the Vid pathway. Therefore, we next tested whether cargo proteins were targeted to actin patches. Wild-type cells expressing FBPase-GFP were starved and re-fed with fresh glucose. Cells were fixed, and the localizations of GFP-tagged proteins with actin patches were visualized by fluorescence microscopy. We first verified this protocol using a wild-type strain expressing Abp1p-GFP that was grown under our conditions. Abp1p is an actin-binding protein that localizes to actin patches. As expected, this protein co-localized with rhodamine-conjugated phalloidin (Fig. 6A). Note that this protocol appeared to compromise the structures of organelles. For example, under our growth conditions, the vacuole usually appears large, and it exhibits indentations that are visible under Nomarski optics. However, the vacuole was not readily seen clearly using this procedure. Nevertheless, this protocol allowed us to study the localization of GFP with actin patches at the same time. When cells were maintained under glucose starvation conditions, the majority of FBPase was not associated with actin patches (Fig. 6B). However, a fraction of FBPase was observed in areas that stained positively with actin patches following the addition of glucose for 15 and 30 min (Fig. 6B). Less co-localization of FBPase and actin patches were seen at t = 60 min of glucose addition.
FIGURE 6.
FBPase and MDH2 associate with actin patches following the addition of glucose. A, wild-type cells expressing Abp1p-GFP were grown in YPKG media for 3 days. The localization of Abp1p-GFP with rhodamine-conjugated phalloidin was examined. B, wild-type cells expressing FBPase-GFP were transferred to media containing fresh glucose for the indicated times. Cells were fixed and processed as described under “Experimental Procedures.” Actin was stained with phalloidin conjugated with rhodamine. FBPase, actin, and cells were visualized by fluorescence microscopy. C, MDH2-GFP was expressed in Δmyo3 cells that were transferred to media containing glucose for 0–60 min. MDH2 and actin localization was examined by fluorescence microscopy.
We next examined the distribution of actin and the MDH2 cargo protein in wild-type cells (supplemental Fig. S1). Transient association of this protein with actin patches was observed following the addition of glucose for 15–30 min. Less co-localization of MDH2 with actin patches was seen at t = 60 min (supplemental Fig. S1). Association of MDH2 with actin patches was also seen in myo3 mutants that block a late step of actin assembly (Fig. 6C).
Sec28p Is Present at the Cell Periphery in Δsla1 and Δrvs167 Mutants
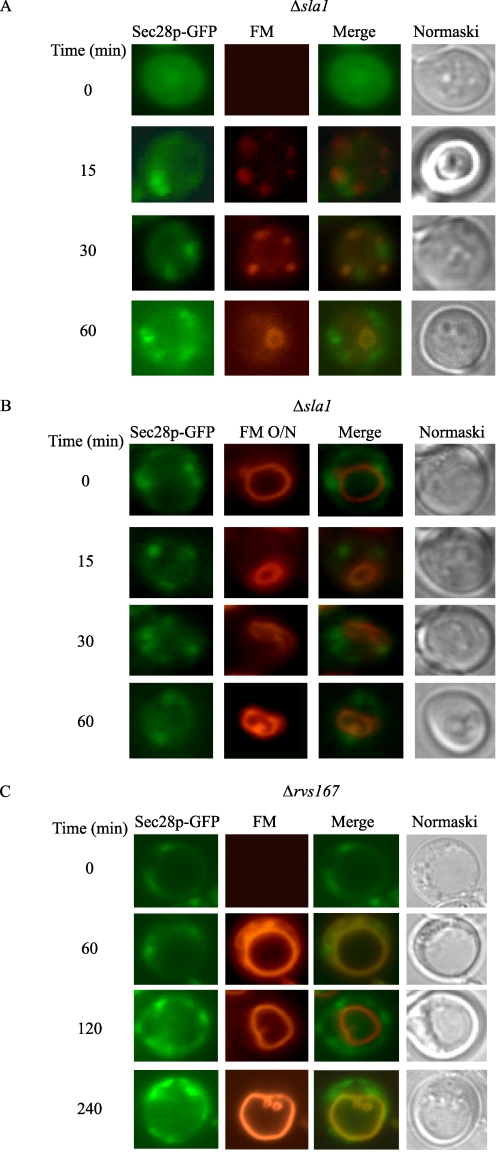
Having established that cargo proteins were targeted to actin patches on the plasma membrane, we next examined whether Vid vesicles were also present on the plasma membrane. If Vid vesicles are on the plasma membrane, then coatomer proteins may also be at this location, because these proteins are components of Vid vesicles. Coatomer proteins have been observed at the Golgi and endosomes (58–65), but there are no reports of these proteins at the cell periphery. One possible explanation is that coatomer proteins are involved in multiple trafficking pathways, and they move quickly from one compartment to the next. Therefore, to enrich for coatomer proteins on the cell periphery, we expressed Sec28p-GFP in Δsla1 and Δrvs167 mutants. When glucose was added to the Δsla1 mutant in the presence of FM, Sec28p was observed in punctate structures (Fig. 7A). Note, however, that they did not show obvious co-localization with FM.
FIGURE 7.
Sec28p is present at the cell periphery in the Δsla1 and Δrvs167mutants. A, Sec28p-GFP was expressed in Δsla1 cells. These cells were re-fed with glucose in the presence of FM. Sec28p localization with FM was examined. B, Δsla1 cells were incubated with FM for 16 h. Cells were then shifted to glucose in the absence of FM. The distribution of Sec28p and FM was examined. C, cells lacking the RVS167 gene were transformed with Sec28p-GFP. Cells were re-fed with glucose in the presence of FM for the indicated times. The distribution of Sec28p and FM was examined.
In a previous study, we observed Sec28p-GFP associated with retrograde vesicles in the ret2-1 mutant when the vacuole was prelabeled with FM (42). Thus, we next determined whether the above Sec28p-GFP punctate structures associated with retrograde vesicles originating from the vacuole. When the vacuoles were prelabeled in the Δsla1 mutant, little association of Sec28p with the vacuole membrane was seen (Fig. 7B). Most of the Sec28p dots appeared to be closely associated with the cell periphery.
We next examined Sec28p-GFP distribution in Δrvs167 mutants that were transferred to media containing high glucose in the presence of FM (Fig. 7C). Again, most of Sec28p was found in punctate structures that appeared to be associated with the cell periphery in this mutant.
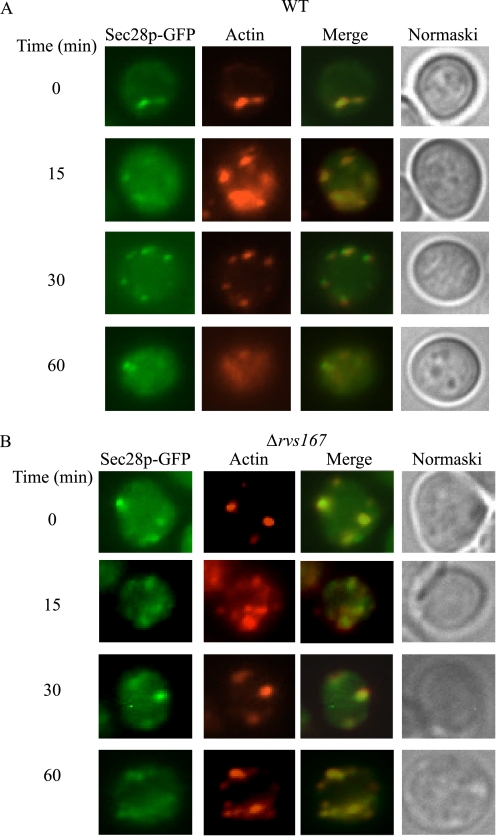
Sec28p Is Associated with Actin Patches
Because Sec28p appeared on the cell periphery in the Δsla1 and Δrvs167 mutants (Fig. 7), we next tested whether Sec28p co-localized with actin patches (Fig. 8A). Wild-type cells expressing Sec28p-GFP were transferred from low glucose- to high glucose-containing media for the indicated times. Sec28p co-localized with actin patches at t = 0 min. Co-localization was also observed following the addition of glucose for 15–30 min. Less co-localization was seen at 60 min.
FIGURE 8.
Sec28p co-localizes with actin patches in wild-type cells. A, wild-type cells expressing Sec28p-GFP were transferred to media containing fresh glucose for 0–60 min. The distribution of Sec28p and actin patches was examined. B, the Δrvs167 mutant was transformed with Sec28p-GFP and grown in low glucose media. Cells were transferred to media containing fresh glucose for the indicated time. The distribution of Sec28p and actin patches was examined.
RVS167 (END6) is involved in the scission of endocytic vesicles from the plasma membrane. Therefore, endocytic vesicles are formed in this mutant, but they are not released from the plasma membrane. If Sec28p vesicles utilized actin polymerization to pinch off from the plasma membrane, then Sec28p association with actin patches may be prolonged in cells lacking this gene. To test this, Sec28p association with actin patches was examined in the Δrvs167 mutant expressing Sec28p-GFP (Fig. 8B). Sec28p co-localized with actin patches at t = 0, 15, and 30 min. A significant amount of Sec28p remained associated with actin patches at t = 60 min.
Vid24p Is Associated with Actin Patches
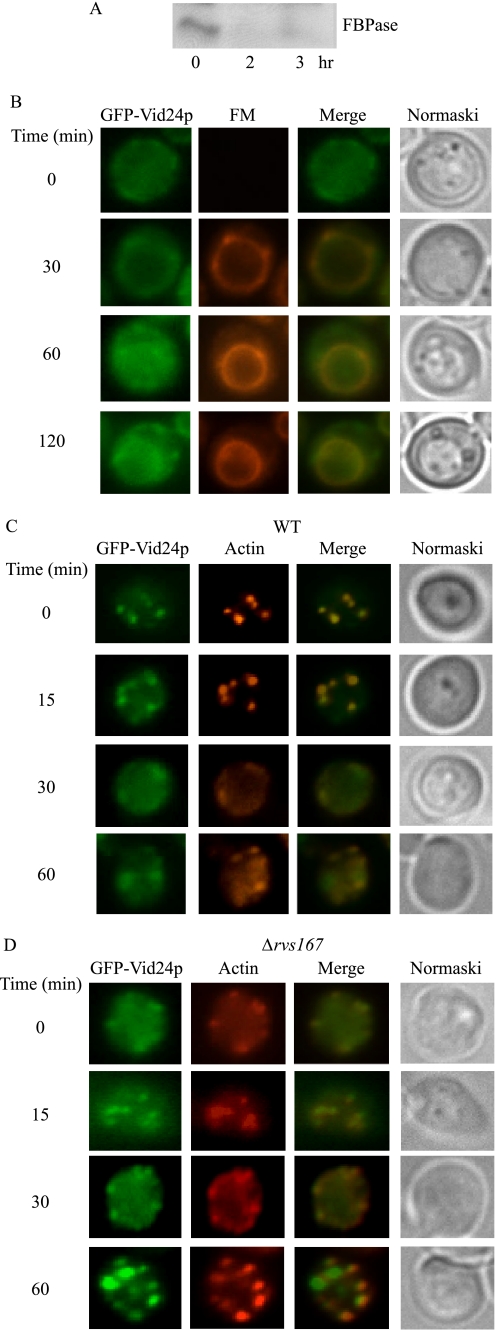
Unlike Sec28p, which is present in multiple locations, Vid24 is a specific Vid vesicle marker (39). Thus, we next examined the localization of Vid24p in wild-type cells expressing a GFP form of this protein. Previously, we attempted to tag GFP or RFP at the C terminus of Vid24p and integrate this into the VID24 gene. Unfortunately, Vid24p-GFP or Vid24p-RFP protein levels were below detection. As an alternative, we produced a construct that contained the VID24 promoter and the open reading frame. GFP was then fused in-frame with Vid24p at the N terminus. This construct was integrated into the promoter of VID24 gene so that the expression of GFP-Vid24p was under the control of the VID24 promoter. GFP-Vid24p was expressed in wild-type cells that were starved for 3 days and refed with fresh glucose. To verify the function of the tagged Vid24p protein, we performed an FBPase degradation experiment. FBPase was degraded normally in these cells (Fig. 9A), indicating that this protein is functional in FBPase degradation.
FIGURE 9.
Vid24p co-localizes with actin patches in wild-type cells. A, GFP-Vid24p was transformed into wild-type cells that were grown in low glucose media for 3 days and re-fed with glucose for 0, 2, and 3 h. FBPase degradation was examined. B, wild-type cells expressing GFP-Vid24p were transferred to media containing fresh glucose in the presence of FM for the indicated times. Vid24p distribution with FM was examined. C, wild-type cells expressing GFP-Vid24p were glucose-starved and then transferred to glucose containing media. The distribution of GFP-Vid24p with actin patches was examined by fluorescence microscopy. D, GFP-Vid24p was expressed in Δrvs167 cells that were grown in low glucose media. Glucose was then added to these cells for 0–60 min. Vid24p and actin patches were examined using fluorescence microscopy.
We next examined the distribution of GFP-Vid24p. Wild-type cells expressing GFP-Vid24p were transferred to media containing fresh glucose in the presence of FM (Fig. 9B). At t = 0 min, Vid24p levels were low, although a portion of this protein was seen at the cell periphery. Following the addition of glucose, a percentage of Vid24p co-localized with FM-containing endosomes. Some of these endosomes appeared to be in a process of fusion with the vacuole. At later time points, Vid24p was seen on the vacuole membrane and also on the plasma membrane.
When Vid24p and actin distribution were examined, a significant amount of Vid24p was associated with actin patches at t = 0, 15, and 30 min (Fig. 9C). Co-localization was less obvious at t = 60 min, suggesting that most of the Vid24p dissociated from actin patches at this time point.
If actin polymerization is needed for endocytic vesicles to pinch off from the plasma membrane, then Vid24p association with actin patches may be prolonged in Δrvs167 cells. GFP-Vid24p was expressed in the Δrvs167 cells, and the co-localization of GFP-Vid24p with actin patches was examined (Fig. 9D). A substantial amount of Vid24p co-localized with actin patches at t = 0, 15, and 30 min. High levels of GFP-Vid24p remained associated with actin patches at t = 60 min. Thus, the absence of this gene prolongs the association of Vid24p with actin patches.
DISCUSSION
In a previous study, we showed that the FBPase degradation pathway utilizes the endocytic pathway (42). However, the exact point at which the Vid pathway and the endocytic pathway converge is unknown. Vid vesicles may merge with endocytic vesicles as they are forming on the plasma membrane. Alternatively, Vid vesicles may merge with endosomes that are released into the cytoplasm. Based on our immuno-EM studies, we suggest that FBPase is targeted to the endocytic pathway at early steps, perhaps at the point of endocytic vesicle formation. Along these lines, FBPase was found associated with regions underneath the plasma membrane. In some cases, FBPase was in compartments that were connected to the periplasm. Our immuno-EM studies indicated that FBPase was also associated with large irregularly shaped structures.
These FBPase-containing organelles were purified using S-1000 column chromatography followed by two sucrose density gradients. When thin sections of these FBPase-containing organelles were incubated with anti-FBPase and anti-HA antibodies, both Vid24p and FBPase were found inside these structures. Furthermore, Vid24p was distributed in multiple areas on the surface of these FBPase-containing structures. Therefore, we suggest that Vid vesicles cluster or aggregate together to form large FBPase-containing structures.
When fractionation studies were conducted using the Δend3 mutant that blocks early steps of endocytosis, low levels of FBPase were in the plasma membrane, endosome, or Vid vesicle fractions. A reduction of the levels of FBPase in the Vid vesicle fraction in this mutant was unexpected. These results suggest that FBPase association with Vid vesicles is linked to early steps of endocytosis.
In yeast, endocytic vesicles use actin polymerization to pinch off from the plasma membrane. We examined a number of mutants that affect actin polymerization at different steps, and all of these blocked FBPase degradation. FBPase was found in the cytosol when early steps of endocytosis were blocked, suggesting actin polymerization is needed for FBPase association with Vid vesicles. Further supporting this idea was our observation that the cargo proteins FBPase and MDH2 were targeted to areas that form actin patches. These cargo proteins showed little association with actin patches at t = 60 min, suggesting that most of them dissociate from actin patches at this time point. In contrast, the co-localization of MDH2 with actin patches was prolonged in the Δmyo3 strain. This suggests that the absence of this gene prolongs the association of MDH2 with actin patches.
A similar pattern of actin association and dissociation was observed with Sec28p and Vid24p. Both of these proteins were associated with actin patches at t = 0, 15, and 30 min. However, less co-localization was seen at t = 60 min, suggesting that Sec28p and Vid24p dissociate from actin patches following the addition of glucose for 60 min. In cells lacking the RVS167 gene, both Sec28p and Vid24p remained co-localized with actin patches at t = 60 min. Thus, this suggests that RVS167 is needed for these proteins and actin patches to dissociate.
Co-localization of Vid24p and Sec28p with actin patches at t = 0 min suggests that these proteins are present in actin patches under glucose starvation conditions. However, cargo proteins were not seen in actin patches until 15–30 min after the addition of glucose. We suggest that Vid vesicles are clustered at actin patches during periods of glucose starvation. Cargo proteins arrived at these sites shortly following the addition of glucose. Then, actin polymerization may be used to pinch these vesicles off from the plasma membrane.
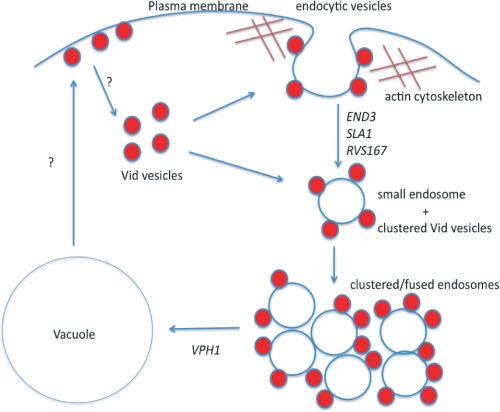
Based upon our results, we propose the following model (Fig. 10). During glucose starvation, Vid24p and Sec28p are present at actin patches where endocytic vesicles are forming from the plasma membrane. Following the addition of glucose, FBPase and MDH2 are then targeted to actin patches. Endocytic vesicles are released into the cytoplasm to become small endosomes. Vid vesicles cluster or aggregate to form large, irregularly shaped structures that also contain the endosomal marker. These endosomes may also receive membrane influx from other trafficking pathways such as the Vps sorting pathway from the Golgi. In the Δvph1 mutant, FM was seen at the periphery of large endosomes. Therefore, large endosomes may be surrounded by a membrane layer. Vid vesicles may attach to endocytic vesicles that are forming on the plasma membrane. Vid vesicles may continue to attach to endosomes that are released into the cytoplasm. It is also possible that Vid vesicles fuse directly with the vacuole. We suggest that the Vid pathway utilizes a specialized endocytic pathway. This pathway may provide an effective way to remove molecules from the media as well as to remove molecules from the interiors.
FIGURE 10.
The Vid pathway utilizes a specialized endocytic pathway. During glucose starvation, Vid vesicle proteins are present at actin patches where endocytic vesicles are forming from the plasma membrane. Following the addition of glucose, cargo proteins arrive at these actin patches. Endocytic vesicles use actin polymerization to pinch off from the plasma membrane. Small endosomes that are released from the plasma membrane may cluster or fuse together to form large endosomes. These large endosomes then fuse with the vacuole. Vid vesicles may attach to endocytic vesicles that are forming from the plasma membrane. Vid vesicles may continue to attach to endosomes that are released into the cytoplasm. It is possible that Vid vesicles fuse directly with the vacuole. We suggest that Vid vesicles are formed from different regions of the plasma membrane. They may be derived from retrograde vesicles coming from the vacuole membrane. These retrograde vesicles travel to the plasma membrane and then become Vid vesicles.
Supplementary Material
Acknowledgments
Electron microscopy was performed at the EM facility at Harvard Medical School and the EM facility at the Pennsylvania State College of Medicine. Primers used in this study were synthesized in the Core Facility at the Pennsylvania State College of Medicine. Pep12p antibodies were obtained from Dr. S. Emr (University of California, San Diego).
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM 59480 (to H.-L. C.). This work was also supported by a grant from the Pennsylvania Tobacco Settlement Fund (to H.-L. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- FBPase
- fructose-1,6-bisphosphatase
- MDH2
- malate dehydrogenase 2
- Vid
- vacuole import and degradation
- GFP
- green fluorescent protein
- RFP
- red fluorescent protein
- HA
- hemagglutinin
- EM
- electron microscopy
- FM
- 4-64[FM-4-64, FM4-64, N-(3 triethylammonium propyl)-4-(4-diethylaminophenylhexatrienyl) pyridinium dibromide].
REFERENCES
- 1.Goldberg A. L. (2007) Biochem. Soc. Trans. 35, 12–17 [DOI] [PubMed] [Google Scholar]
- 2.Hanna J., Finley D. (2007) FEBS Lett. 581, 2854–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varshavsky A. (2005) Trends Biochem. Sci. 30, 283–286 [DOI] [PubMed] [Google Scholar]
- 4.Seglen P. O., Gordon P. B., Holen I. (1990) Semin. Cell Biol. 1, 441–448 [PubMed] [Google Scholar]
- 5.Neely A. N., Cox J. R., Fortney J. A., Schworer C. M., Mortimore G. E. (1977) J. Biol. Chem. 252, 6948–6954 [PubMed] [Google Scholar]
- 6.Mortimore G. E., Hutson N. J., Surmacz C. A. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 2179–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortimore G. E., Neely A. N., Cox J. R., Guinivan R. A. (1973) Biochem. Biophys. Res. Commun. 54, 89–95 [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N., Klionsky D. J. (2007) Annu. Rev. Nutr. 27, 19–40 [DOI] [PubMed] [Google Scholar]
- 10.Cecconi F., Levine B. (2008) Dev. Cell 15, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang H. L., Dice J. F. (1988) J. Biol. Chem. 263, 6797–6805 [PubMed] [Google Scholar]
- 12.Chiang H. L., Terlecky S. R., Plant C. P., Dice J. F. (1989) Science 246, 382–385 [DOI] [PubMed] [Google Scholar]
- 13.Cuervo A. M., Dice J. F. (1996) Science 273, 501–503 [DOI] [PubMed] [Google Scholar]
- 14.Dice J. F. (2007) Autophagy 3, 295–299 [DOI] [PubMed] [Google Scholar]
- 15.Bryant N. J., Stevens T. H. (1998) Microbiol. Mol. Biol. Rev. 62, 230–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones E. W. (1991) J. Biol. Chem. 266, 7963–7966 [PubMed] [Google Scholar]
- 17.Klionsky D. J., Herman P. K., Emr S. D. (1990) Microbiol. Rev. 54, 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothman J. H., Stevens T. H. (1986) Cell 47, 1041–1051 [DOI] [PubMed] [Google Scholar]
- 20.Klionsky D. J. (2005) J. Cell Sci. 118, 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W. P., Klionsky D. J. (2002) Cell. Struct. Funct. 27, 409–420 [DOI] [PubMed] [Google Scholar]
- 22.Reggiori F., Klionsky D. J. (2005) Curr. Opin. Cell Biol. 17, 415–422 [DOI] [PubMed] [Google Scholar]
- 23.Wang C. W., Klionsky D. J. (2003) Mol. Med. 9, 65–76 [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn W. A., Jr. (1990) J. Cell Biol. 110, 1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuttle D. L., Dunn W. A., Jr. (1995) J. Cell Sci. 108, 25–35 [DOI] [PubMed] [Google Scholar]
- 26.Rubinsztein D. C. (2006) Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 27.Sarkar S., Rubinsztein D. C. (2008) FEBS J. 275, 4263–4270 [DOI] [PubMed] [Google Scholar]
- 28.Cuervo A. M. (2004) Trends Cell Biol. 14, 70–77 [DOI] [PubMed] [Google Scholar]
- 29.Cuervo A. M., Dice J. F. (2000) J. Biol. Chem. 275, 31505–31513 [DOI] [PubMed] [Google Scholar]
- 30.Rajawat Y. S., Hilioti Z., Bossis I. (2009) Ageing Res. Rev. 8, 199–213 [DOI] [PubMed] [Google Scholar]
- 31.Chiang H. L., Schekman R. (1991) Nature 350, 313–318 [DOI] [PubMed] [Google Scholar]
- 32.Chiang H. L., Schekman R., Hamamoto S. (1996) J. Biol. Chem. 271, 9934–9941 [DOI] [PubMed] [Google Scholar]
- 33.Shieh H. L., Chiang H. L. (1998) J. Biol. Chem. 273, 3381–3387 [DOI] [PubMed] [Google Scholar]
- 34.Brown C. R., McCann J. A., Hung G. G., Elco C. P., Chiang H. L. (2002) J. Cell Sci. 115, 655–666 [DOI] [PubMed] [Google Scholar]
- 35.Hoffman M., Chiang H. L. (1996) Genetics 143, 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown C. R., McCann J. A., Chiang H. L. (2000) J. Cell Biol. 150, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown C. R., Cui D. Y., Hung G. G., Chiang H. L. (2001) J. Biol. Chem. 276, 48017–48026 [DOI] [PubMed] [Google Scholar]
- 38.Shieh H. L., Chen Y., Brown C. R., Chiang H. L. (2001) J. Biol. Chem. 276, 10398–10406 [DOI] [PubMed] [Google Scholar]
- 39.Chiang M. C., Chiang H. L. (1998) J. Cell Biol. 140, 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hung G. C., Brown C. R., Wolfe A. B., Liu J., Chiang H. L. (2004) J. Biol. Chem. 279, 49138–49150 [DOI] [PubMed] [Google Scholar]
- 41.Huang P. H., Chiang H. L. (1997) J. Cell Biol. 136, 803–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown C. R., Wolfe A. B., Cui D., Chiang H. L. (2008) J. Biol. Chem. 283, 26116–26127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toret C. P., Drubin D. G. (2006) J. Cell Sci. 119, 4585–4587 [DOI] [PubMed] [Google Scholar]
- 44.Engqvist-Goldstein A. E., Drubin D. G. (2003) Annu. Rev. Cell Dev. Biol. 19, 287–332 [DOI] [PubMed] [Google Scholar]
- 45.Dewar H., Warren D. T., Gardiner F. C., Gourlay C. G., Satish N., Richardson M. R., Andrews P. D., Ayscough K. R. (2002) Mol. Biol. Cell 13, 3646–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren D. T., Andrews P. D., Gourlay C. W., Ayscough K. R. (2002) J. Cell Sci. 115, 1703–1715 [DOI] [PubMed] [Google Scholar]
- 47.Howard J. P., Hutton J. L., Olson J. M., Payne G. S. (2002) J. Cell Biol. 157, 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaksonen M., Sun Y., Drubin D. G. (2003) Cell 115, 475–487 [DOI] [PubMed] [Google Scholar]
- 49.Adams A. E., Pringle J. R. (1984) J. Cell Biol. 98, 934–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang H. C., Pon L. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evangelista M., Klebl B. M., Tong A. H., Webb B. A., Leeuw T., Leberer E., Whiteway M., Thomas D. Y., Boone C. (2000) J. Cell Biol. 148, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toshima J. Y., Toshima J., Kaksonen M., Martin A. C., King D. S., Drubin D. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5793–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geli M. I., Riezman H. (1996) Science 272, 533–535 [DOI] [PubMed] [Google Scholar]
- 54.Sun Y., Martin A. C., Drubin D. G. (2006) Dev. Cell 11, 33–46 [DOI] [PubMed] [Google Scholar]
- 55.Kaksonen M., Toret C. P., Drubin D. G. (2005) Cell 123, 305–320 [DOI] [PubMed] [Google Scholar]
- 56.Kübler E., Riezman H. (1993) EMBO J. 12, 2855–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vida T. A., Emr S. D. (1995) J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wieland F., Harter C. (1999) Curr. Opin. Cell Biol. 11, 440–446 [DOI] [PubMed] [Google Scholar]
- 59.Duden R. (2003) Mol. Membr. Biol. 20, 197–207 [DOI] [PubMed] [Google Scholar]
- 60.Whitney J. A., Gomez M., Sheff D., Kreis T. E., Mellman I. (1995) Cell 83, 703–713 [DOI] [PubMed] [Google Scholar]
- 61.Piguet V., Gu F., Foti M., Demaurex N., Gruenberg J., Carpentier J. L., Trono D. (1999) Cell 97, 63–73 [DOI] [PubMed] [Google Scholar]
- 62.Daro E., Sheff D., Gomez M., Kreis T., Mellman I. (1997) J. Cell Biol. 139, 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu F., Aniento F., Parton R. G., Gruenberg J. (1997) J. Cell Biol. 139, 1183–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aniento F., Gu F., Parton R. G., Gruenberg J. (1996) J. Cell Biol. 133, 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gabriely G., Kama R., Gerst J. E. (2007) Mol. Cell. Biol. 27, 526–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.