Abstract
Large T antigen (TAg) of the human polyomavirus JC virus (JCV) possesses DNA binding and helicase activities, which, together with various cellular proteins, are required for replication of the viral genome. We now show that JCV-infected cells expressing TAg accumulate in the G2 phase of the cell cycle as a result of the activation of ATM- and ATR-mediated G2 checkpoint pathways. Transient transfection of cells with a TAg expression vector also induced G2 checkpoint signaling and G2 arrest. Analysis of TAg mutants with different subnuclear localizations suggested that the association of TAg with cellular DNA contributes to the induction of G2 arrest. Abrogation of G2 arrest by inhibition of ATM and ATR, Chk1, and Wee1 suppressed JCV genome replication. In addition, abrogation of the G2-M transition by Cdc2 depletion disabled Wee1 depletion-induced suppression of JCV genome replication, suggesting that JCV replication is facilitated by G2 arrest resulting from G2 checkpoint signaling. Moreover, inhibition of ATM and ATR by caffeine suppressed JCV production. The observation that oligodendrocytes productively infected with JCV in vivo also undergo G2 arrest suggests that G2 checkpoint inhibitors such as caffeine are potential therapeutic agents for JCV infection.
Keywords: Cell/Checkpoint, Cell/Cycle, DNA/Damage, Viruses/DNA, JC Virus, Large T Antigen
Introduction
The human polyomavirus JC virus (JCV)2 is the causative agent of progressive multifocal leukoencephalopathy (PML), a fatal demyelinating disease of the central nervous system. JCV infection usually occurs during childhood, but remains subclinical. However, opportunistic reactivation of JCV results in the development of PML in individuals with a compromised immune system, such as those with acquired immunodeficiency syndrome (AIDS) or advanced-stage malignant tumors, or those recently having undergone organ transplantation with immunosuppressive therapy (1). There is currently no effective or specific therapy for PML, even though the incidence of this condition is increasing with the growing number of individuals with AIDS.
JCV is a small virus with a double-stranded DNA genome that encodes early proteins (large T antigen, small t antigen, and T′ antigen) and late proteins (VP1, VP2, VP3, and agnoprotein) (2). The entry of JCV into the nucleus of an infected cell is followed by transcription of the early genes and the production of large T antigen (TAg). The replication of JCV DNA progresses in association with the accumulation of TAg, which subsequently stimulates transcription of late genes and represses that of the early genes (3). JCV TAg shares 72% amino acid sequence identity with TAg of simian virus 40 (SV40), another primate polyomavirus, and, like SV40 TAg, it is necessary for replication of the viral genome as a result of its DNA binding and helicase activities. The replication of polyomaviruses also requires DNA replication proteins of the host cell, such as DNA polymerase α, topoisomerases, and replication protein A (RPA) (4, 5), with such host cell factors being thought to serve as determinants of host specificity (6).
The utilization of such cellular proteins requires that polyomaviruses replicate in a phase of the cell cycle in which they are available. Polyomavirus TAg thus modulates cellular signaling pathways to induce quiescent cells to enter S phase, in which cellular DNA is replicated (7). A key event in this process is the interaction of TAg with members of the retinoblastoma protein family, which results in inactivation of retinoblastoma protein and in consequent progression of the cell cycle (8). Moreover, polyomaviruses are thought to take advantage of the DNA damage response to enhance viral replication. SV40 lytic infection thus triggers the DNA damage checkpoint, resulting in a bypass of mitosis and additional replication of cellular and viral DNA (9), and murine polyomavirus-infected cells accumulate in S and G2 phases (10). Replication of viral DNA in cells infected with SV40 or murine polyomavirus has also been recently shown to activate the ataxia telangiectasia mutated (ATM)-mediated signaling pathway and thereby promote viral replication (11, 12). However, it is not clear how polyomaviruses trigger DNA damage signaling.
DNA damage checkpoints are biochemical pathways that delay or arrest cell cycle progression in response to DNA damage or replication stress. In mammalian cells, the DNA damage checkpoint is usually activated by ATM or ATM- and Rad3-related (ATR), both of which are members of the phosphatidylinositol 3-kinase-like kinase family. ATM is recruited to DNA double strand breaks by the Mre11-Rad50-Nbs1 (MRN) complex (13), whereas ATR is recruited by ATR-interacting protein to RPA-coated single-stranded DNA (ssDNA) that accumulates at stalled DNA replication forks, or is generated by the processing of the initial DNA damage (14). The Rad9-Rad1-Hus1 (9:1:1) complex is also recruited to RPA-coated ssDNA and brings the ATR activator, topoisomerase-binding protein-1. After their recruitment to sites of DNA damage, ATM and ATR phosphorylate various substrate proteins, including Chk1 and Chk2, which in turn phosphorylate downstream factors to induce cell cycle arrest and facilitate DNA repair (16, 17). The G2-M cell cycle transition is controlled by the cyclin-dependent kinase Cdc2 (also known as Cdk1), which is positively regulated by cyclin B and negatively regulated by tyrosine phosphorylation and cyclin-dependent kinase inhibitors (18). In response to ATM- or ATR-mediated activation of the DNA damage checkpoint, the inhibitory phosphorylation of Cdc2 is maintained by the kinase Wee1 and by inactivation of the phosphatase Cdc25 to arrest cells at the G2-M transition and to block entry into mitosis (19). ATM and ATR also phosphorylate p53 at Ser15 (20), resulting in inhibition of the interaction of p53 with Mdm2 (21) and in p53 stabilization. Cdc2 is inhibited by the products of p53 target genes, including the cyclin-dependent kinase inhibitor p21, Gadd45, and 14-3-3σ (22–24). Cells arrested at G2-M in this manner are able to resume cell cycle progression after abrogation of the checkpoint response subsequent to completion of DNA repair, unless the DNA damage is irreparable and leads to apoptosis (25).
We have now investigated the relationship between JCV replication and the host cell cycle to identify the cellular machinery required for JCV replication. We found that JCV TAg triggers the ATM- and ATR-mediated G2 checkpoint pathways and consequent G2 arrest during viral DNA replication. This function of TAg was shown to correlate with its association with cellular DNA. We also show that caffeine, an inhibitor of ATM and ATR function, markedly suppressed JCV replication and propagation after establishment of infection in host cells.
EXPERIMENTAL PROCEDURES
Cells and Virus Infection
Human neuroblastoma IMR-32 cells (HSRRB, Osaka, Japan) were maintained under an atmosphere of 5% CO2 at 37 °C in Dulbecco's minimum essential medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, 2 mm l-glutamine, and 0.1 mm nonessential amino acids. For virus infection, IMR-32 cells were inoculated with JCV (200 HAU/ml), prepared from JCV-producing JCI cells (26) in growth culture medium for 24 h and then transferred to fresh medium without virus.
Plasmids
The JCV TAg expression vectors pCXN2-JCV TAg and pCMV-FLAG-JCV TAg were constructed from pBR-Mad1. The ΔOBD, E461D, E461A, R541K, and K617R mutants of JCV TAg were generated by polymerase chain reaction-mediated mutagenesis of pCMV-FLAG-JCV TAg with appropriate primers. All constructs were verified by sequencing and subcloned into pCXN2. The pCXN2-GFP and pCXN2-red fluorescent protein vectors were also generated as controls. For the assays of viral DNA replication, the full-length transcriptional control region of JCV Mad1 was subcloned into pBluescript II SK(+), and the resulting construct was designated pBS-JCori (27).
Reagents and Antibodies
Caffeine and KU55933 (both from Sigma) were dissolved at 100 and 10 mm, respectively, in deionized water and dimethyl sulfoxide. UCN-01 (Calbiochem, San Diego, CA) was dissolved in 1 mm dimethyl sulfoxide. A mouse monoclonal antibody to SV40 TAg (pAb416), which was previously shown to cross-react with JCV TAg (28), was obtained from Calbiochem. Rabbit polyclonal antibodies to JCV TAg (JCT652), JCV VP1, and JCV Agnoprotein were generated as described (28–30). Rabbit polyclonal antibodies to the Thr21-phosphorylated form of RPA32 were prepared by Immuno-Biological Laboratories (Gunma, Japan) with a thyroglobulin-conjugated synthetic phosphopeptide corresponding to amino acid residues 16 to 29 of the human RPA32 (GAGGYpTQSPGGFGC). The immunoglobulin G fraction specific for the peptide was isolated from serum by chromatography on a column of antigen-coupled activated thiol-Sepharose 4B (GE Healthcare). Antibodies to Chk1, Ser317-phosphorylated Chk1, Chk2, Thr68-phosphorylated Chk2, Tyr15-phosphorylated Cdc2, and Ser15-phosphorylated p53 were obtained from Cell Signaling Technology (Beverly, MA). Additional antibodies included those to ATR (N-19), Wee1 (B-11) and lamin A/C (Santa Cruz Biotechnology, Santa Cruz, CA), Ser139-phosphorylated histone H2A.X (Upstate, Lake Placid, NY), Cdc2 and cyclin B (BD Biosciences), cyclin B1 (Novocastra, Newcastle Upon Tyne, UK), p53 (DO-7; Dako, Carpinteria, CA), topoisomerase-binding protein-1, actin, and HP1α (Chemicon Millipore, Billerica, MA), RPA32 (Neomarkers, Fremont, CA), and FLAG M2 (Sigma).
Flow Cytometry and Cell Cycle Analysis
IMR-32 cells seeded onto 3.5-cm dishes were transfected with the indicated constructs by using Lipofectamine 2000 (Invitrogen) 3 days before collection. Virus-infected cells were harvested at 2 weeks after inoculation with JCV. Cells were harvested by exposure to trypsin, washed three times with phosphate-buffered saline (PBS), and fixed overnight at −20 °C with 70% ethanol. For flow cytometric analysis of TAg or VP1 expression, cells were washed with PBS containing 1% bovine serum albumin and then incubated for 2 h at room temperature with fluorescein isothiocyanate (FITC)-conjugated antibodies to TAg (pAb108, BD Biosciences) and antibodies to VP1 and allophycocyanin-labeled secondary antibodies. For cell cycle analysis, DNA content was measured by staining the cells for 30 min at room temperature with propidium iodide (PI) (10 μg/ml) in PBS containing RNase A (0.1 mg/ml). Flow cytometry was performed with a FACSCanto system (BD Biosciences), and data were analyzed with Flowjo software (Tree Star, Ashland, OR). All analysis was performed with the PI-A (area) versus PI-W (width) subset to exclude doublet cells.
Immunocytofluorescence Analysis
IMR-32 cells seeded onto glass-bottom dishes or chamber slides were transfected with the indicated constructs at 3 days before fixation. Virus-infected cells were seeded onto poly-l-lysine-coated slide glasses and fixed at 2 weeks after inoculation with JCV. Cells were fixed for 3 min in 100% methanol at −20 °C, exposed to 1% bovine serum albumin to block nonspecific sites, and incubated with primary antibodies overnight at 4 °C. Immune complexes were visualized by incubation with Alexa Fluor 488-conjugated secondary antibodies (Invitrogen) for 1 h at room temperature. For sequential double immunostaining of TAg, the cells were incubated with pAb416 or JCT652 antibodies to Tag, and then with Alexa Fluor 594-conjugated secondary antibodies (Invitrogen). Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Fluorescence signals were detected with an FV-1000 laser-scanning confocal microscope (Olympus, Tokyo, Japan).
For visualization of chromatin- or nuclear matrix-associated proteins, HeLa cells were seeded onto chamber slide glasses and transfected with the indicated constructs. The cells were then washed with PBS, incubated for 5 min on ice in isotonic Triton buffer (0.5% Triton X-100, 20 mm HEPES-NaOH (pH 7.4), 50 mm NaCl, 3 mm MgCl2, 300 mm sucrose) to remove cytoplasmic and soluble nuclear proteins, washed three times with PBS, and fixed in ice-cold 100% methanol for 3 min. For preparation of the nuclear matrix, after treatment of cells with isotonic Triton buffer, chromatin was digested for 20 min at room temperature with DNase I (100 μg/ml, Sigma) in isotonic buffer (20 mm HEPES-NaOH (pH 7.4), 50 mm NaCl, 6 mm MgCl2, 300 mm sucrose) and then extracted for 2 min at room temperature with a solution containing 0.25 m ammonium sulfate, 20 mm HEPES-NaOH (pH 7.4), and 0.2 mm MgCl2. The remaining nuclear matrix was washed with PBS and fixed with ice-cold methanol for 3 min. Fluorescence signals were detected with an inverted fluorescence and phase-contrast microscope (IX70, Olympus), and images were collected with a charge-coupled device (CCD) camera with the use of DP controller software (Olympus).
5-Ethynyl-2′-deoxyuridine (EdU) Incorporation
Cells were exposed to 10 μm EdU (Invitrogen) in culture medium for 45 min. For flow cytometric analysis of EdU incorporation and TAg expression, cells were fixed overnight with 70% ethanol at −20 °C. After staining with the FITC-labeled antibody to TAg as described above, EdU was detected with Alexa Fluor 647-azide with the use of a Click-iT EdU Flow Cytometry Assay Kit (Invitrogen). Flow cytometry was performed as described above. For immunocytofluorescence analysis, cells were fixed with 100% methanol for 3 min at −20 °C before detection of EdU with Alexa Fluor 647-azide. Sequential double immunostaining was performed with a monoclonal antibody to cyclin B and polyclonal antibodies to JCV Tag, and immune complexes were visualized with Alexa Fluor 405- or Alexa Fluor 488-conjugated secondary antibodies (Invitrogen), respectively.
Immunoblot and Immunoprecipitation Analysis
IMR-32 cells seeded onto 12-well dishes were transfected with the indicated constructs 3 days before collection. Virus-infected cells were collected at the indicated times after inoculation with JCV. For induction of DNA damage, cells were exposed to UV (10 J/m2) with the use of a UV Cross-linker (UVP, Upland, CA) for 2 h prior to collection. Cells were suspended in RIPA buffer (1% Triton X-100, 150 mm NaCl, 0.1% SDS, 1% deoxycholic acid, 10 mm Tris-HCl (pH 7.5), 5 mm EDTA, 10% glycerol, 50 mm sodium fluoride, 1 mm phenylmethylsulfonyl fluoride, complete protease inhibitor mixture (Roche, Basel, Switzerland), and phosphatase inhibitor mixture 1 (Sigma)). Lysate proteins (10 μg) were fractionated by SDS-polyacrylamide gel electrophoresis and subjected to immunoblot analysis with the indicated antibodies. Immune complexes were detected with horseradish peroxidase-conjugated secondary antibodies (BIOSOURCE International, Camarillo, CA), Immobilon Western HRP Substrate (Millipore), and a LAS-1000 Plus system (Fujifilm, Tokyo, Japan). For immunoprecipitation, cells were lysed in immunoprecipitation lysis buffer (1% Nonidet P-40, 150 mm NaCl, 10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 50 mm sodium fluoride, 1 mm phenylmethylsulfonyl fluoride, complete protease inhibitor mixture, and phosphatase inhibitor mixture 1). Lysed protein was immunoprecipitated with Dynabeads Protein G (Invitrogen) for 2 h at 4 °C after coating with FLAG M2 antibody. Protein complexes were eluted with SDS sample buffer (125 mm Tris-HCl (pH 6.8), 10% 2-mercaptoethanol, 4% SDS, 10% sucrose, and 0.04% bromphenol blue) after washing with immunoprecipitation lysis buffer.
Fractionation of Cellular Proteins
For nuclear fractionation, IMR-32 cells transfected with the indicated constructs were subjected to sequential extraction as described previously (31, 32), with some modifications. One-tenth of the harvested cells were lysed with RIPA buffer to obtain a whole cell extract. The remaining cells were suspended in buffer A (0.05% Triton X-100, 10 mm HEPES-NaOH (pH 7.8), 1.5 mm MgCl2, 10 mm KCl, 1 mm phenylmethylsulfonyl fluoride, and complete protease inhibitor mixture) and centrifuged at 700 × g for 5 min at 4 °C to yield the soluble cytoplasmic extract. The nuclear pellet was washed with buffer A, suspended, and incubated for 5 min on ice in isotonic Triton buffer supplemented with 1 mm phenylmethylsulfonyl fluoride and complete protease inhibitor mixture, then centrifuged at 700 × g for 5 min at 4 °C to yield the nuclear soluble extract. The associated nuclear pellet was washed with isotonic Triton buffer and further extracted consecutively with increasing concentrations of NaCl (0.25, 0.5, and 1 m) in a solution containing 10 mm Tris-HCl (pH 7.4) and 0.2 mm MgCl2 to yield chromatin-associated proteins. The final nuclear residue (NR) consisting of DNA and nuclear matrix was dissolved by ultrasonic treatment in RIPA buffer. Equal amounts of protein for each fraction were then subjected to immunoblot analysis with the indicated antibodies.
The nuclear matrix was also prepared as described previously (33). After treatment of the initial nuclear pellet with isotonic Triton buffer, as described above, chromatin was digested for 30 min at room temperature with DNase I (100 μg/ml) in the same buffer and then extracted by incubation with 0.25 m ammonium sulfate for 5 min at room temperature. The mixture was centrifuged at 10,000 × g for 5 min at 4 °C to yield a supernatant containing chromatin-associated proteins and a pellet containing insoluble and nuclear matrix proteins. The pellet was suspended in a solution containing 8 m urea, 100 mm NaH2PO4, and 10 mm Tris-HCl (pH 8.0). Equal amounts of protein for each fraction were then subjected to immunoblot analysis.
Small Interfering RNA (siRNA) Transfection
The siRNA constructs were obtained as the siGENOME SMARTpool, ATR (M-003202), WEE1 (M-005050; Dharmacon, Thermo Scientific, Lafayette, CO), and CDC2 HP Validated siRNA (SI00299726; Qiagen, Valencia, CA). The non-targeting control siRNA was obtained from Qiagen. IMR-32 cells were transfected with 20 nm siRNA diluted in Opti-MEM medium (Invitrogen) with the use of Lipofectamine RNAi MAX reagent (Invitrogen) according to the reverse transfection protocol of the manufacturer. After 24 h, the cells were transfected with pCXN2-JCV TAg or the empty vector, or pCXN2-JCV TAg and pBS-JCori for replication assay, with the use of Lipofectamine 2000, and cultured for 2 or 3 days. KU55933 (20 μm) or dimethyl sulfoxide (control) was added to the cells 15 h before collection.
DpnI Replication Assay
IMR-32 cells or siRNA-transfected IMR-32 cells were seeded onto 3.5-cm dishes and transfected with pBS-JCori and pCXN2-JCV TAg. One day after transfection, caffeine (2.5 mm), UCN-01 (50 nm), or dimethyl sulfoxide was added to the cells and cultured for an additional 24 h. Low molecular weight DNA was isolated from the cells as described previously (34), incubated for 1 h at 56 °C with proteinase K (0.5 mg/ml), and precipitated with phenol/chloroform/isoamyl alcohol/ethanol. The DNA was suspended in TE buffer (10 mm Tris (pH 7.5), 1 mm EDTA) containing RNase A (75 μg/ml), and a 5-μg portion was digested with EcoRI and DpnI, the latter of which selectively digests transfected DNA that has been methylated during prokaryotic replication. The products of digestion were fractionated by electrophoresis on a 1% agarose gel, and DNA fragments were transferred to a Hybond N+ membrane (GE Healthcare Bio-Sciences) by capillary transfer in ×20 standard saline citrate and then exposed to a digoxigenin-labeled DNA probe targeted to a vector-derived, ampicillin-resistant DNA sequence (DIG High Prime DNA Labeling and Detection Starter Kit II, Roche). Hybridization signals were detected with the LAS-1000 Plus system, and replication activities were determined by quantifying the intensity of the bands with the use of Multi Gauge software (Fujifilm).
Hemagglutin (HA) Assay
Measurement of JCV titer was based on HA of human type O erythrocytes as described previously (35). Human blood was supplied from the Hokkaido Red Cross Blood Center. For analysis of viral proliferation, caffeine (1 or 2 mm) was added to JCV-infected IMR-32 cells at 7, 10, and 13 days postinoculation (d.p.i.) and the cells were collected at 13 or 16 d.p.i. The JCV in 5 × 106 cells was extracted by exposing the cells to 3 freeze-thaw cycles in 150 μl of Tris-HCl (pH 7.5) containing 0.2% bovine serum albumin. The extract was incubated for 15 h at 37 °C with neuraminidase (0.05 units/ml), and, after inactivation of neuraminidase for 30 min at 56 °C, it was centrifuged at 600 × g for 10 min. The resulting supernatant was then subjected to the HA assay. 2-Fold serial dilutions of the supernatant (25 μl) in PBS (adjusted to pH 7.15) containing 0.2% bovine serum albumin were added to the wells of a 96-well, V-bottomed microplate, and an equal volume of 0.5% red blood cells in PBS (pH 7.15) was added to each well. The mixture was incubated for 3 h at 4 °C, and the HA titer of the virus was then determined as the highest dilution resulting in HA.
Immunohistofluorescence Analysis
A surgical specimen was obtained from the brain of an individual with PML positive for JCV VP1 immunostaining. This aspect of the study was approved by the Ethical Committee of Hokkaido University Graduate School of Medicine. The tissue was fixed in formalin, embedded in paraffin, and sectioned. Paraffin was then removed from the sections, and antigen retrieval was performed in 10 mm citrate buffer (pH 6.0) by placing the sections in a pressure cooker for 3 min. The sections were exposed to 10% normal goat serum and then incubated overnight at 4 °C with a monoclonal antibody to cyclin B1 or with polyclonal antibodies to Ser15-phosphorylated p53 or Thr21-phosphorylated RPA32. Immune complexes were detected by incubation for 90 min at room temperature with Alexa Fluor 488-conjugated secondary antibodies. The sections were then incubated with polyclonal antibodies to JCV TAg or monoclonal antibodies to TAg, and immune complexes were detected with Alexa Fluor 594-conjugated secondary antibodies. Nuclei were counterstained with DAPI, and the sections were then stained with 1% Sudan black B for 5 min to quench the autofluorescence of brain tissue. Fluorescence was then observed with an inverted fluorescence and phase-contrast microscope, and images were obtained as described above.
RESULTS
TAg-expressing Cells Accumulate at G2 Phase
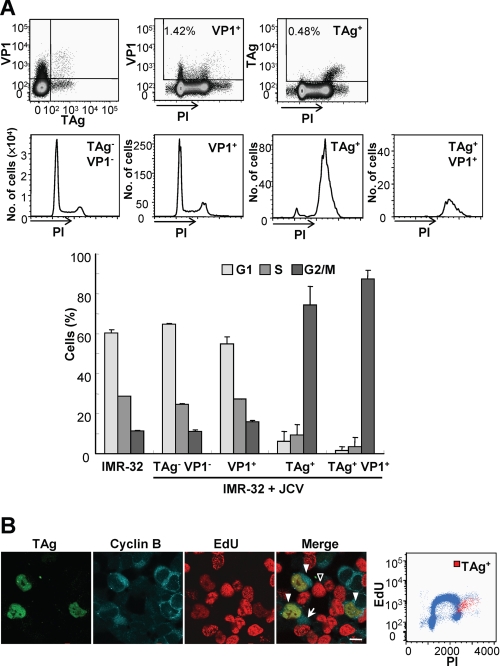
We first investigated the cell cycle in human neuroblastoma IMR-32 cells, which are permissive for JCV replication, expressing JCV early (TAg) or late (VP1) proteins 2 weeks after virus inoculation. Flow cytometric analysis of cells stained for DNA with PI revealed that those expressing TAg (TAg+ and TAg+VP1+ cells) accumulated at the G2/M phase, whereas those expressing VP1 (VP1+ cells) showed a cell cycle profile similar to that of noninfected cells or cells not expressing TAg or VP1 (TAg−VP1− cells) (Fig. 1A). Given that TAg is indispensable for viral DNA replication, the JCV genome would be expected to be replicated in TAg-expressing cells. We next examined DNA synthesis in JCV-infected cells by measurement of EdU incorporation. TAg-expressing cells incorporated less EdU than cells in S phase, even though they expressed cyclin B in the cytoplasm (36) (Fig. 1B), indicating that the cells synthesized viral DNA in the G2 phase. Flow cytometric analysis also provided evidence of additional viral DNA replication in the G2 phase (Fig. 1B). These results indicate that expression of TAg is related to G2 arrest in JCV-infected cells, and that viral DNA is replicated during G2 arrest.
FIGURE 1.
Accumulation of JCV-infected cells in G2 of the cell cycle. A, JCV-infected IMR-32 cells at 14 d.p.i. were stained with PI for DNA content, with antibodies to VP1, and allophycocyanin-labeled secondary antibodies, as well as with FITC-labeled antibodies to TAg. The cells were then subjected to flow cytometry, with TAg+VP1+, TAg−VP1−, TAg+, and VP1+ cell subsets indicated in the dot plots being shown in the histograms of PI fluorescence intensity. The bar graph indicates the percentage of cells in each phase of the cell cycle for noninfected cells and for the various subsets of JCV-infected cells; data are mean ± S.D. of triplicates from a representative experiment. B, JCV-infected IMR-32 cells (14 d.p.i.) were exposed to EdU for 45 min, after which EdU (red), TAg (green), and cyclin B (blue) were detected in the cells by fluorescence microscopy. In the merged image, TAg and EdU were detected in G2 phase cells that express cyclin B1 in the cytoplasm (solid arrowheads). Cells in S phase are EdU positive (open arrowhead), and cells in M phase show a diffuse pattern of cyclin B1 staining (arrow). Scale bar, 10 μm. The EdU-labeled cells were also subjected to flow cytometric analysis of EdU incorporation, DNA content, and TAg expression. The dot plot of EdU incorporation versus DNA content (PI fluorescence intensity) reveals a population of TAg+ cells (red dots) among the cells that had incorporated EdU.
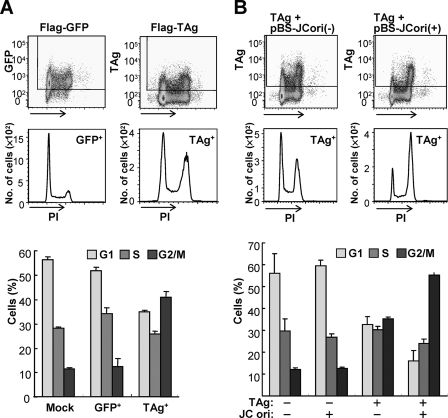
To test whether the G2 arrest in TAg-expressing cells is induced by TAg itself or by viral DNA replication, we analyzed the cell cycle profile of cells transfected with an expression vector for the FLAG epitope-tagged TAg. Cells transiently expressing TAg also accumulated in G2 phase compared with control cells expressing FLAG-tagged green fluorescent protein (GFP), suggesting that TAg triggers G2 arrest in the absence of viral DNA (Fig. 2A). G2 arrest of TAg-expressing cells was observed at 2 or 3 days post-transfection; thereafter, G2 arrest was abrogated with a decrease in the levels of TAg expression (supplemental Fig. S1). Interestingly, in comparison with other polyomaviruses, G2 arrest in IMR-32 cells expressing SV40 TAg was greater than that in JCV TAg expressing cells, whereas the extent of G2 arrest in Vero cells (which are derived from African green monkey kidney epithelium) expressing SV40 or JCV TAg was equivalent (supplemental Fig. S2). In addition, IMR-32 cells expressing murine polyomavirus TAg accumulated in S and G2 phases. Because G2 arrest was more striking in JCV-infected cells than in cells transiently expressing TAg (Figs. 1A and 2A), we also examined the cell cycle profile of TAg-expressing cells in the presence of a plasmid containing the JCV origin (pBS-JCori). We found that G2 arrest in TAg-expressing cells was enhanced by the presence of a plasmid containing the JCV origin, indicating that viral DNA replication performed by TAg leads to further accumulation of cells at the G2 phase (Fig. 2B).
FIGURE 2.
Accumulation of TAg-expressing cells in G2 of the cell cycle. IMR-32 cells transfected with expression vectors for FLAG-TAg or FLAG-GFP, or with the corresponding empty vector (Mock) 3 days before analysis (A); and IMR-32 cells transfected with FLAG-TAg and pBS-JCori(+), or FLAG-TAg and pBS-JCori(−) 3 days before analysis (B), were stained with PI and FITC-labeled antibodies to FLAG. The cells were then subjected to flow cytometry with the TAg+ or GFP+ cell subsets indicated in the dot plots being shown in the histograms of PI fluorescence intensity. The bar graph indicates the percentage of cells in each phase of the cell cycle for mock-transfected cells and for the TAg+, TAg−, and GFP+ cell subsets; data are mean ± S.D. of triplicates from a representative experiment.
TAg Induces ATM- and ATR-dependent G2 Checkpoint Signaling
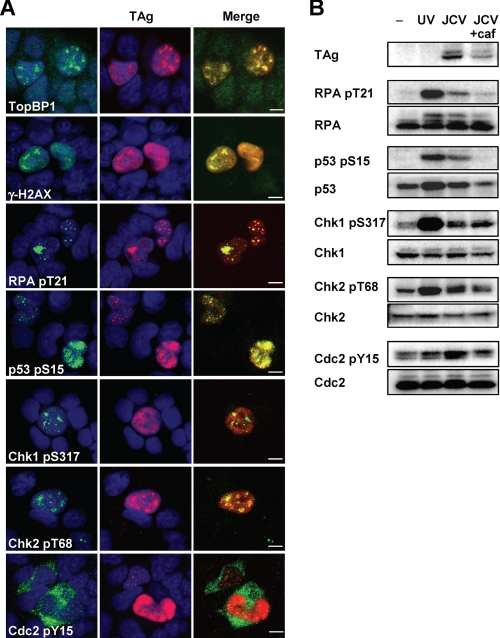
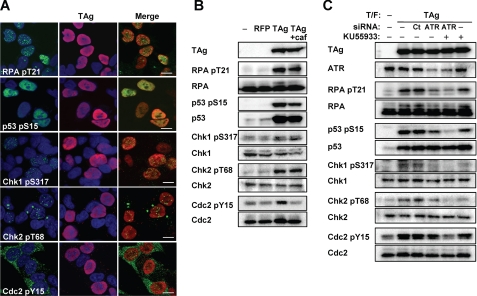
In mammalian cells, G2 progression is arrested by G2 checkpoint signaling in response to DNA damage or replication block. The major regulators of the G2 checkpoint pathway, ATM and ATR, phosphorylate various proteins, including Chk1, Chk2, p53, histone H2A.X, and RPA32, and thereby inactivate Cdc2-cyclin B. To investigate whether TAg-induced G2 arrest is mediated by such signaling pathways, we examined the phosphorylation status of ATM and ATR substrates by immunofluorescence and immunoblot analyses. We found that TAg colocalized with phosphorylated forms of the ATM and ATR substrates, and also with the ATR activator, topoisomerase-binding protein-1, in the nuclei of JCV-infected IMR-32 cells (Fig. 3A). Furthermore, Tyr15-phosphorylated (inactivated) Cdc2 was detected predominantly in the cytoplasm of the infected cells (Fig. 3A). The phosphorylation of these various proteins in JCV-infected cells was confirmed by immunoblot analysis (Fig. 3B). Although only ∼0.5% of infected cells expressed TAg (Fig. 1A), we found phosphorylation of checkpoint proteins in JCV-infected cells similar to that in cells with ultraviolet radiation (UV)-induced DNA damage (Fig. 3B). The JCV-induced phosphorylation of RPA32 on Thr21, p53 on Ser15, and Cdc2 on Tyr15 was abrogated by the ATM-ATR inhibitor caffeine, whereas that of Chk1 on Ser317 and Chk2 on Thr68 were not affected (Fig. 3B). Caffeine inhibited consequent G2 arrest, and led the cells to enter mitosis (supplemental Fig. S3). We obtained similar results for immunofluorescence and immunoblot analyses of cells transiently expressing TAg (Fig. 4, A and B). To exclude the possibility that plasmid transfection itself or overexpression of any exogenous protein might induce G2 checkpoint signaling, we confirmed that the checkpoint proteins were not phosphorylated in cells transfected with an expression vector for red fluorescent protein (Figs. 4B and supplemental S4). These results thus indicated that TAg activates G2 checkpoint signaling. Given that caffeine did not block the phosphorylation of Chk1 and Chk2, we further examined the role of ATM or ATR in the checkpoint signaling induced by TAg with the use of a specific inhibitor of ATM (KU55933) and siRNAs specific for ATR mRNA. The level of phosphorylation of checkpoint proteins, including Chk1 and Chk2, in cells treated with both KU55933 and the ATR siRNAs was lower than that in those treated with either of these agents alone (Fig. 4C), suggesting that TAg induces G2 checkpoint signaling by both ATM- and ATR-mediated pathways.
FIGURE 3.
Induction of G2 checkpoint signaling in JCV-infected IMR-32 cells. A, JCV-infected IMR-32 cells (14 d.p.i.) were subjected to double immunofluorescence staining with antibodies to the indicated phosphorylated forms of checkpoint proteins (green) and with antibodies to TAg (red). Cell nuclei were stained with DAPI (blue). TopBP1 indicates topoisomerase-binding protein-1; γ-H2A.X indicates phosphorylated histone H2A.X; RPA indicates RPA32. pS, phosphoserine; pY, phosphotyrosine; pT, phosphothreonine. Scale bars, 5 μm. B, JCV-infected IMR-32 cells (14 d.p.i.) incubated in the absence or presence of 2.5 mm caffeine (caf) for 15 h, as well as UV-treated or control (−) IMR-32 cells, were subjected to immunoblot analysis with antibodies to phosphorylated or total forms of the indicated proteins.
FIGURE 4.
Induction of ATM- and ATR-dependent G2 checkpoint signaling in TAg-expressing IMR-32 cells. A, IMR-32 cells were transfected with an expression vector for FLAG-TAg 3 days before double immunofluorescence staining with antibodies to the indicated phosphorylated forms of checkpoint proteins (green) and antibodies to TAg (red). Cell nuclei were stained with DAPI (blue). pS, phosphoserine; pY, phosphotyrosine; pT, phosphothreonine. Scale bars, 10 μm. B, IMR-32 cells transfected with expression vectors for FLAG-red fluorescent protein or FLAG-TAg, or with the corresponding empty vector (−), were incubated in the absence or presence of 2.5 mm caffeine (caf) for 15 h and then subjected to immunoblot analysis with antibodies to phosphorylated or total forms of the indicated proteins. C, IMR-32 cells transfected (T/F) with control (CT) or ATR siRNAs, as well as with an expression vector for FLAG-TAg or the corresponding empty vector (−), were incubated for 15 h in the absence or presence of the ATM inhibitor KU55933 (20 μm) and then subjected to immunoblot analysis with antibodies to phosphorylated or total forms of the indicated proteins.
The Induction of G2 Arrest Is Associated with Localization of TAg to Cellular DNA
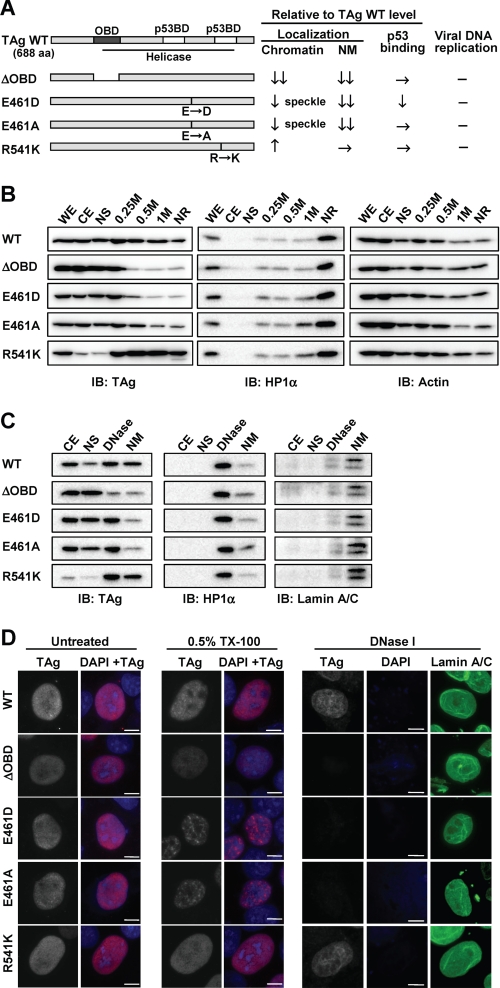
TAg contains an origin DNA binding domain (OBD) and a helicase domain that mediates the initiation of polyomavirus DNA replication. The OBD contributes to sequence-specific initiation of viral DNA replication through its binding to the viral core origin (37). SV40 TAg is also capable of binding nonspecifically to double-stranded DNA and ssDNA, and such nonspecific contacts with DNA are also required during viral DNA replication (38, 39). We examined whether this nonspecific DNA binding activity of TAg might affect the replication of cellular DNA by generating various mutants of JCV TAg expected to possess different levels of such activity on the basis of previous in vitro studies of SV40 TAg (39–41). The OBD mutant (ΔOBD) of JCV TAg lacks amino acids 147 to 251, which include not only the origin-specific binding domain, but also one of the nonspecific double-stranded DNA binding domains identified in SV40 TAg (42). We also generated point mutants of JCV TAg in which Glu at position 461 is replaced with Asp (E461D) or Ala (E461A); the corresponding mutants of SV40 TAg (residue 460) failed to bind nonspecifically to double-stranded DNA or ssDNA, respectively (39–41). In addition, we constructed R541K of JCV TAg, the corresponding mutations of SV40 TAg (residues 540) having been shown to increase ssDNA binding activity (41) (Fig. 5A). To determine whether JCV TAg is actually bound to cellular DNA in the nucleus, we examined the association of TAg with chromatin by subcellular fractionation of cell lysates prepared from IMR-32 cells expressing wild-type (WT) or mutant forms of JCV TAg (Fig. 5B). Fractionation of the cell lysates with detergent and sequential salt extraction revealed WT TAg to be present in all fractions, including the high-salt nuclear extract (1 m) and NR fractions, indicating that a portion of TAg is tightly bound to chromatin. Most of the R541K mutant was detected in chromatin protein (0.25, 0.5, and 1 m) and NR fractions, whereas only small amounts of the ΔOBD, E461D, and E461A mutants remained in the high-salt (0.5 and 1 m) and NR fractions. The NR fraction contains nuclear matrix, DNA, and proteins tightly bound to DNA such as heterochromatin protein 1α (HP1α) (Fig. 5B). We therefore applied DNase I digestion followed by extraction with 0.25 m ammonium sulfate to separate the nuclear matrix and DNA-bound proteins (Fig. 5C). Whereas the WT and R541K forms of TAg were detected in both chromatin protein (DNase) and nuclear matrix fractions, only small amounts of ΔOBD, E461D, or E461A were found to be associated with the nuclear matrix. To further investigate the association of TAg with cellular chromatin and the nuclear matrix in situ, we performed immunofluorescence staining of cells after detergent and DNase I treatment. We used HeLa cells for this analysis because IMR-32 cells detached from slide glasses after detergent treatment. Transient expression of TAg resulted in G2 arrest in HeLa cells (supplemental Fig. S6) as it did in IMR-32 cells. HeLa cells expressing WT or mutant forms of TAg were fixed with methanol directly or after extraction with 0.5% Triton X-100 to remove soluble nuclear proteins followed (or not) by treatment with DNase I and ammonium sulfate extraction to remove chromatin. Whereas the amounts of WT or R541K forms of TAg in the nucleus of detergent-treated cells were similar to those in nontreated cells, the amounts of ΔOBD, E461D, or E461A mutants in the nucleus were reduced by detergent treatment (Fig. 5D). The E461D and E461A proteins remaining in the nucleus of detergent-treated cells manifested a punctate staining pattern. Further treatment of the cells with DNase I and ammonium sulfate resulted in the removal of most DNA, as revealed by DAPI staining, whereas staining for lamin A/C, a component of the nuclear lamina, remained apparent (Fig. 5D). Both WT and R541K forms of TAg, but not the ΔOBD, E461D, or E461A mutants, were still detected in the nucleus of such cells, suggesting that WT TAg and the R541K mutant associate with the nuclear matrix as well as with chromatin. Because amino acid residues 461 and 541 of JCV TAg are located in a helicase/ATPase domain and 541 in a p53 binding domain, we confirmed replication activity and the p53 binding ability of mutant forms of TAg. All mutant forms of TAg lost replication activity, whereas the p53 binding ability of the mutants was maintained except for a slight decrease in that of the E461D mutant (Figs. 5A and supplemental S5, A and B).
FIGURE 5.
Subcellular localization of WT and mutant forms of TAg. A, schematic representation and summary of intracellular properties of the mutant forms of JCV TAg. Predicted functional domains of JCV TAg were shown in TAg wild-type (WT, 688 amino acid (aa)). OBD, ori DNA binding domain (dark gray, 138–247 aa); p53BD, p53 binding domain (light gray, 351–450 and 534–628 aa); Helicase domain (under bar, 132–628 aa). ΔOBD lacks residues of WT 147–251 aa. The positions of amino acid point mutations for E461D, E461A, and R541K are indicated by black bars. The results of subcellular localization as shown in B–D, and p53 binding and the viral DNA replication assay for TAg mutants as shown in supplemental Fig. 6 were summarized as relative to those of TAg WT; decrease (↓), high decrease (↓↓), increase (↑), equivalence (→), and no activity (−). B, IMR-32 cells expressing FLAG-tagged WT or mutant forms of JCV TAg were subjected to sequential subcellular fractionation and salt extraction of insoluble nuclear material as described under “Experimental Procedures.” WE, whole cell extract; CE, cytoplasmic extract; NS, nuclear soluble extract; 0.25 m, 0.25 m NaCl nuclear extract; 0.5 m, 0.5 m NaCl nuclear extract; 1 m, 1 m NaCl nuclear extract; IB, immunoblot. Equal amounts of protein for each fraction were subjected to immunoblot analysis with antibodies to FLAG (for detection of TAg), HP1α, and actin (loading control). C, IMR-32 cells expressing WT or mutant forms of TAg were subjected to sequential subcellular fractionation followed by DNase I treatment and ammonium sulfate extraction to separate chromatin-associated proteins (DNase) from nuclear matrix-associated proteins (NM). Equal amounts of protein for each fraction were subjected to immunoblot analysis with antibodies to FLAG, HP1α, and lamin A/C. D, HeLa cells expressing WT or mutant forms of TAg were fixed with methanol either directly (nontreated, NT) or after extraction with 0.5% Triton X-100 without (0.5% TX-100) or with (DNase I) subsequent DNase I treatment and ammonium sulfate extraction. The cells were then subjected to immunofluorescence staining with antibodies to TAg (gray scale or red) or lamin A/C (green). DNA was also stained with DAPI (blue). The gray-scale images were obtained under the same conditions. Scale bars, 10 μm.
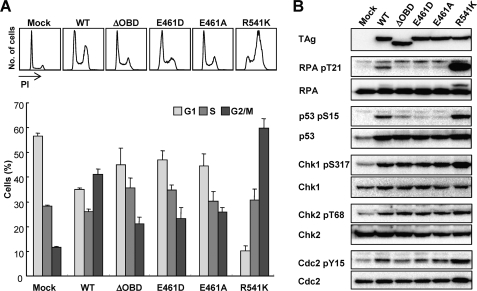
To test whether the association of TAg with cellular DNA affects induction of G2 arrest by TAg, we analyzed the cell cycle profile of IMR-32 cells transiently expressing each TAg mutant. As expected, the G2 population of cells expressing ΔOBD, E461D, or E461A was smaller than that of cells expressing WT TAg (Fig. 6A). In contrast, the proportion of cells expressing the R541K mutant in the G2 phase was markedly increased compared with that for those expressing the WT protein. However, the replication activity and p53 binding ability of mutant forms of TAg did not correlate with the induction of G2 arrest (Fig. 5A). These results indicate that the localization of TAg to cellular DNA is associated with G2 arrest induced by TAg itself. To confirm that these differences in the extent of G2 arrest reflected G2 checkpoint signaling, we examined the phosphorylation status of checkpoint proteins (Fig. 6B). We found a correlation between the induction of G2 arrest and the phosphorylation of RPA32 on Thr21 and p53 on Ser15. The phosphorylation levels of Chk1 and Chk2 in cells expressing the ΔOBD, E461D, or E461A mutants were similar to those in cells expressing the WT protein. Given that the ΔOBD, E461D, and E461A mutants each induced a small increase in the proportion of cells in the G2 phase compared with that for mock-transfected cells (Fig. 6A), it is likely that TAg triggers at least two pathways leading to G2 arrest and that these mutants activate one pathway mediated by Chk1 and Chk2, but not the other pathway mediated by RPA32 and p53. Together, these results indicate that the localization of TAg to cellular DNA is one of the factors in the induction of G2 arrest.
FIGURE 6.
Effects of mutation of JCV TAg on the induction of G2 arrest. A, flow cytometric analysis of the cell cycle profile either of the TAg+ cell subset of IMR-32 cells transfected with expression vectors for FLAG-tagged WT or mutant forms of TAg, or of the total cells transfected with the corresponding empty vector (mock). The histograms indicate DNA content as revealed by staining with PI and FITC-labeled antibodies to TAg. The bar graph shows the corresponding percentages of cells in each phase of the cell cycle; data are mean ± S.D. of triplicates from a representative experiment. B, immunoblot analysis of phosphorylated or total forms of the indicated proteins in IMR-32 cells transfected as in A.
Abrogation of G2 Arrest Suppresses JCV Genome Replication
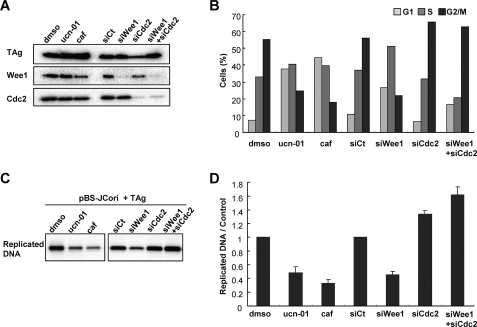
To determine whether TAg-induced G2 checkpoint signaling and/or G2 arrest affect viral genome replication efficiency, we examined viral DNA replication in the presence of inhibitors of G2 checkpoint signaling and G2-M transition. We first confirmed the effects of caffeine, ucn-01 (a Chk1 inhibitor), and siRNA against Wee1 and Cdc2 on the cell cycle profile of cells transfected with pBS-JCori and an expression vector for TAg. Depletion of Wee1 and Cdc2 proteins in these cells were confirmed by immunoblotting (Fig. 7A). As expected, TAg-induced G2 arrest of the cells was abrogated by inhibition of ATM and ATR, Chk1, and Wee1, whereas G2 arrest was maintained by Cdc2 depletion (Fig. 7B). We next assessed the viral replication activity in these cells by the DpnI replication assay. The extent of viral replication was indeed reduced by not only caffeine and ucn-01, but also Wee1 depletion compared with that apparent in the control cells (Fig. 7, C and D). However, Cdc2 depletion abrogated the reduction of viral replication in cells with Wee1 depletion (Fig. 7, C and D). These results indicate that viral replication efficiency is affected by the proportion of cells in G2 phase, not by kinase activities involved in G2 checkpoint signaling.
FIGURE 7.
Suppression of JCV genome replication by abrogation of G2 arrest. Effect of inhibition of G2 checkpoint signaling on JCV DNA replication as determined by the DpnI replication assay. IMR-32 cells transfected with pBS-JCori and an expression vector for TAg were incubated for 24 h before addition of dimethyl sulfoxide (DMSO) (control), 50 nm ucn-01, or 2.5 mm caffeine (caf). The siRNA against non-targeting control (Ct) or Wee1 or/and Cdc2 were transfected 24 h before transfection of the plasmids. The collected cells were subjected to immunoblot analysis with antibodies to FLAG-TAg, Wee1, and Cdc2 (A), flow cytometric analysis of cell cycle profile for the TAg+ cell subset (B), and DpnI replication assay (C and D). Replicated DNA extracted from the cells was detected by Southern blot analysis with a DNA probe specific for pBS-JCori (C). The intensity of DNA bands was quantified and is indicated in the bar graph relative to the value of dimethyl sulfoxide (for ucn-01 and caffeine) or that of siCt (for siWee1, siCdc2, and siWee+siCdc2) (D); data are mean ± S.D. of triplicates from a representative experiment.
Inhibition of ATM and ATR by Caffeine Suppresses JCV Proliferation
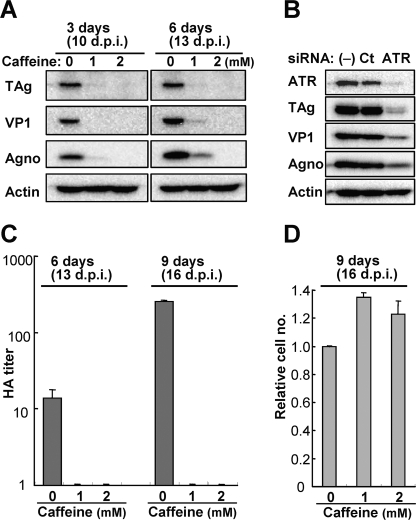
Because the JCV genome replication was greatly diminished by caffeine (Fig. 7D), we further examined the effect of caffeine on virus proliferation in JCV-infected cells. JCV-infected IMR-32 cells (7 d.p.i.) were cultured in the absence or presence of caffeine for 3 to 9 days. Immunoblot analysis revealed little or no expression of early (TAg) or late (VP1 and Agno) viral proteins in cells treated with 1 or 2 mm caffeine for 3 or 6 days, whereas control cells expressed substantial amounts of these viral proteins (Fig. 8A). Furthermore, we did not detect a HA titer in cells treated with caffeine for up to 9 days (16 d.p.i.) (Fig. 8C). In addition, the growth of cells treated with 1 or 2 mm caffeine for 9 days was greater than that of control cells (Fig. 8D), likely as a result of the prevention of virus-induced cytopathic effects (27). However, given that caffeine has many other pharmacological effects in addition to inhibition of ATM and ATR, it remained possible that these other activities might contribute to the observed effect on JCV proliferation. To confirm that suppression of viral proliferation by caffeine was attributable to inhibition of ATM or ATR, we examined the expression of viral proteins in cells transfected with ATR siRNAs. The expression of viral proteins was also inhibited in the ATR-depleted cells compared with that in control cells (Fig. 8B). These results show that at least ATR is required for JCV proliferation. Inhibition of ATM- and ATR-mediated G2 arrest by caffeine thus markedly suppressed viral propagation in JCV-infected cells.
FIGURE 8.
Suppression of JCV production by inhibition of ATM and ATR. A, JCV-infected IMR-32 cells (7 d.p.i.) were incubated in the absence or presence of caffeine (1 or 2 mm) for 3 or 6 days and then subjected to immunoblot analysis of the indicated viral proteins. B, JCV-infected IMR-32 cells were transfected (or not) with control (Ct) or ATR siRNAs at 7 and 10 d.p.i., harvested at 13 d.p.i., and subjected to immunoblot analysis of viral proteins. C and D, JCV-infected IMR-32 cells (7 d.p.i.) were incubated in the absence or presence of caffeine (1 or 2 mm) for 6 or 9 days, after which the HA titer of JCV in cell extracts was determined (C), and the number of cells was counted with the use of a hemocytometer and expressed relative to the value for those incubated in the absence of caffeine (D). Data are mean ± S.D. of triplicates from a representative experiment.
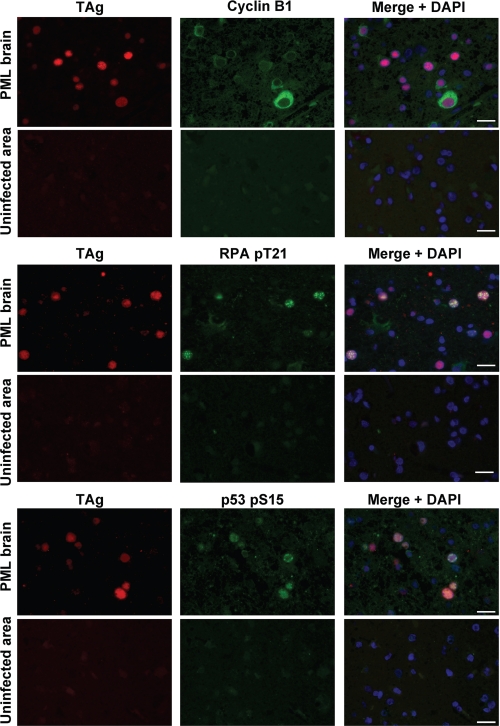
G2 Checkpoint Signaling Is Induced in JCV-infected Cells of the PML Brain
PML results from the productive infection of oligodendrocytes by JCV and consequent central nervous system demyelination. We examined whether the induction of G2 arrest also occurs in JCV-infected oligodendrocytes in vivo. Immunohistofluorescence analysis revealed the expression of cyclin B1, Thr21-phosphorylated RPA32, and Ser15-phosphorylated p53 in a surgical specimen of the brain from an individual with PML. Double immunostaining for TAg and each of these proteins showed that TAg-positive oligodendrocytes specifically expressed cyclin B1 in the cytoplasm as well as phosphorylated RPA32 and p53 in the nucleus (Fig. 9). These results thus suggested that TAg induces G2 checkpoint signaling and subsequent G2 arrest in oligodendrocytes productively infected with JCV in vivo.
FIGURE 9.
TAg-expressing oligodendrocytes in the PML brain are arrested in G2 phase. Sections of brain tissue from an individual with PML were subjected to double immunofluorescence staining with antibodies to TAg (red) as well as with those to cyclin B1, Thr21-phosphorylated RPA32, or Ser15-phosphorylated p53 (green). Cell nuclei were counterstained with DAPI (blue). Images were obtained from JCV-infected and uninfected areas of brain tissue. Scale bars, 20 μm.
DISCUSSION
We have shown that JCV TAg promotes viral DNA replication by arresting the cell cycle at the G2 phase as the result of induction of ATM- and ATR-mediated G2 checkpoint pathways. The induction of G2 arrest is caused by not only viral DNA replication, but also TAg itself (supplemental Fig. S7). The potential benefit to JCV replication of activation of the G2 checkpoint is likely to be the maintenance of cellular replication machinery in the G2 phase by prevention of mitosis, rather than the use of kinase activity involved in G2 checkpoint signaling. In infected cells, the G2 arrest is limited to the early phase of the JCV infection cycle associated with TAg expression; it does not occur during the late phase associated with expression of the late protein VP1. Inhibition of the checkpoint pathways and the consequent release of cells from G2 arrest resulted in cell cycle progression to mitosis and a marked reduction in the extent of JCV DNA replication. These results suggest that JCV DNA is replicated in G2-arrested cells, with the recovery from such arrest occurring as the abundance of TAg declines. In contrast to SV40 (43), infection with JCV does not appear to induce abnormal replication of cellular DNA resulting in a DNA content of >4 n. The replication of JCV DNA is thus accomplished during G2 arrest without a second round of cellular DNA synthesis.
Although a variety of viruses have been shown to induce G2 arrest in host cells, the mechanisms by which such an arrest is achieved appear diverse (44, 45). In the case of JCV infection, our data show that TAg itself triggers the ATM- and ATR-mediated G2 checkpoint pathways. Moreover, viral DNA replication caused by TAg enhanced G2 arrest in TAg-expressing cells by mechanism(s) independent from the G2 arrest triggered by TAg itself. One of the mechanisms of the induction of G2 arrest by TAg is associated with localization of TAg to cellular DNA. The amount of TAg associated with chromatin was thus correlated both with the level of phosphorylation of RPA32 on Thr21 and that of p53 on Ser15, as well as with the extent of cell accumulation in the G2 phase. These data suggest that the association of TAg with cellular DNA affects the cellular replication machinery, resulting in the induction of replication stress and consequent G2 arrest. In addition, TAg localized to the nuclear matrix, which is the site of nuclear foci associated with transcription and DNA replication (46, 47). It therefore seems likely that association of TAg with the nuclear matrix disturbs DNA replication as a result of the DNA binding activity of TAg or its interaction with a protein involved in DNA replication. Further investigation is required to reveal the mechanism by which TAg triggers DNA damage signaling.
A direct link between TAg-induced G2 arrest and the phosphorylation of RPA32 and p53 remains to be demonstrated. Although RPA-coated ssDNA is thought to play a role in the initiation of ATM- and ATR-mediated checkpoint pathways (14, 48), the potential roles of DNA damage- or replication stress-induced hyperphosphorylation of RPA by these kinases have not been defined. Furthermore, although the phosphorylation of p53 on Ser15 was induced in cells expressing WT or R541K mutant forms of JCV TAg, we did not detect the induction of apoptosis or the expression of p21WAF1/CIP1, a cyclin-dependent kinase inhibitor encoded by a p53 target gene, in such cells (data not shown). The phosphorylated p53 associated with activation of the G2 checkpoint in TAg-expressing cells thus appears likely to be inactivated as a result of its binding to TAg.
We also found that caffeine had a pronounced antiviral effect in cells infected with JCV. It thus almost completely blocked viral propagation after establishment of infection. However, we cannot rule out the possibility that caffeine also suppresses another step of the JCV life cycle in addition to viral DNA replication, or that it affects JCV propagation not only by acting as an inhibitor of ATM and ATR, but also through its other biological effects (such as antagonism of adenosine receptors). Given that we obtained evidence that JCV-infected oligodendrocytes were arrested at the G2 phase in a brain of an individual with PML, ATM-ATR inhibitors such as caffeine might prove clinically effective against JCV infection. Although it is difficult to assess the antiviral effect of caffeine at a nontoxic dose in vivo because of the lack of an animal model of JCV infection, recent studies have shown that the administration of caffeine in drinking water resulted in the inhibition of ATM or ATR signaling pathways in mice (49, 50). In addition, given that it readily crosses the blood-brain barrier, caffeine is likely to have access to target cells in central nervous system diseases such as PML.
Supplementary Material
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) and the Ministry of Health, Labor, and Welfare of Japan, the Japan Health Science Foundation, and the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, MEXT.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- JCV
- human polyomavirus JC virus
- PML
- progressive multifocal leukoencephalopathy
- TAg
- large T antigen
- SV40
- simian virus 40
- RPA
- replication protein A
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM- and Rad3-related
- ssDNA
- single-stranded DNA
- PBS
- phosphate-buffered saline
- siRNA
- small interfering RNA
- FITC
- fluorescein isothiocyanate
- PI
- propidium iodide
- DAPI
- 4′,6-diamidino-2-phenylindole
- EdU
- 5-ethynyl-2′-deoxyuridine
- HA
- hemagglutination
- d.p.i.
- days postinoculation
- GFP
- green fluorescent protein
- OBD
- origin DNA binding domain
- WT
- wild-type
- NR
- nuclear residue
- HP1α
- heterochromatin protein 1α.
REFERENCES
- 1.Berger J. R. (2007) Curr. Neurol. Neurosci. Rep. 7, 461–469 [DOI] [PubMed] [Google Scholar]
- 2.Frisque R. J., Bream G. L., Cannella M. T. (1984) J. Virol. 51, 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N. N., Khalili K. (1995) J. Virol. 69, 5843–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullock P. A., Seo Y. S., Hurwitz J. (1991) Mol. Cell. Biol. 11, 2350–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fairman M. P., Stillman B. (1988) EMBO J. 7, 1211–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider C., Weisshart K., Guarino L. A., Dornreiter I., Fanning E. (1994) Mol. Cell. Biol. 14, 3176–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickmanns A., Zeitvogel A., Simmersbach F., Weber R., Arthur A. K., Dehde S., Wildeman A. G., Fanning E. (1994) J. Virol. 68, 5496–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caracciolo V., Reiss K., Khalili K., De Falco G., Giordano A. (2006) Oncogene 25, 5294–5301 [DOI] [PubMed] [Google Scholar]
- 9.Okubo E., Lehman J. M., Friedrich T. D. (2003) J. Virol. 77, 1257–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehman J. M., Laffin J., Friedrich T. D. (1994) Cytometry 16, 138–143 [DOI] [PubMed] [Google Scholar]
- 11.Dahl J., You J., Benjamin T. L. (2005) J. Virol. 79, 13007–13017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y., Dodson G. E., Shaikh S., Rundell K., Tibbetts R. S. (2005) J. Biol. Chem. 280, 40195–40200 [DOI] [PubMed] [Google Scholar]
- 13.Lee J. H., Paull T. T. (2005) Science 308, 551–554 [DOI] [PubMed] [Google Scholar]
- 14.Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 15.Lee J., Kumagai A., Dunphy W. G. (2007) J. Biol. Chem. 282, 28036–28044 [DOI] [PubMed] [Google Scholar]
- 16.Chaturvedi P., Eng W. K., Zhu Y., Mattern M. R., Mishra R., Hurle M. R., Zhang X., Annan R. S., Lu Q., Faucette L. F., Scott G. F., Li X., Carr S. A., Johnson R. K., Winkler J. D., Zhou B. B. (1999) Oncogene 18, 4047–4054 [DOI] [PubMed] [Google Scholar]
- 17.Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. (2000) Genes Dev. 14, 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 18.Nurse P. (1990) Nature 344, 503–508 [DOI] [PubMed] [Google Scholar]
- 19.Raleigh J. M., O'Connell M. J. (2000) J. Cell Sci. 113, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 20.Banin S., Moyal L., Shieh S., Taya Y., Anderson C. W., Chessa L., Smorodinsky N. I., Prives C., Reiss Y., Shiloh Y., Ziv Y. (1998) Science 281, 1674–1677 [DOI] [PubMed] [Google Scholar]
- 21.Shieh S. Y., Ikeda M., Taya Y., Prives C. (1997) Cell 91, 325–334 [DOI] [PubMed] [Google Scholar]
- 22.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 23.Hermeking H., Lengauer C., Polyak K., He T. C., Zhang L., Thiagalingam S., Kinzler K. W., Vogelstein B. (1997) Mol. Cell 1, 3–11 [DOI] [PubMed] [Google Scholar]
- 24.Wang X. W., Zhan Q., Coursen J. D., Khan M. A., Kontny H. U., Yu L., Hollander M. C., O'Connor P. M., Fornace A. J., Jr., Harris C. C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3706–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoo H. Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. (2004) Cell 117, 575–588 [DOI] [PubMed] [Google Scholar]
- 26.Nukuzuma S., Yogo Y., Guo J., Nukuzuma C., Itoh S., Shinohara T., Nagashima K. (1995) J. Med. Virol. 47, 370–377 [DOI] [PubMed] [Google Scholar]
- 27.Orba Y., Sunden Y., Suzuki T., Nagashima K., Kimura T., Tanaka S., Sawa H. (2008) Virology 370, 173–183 [DOI] [PubMed] [Google Scholar]
- 28.Okada Y., Sawa H., Endo S., Orba Y., Umemura T., Nishihara H., Stan A. C., Tanaka S., Takahashi H., Nagashima K. (2002) Acta Neuropathol. 104, 130–136 [DOI] [PubMed] [Google Scholar]
- 29.Komagome R., Sawa H., Suzuki T., Suzuki Y., Tanaka S., Atwood W. J., Nagashima K. (2002) J. Virol. 76, 12992–13000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunden Y., Suzuki T., Orba Y., Umemura T., Asamoto M., Nagashima K., Tanaka S., Sawa H. (2006) Acta Neuropathol. 111, 379–387 [DOI] [PubMed] [Google Scholar]
- 31.Wang Q. E., Zhu Q., Wani G., Chen J., Wani A. A. (2004) Carcinogenesis 25, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 32.Rapić-Otrin V., McLenigan M. P., Bisi D. C., Gonzalez M., Levine A. S. (2002) Nucleic Acids Res. 30, 2588–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He D. C., Nickerson J. A., Penman S. (1990) J. Cell Biol. 110, 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirt B. (1967) J. Mol. Biol. 26, 365–369 [DOI] [PubMed] [Google Scholar]
- 35.Suzuki S., Sawa H., Komagome R., Orba Y., Yamada M., Okada Y., Ishida Y., Nishihara H., Tanaka S., Nagashima K. (2001) Virology 286, 100–112 [DOI] [PubMed] [Google Scholar]
- 36.Toyoshima F., Moriguchi T., Wada A., Fukuda M., Nishida E. (1998) EMBO J. 17, 2728–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean F. B., Bullock P., Murakami Y., Wobbe C. R., Weissbach L., Hurwitz J. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 16–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin H. J., Upson R. H., Simmons D. T. (1992) J. Virol. 66, 5443–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C., Roy R., Simmons D. T. (2001) J. Virol. 75, 2839–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiao J., Simmons D. T. (2003) J. Virol. 77, 12720–12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mu J. J., Wang Y., Luo H., Leng M., Zhang J., Yang T., Besusso D., Jung S. Y., Qin J. (2007) J. Biol. Chem. 282, 17330–17334 [DOI] [PubMed] [Google Scholar]
- 42.Simmons D. T., Wun-Kim K., Young W. (1990) J. Virol. 64, 4858–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehman J. M., Laffin J., Friedrich T. D. (2000) Exp. Cell Res. 258, 215–222 [DOI] [PubMed] [Google Scholar]
- 44.Davy C., Doorbar J. (2007) Virology 368, 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao R. Y., Elder R. T. (2005) Cell Res. 15, 143–149 [DOI] [PubMed] [Google Scholar]
- 46.Ciejek E. M., Tsai M. J., O'Malley B. W. (1983) Nature 306, 607–609 [DOI] [PubMed] [Google Scholar]
- 47.Nelson W. G., Pienta K. J., Barrack E. R., Coffey D. S. (1986) Annu. Rev. Biophys. Biophys. Chem. 15, 457–475 [DOI] [PubMed] [Google Scholar]
- 48.Robison J. G., Bissler J. J., Dixon K. (2007) Cell Cycle 6, 2408–2416 [DOI] [PubMed] [Google Scholar]
- 49.Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L. V., Kolettas E., Niforou K., Zoumpourlis V. C., Takaoka M., Nakagawa H., Tort F., Fugger K., Johansson F., Sehested M., Andersen C. L., Dyrskjot L., Ørntoft T., Lukas J., Kittas C., Helleday T., Halazonetis T. D., Bartek J., Gorgoulis V. G. (2006) Nature 444, 633–637 [DOI] [PubMed] [Google Scholar]
- 50.Paulson Q. X., Pusapati R. V., Hong S., Weaks R. L., Conti C. J., Johnson D. G. (2008) Oncogene 27, 4954–4961, [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.