Abstract
The box H/ACA ribonucleoproteins (RNPs) are protein-RNA complexes responsible for pseudouridylation, the most abundant post-transcriptional modification of cellular RNAs. Integrity of its box H/ACA domain is also essential for assembly and stability of the human telomerase RNP. The recent publication of the complete box H/ACA RNP structures combined with the previously reported structures of the protein and RNA components makes it possible to deduce the structural accommodation that accompanies assembly of the full particle. This analysis reveals how the protein components distort the RNA component of the RNP, enabling productive docking of the substrate RNA into the enzymatic active site.
Keywords: Crystal Structure, Protein-Nucleic Acid Interaction, Protein Structure, Ribonucleoprotein (RNP), RNA Modification, RNA Structure, Box H/ACA snoRNP, Pseudouridine Synthase
Introduction
Cellular RNAs undergo numerous site-specific post-transcriptional modifications. Over 100 chemically distinct modified nucleotides have been identified (the RNA Modification Database (1) and the Modomics Database (2)). The most abundant is Ψ,2 the C-5 glycoside isomer of uridine. The isomerization of U residues into Ψ is performed by universally distributed enzymes called Ψ synthases, which can be classified into two groups depending on their substrate recognition mechanism. First, a number of Ψ synthases composed of a single polypeptide recognize their substrate RNAs with high specificity and also catalyze the isomerization reaction. Second, the Ψ synthase Cbf5/dyskerin associates with three additional polypeptides and a guide RNA (which is responsible for site specificity) to form an RNP. These guide RNAs are characterized by a conserved secondary structure and two closely related sequence elements called “box H” and “box ACA,” hence the name of the RNP (reviewed in Refs. 3–5).
Box H/ACA guide RNAs were first identified in the nucleoli of eukaryotes (reviewed in Ref. 6). Subsequently, they were also found in archaea and in the Cajal bodies of eukaryotic nuclei. Depending on their source, these box H/ACA RNAs are denoted snoRNA (small nucleolar), sRNA (small), or scaRNA (small Cajal), respectively. Regardless of their cellular location, these RNAs are composed of one or more stem-loops separated by the box H and ACA sequences. Each stem-loop is interrupted by an internal bulge that is complementary in sequence to nucleotides flanking the pseudouridylation site in a substrate RNA (the bulge is also known as the Ψ pocket). This complementarity is sufficient for targeting the box H/ACA RNP to any cellular substrate, as demonstrated by introduction of Ψ to previously unmodified loci in cellular RNAs using recombinant box H/ACA guides (7). The use of guide RNAs endows the box H/ACA RNP with much greater substrate versatility than that displayed by the single-polypeptide Ψ synthases, which typically modify one RNA substrate or several substrates that share structural and sequence similarity (8–10). Although the biological function of Ψ is likely to be different for each of its occurrences, its importance for correct function of tRNA, the ribosome, and the spliceosome is well documented (reviewed in Refs. 11–14).
Some box H/ACA RNPs have functions unrelated to Ψ (reviewed in Refs. 6 and 15). The guide RNA snR30 (yeast nomenclature) is required for cleavage of the 35 S precursor to 18 S rRNA (although it is not itself the nuclease) but is not known to introduce Ψ to any cellular RNA. Predictably, snR30 and its orthologs are essential. Vertebrate telomerase RNA contains an H/ACA domain that is important for telomerase RNP assembly and activity. This domain is not known to introduce Ψ into any cellular RNA either. Mutations in the H/ACA domain of the telomerase RNA as well as in the protein components of the H/ACA RNP are associated with the human bone marrow failure syndrome dyskeratosis congenita. Cells of patients have been shown to be deficient in telomerase activity and to have shorter telomeres than healthy cells (reviewed in Refs. 16 and 17).
Numerous studies have been undertaken over the past decade to elucidate the molecular basis for the functions of box H/ACA RNPs and of their protein and RNA constituents. Catalytically active H/ACA RNPs have been reconstituted in vitro with recombinant archaeal proteins and synthetic RNA (18, 19), giving impetus to crystallographic studies. This year, two groups reported crystal structures of archaeal H/ACA RNPs with substrate RNAs docked in their active sites (20, 21). These structures, together with the previously determined structures of the constituents and subcomplexes of the RNP (22–31), now reveal structural changes that accompany complex formation. From this comparison, it is possible to deduce the roles of the different protein components in assembly of the RNP and also to delineate structural motifs whose importance was previously not recognized. These studies of the box H/ACA RNP parallel recent progress in the structural study of other RNPs, such as the ribosome, the spliceosome, the signal recognition particle, the exon junction complex, and the box C/D RNP (reviewed in Refs. 5 and 32–35).
Overall Structure of the Box H/ACA RNP
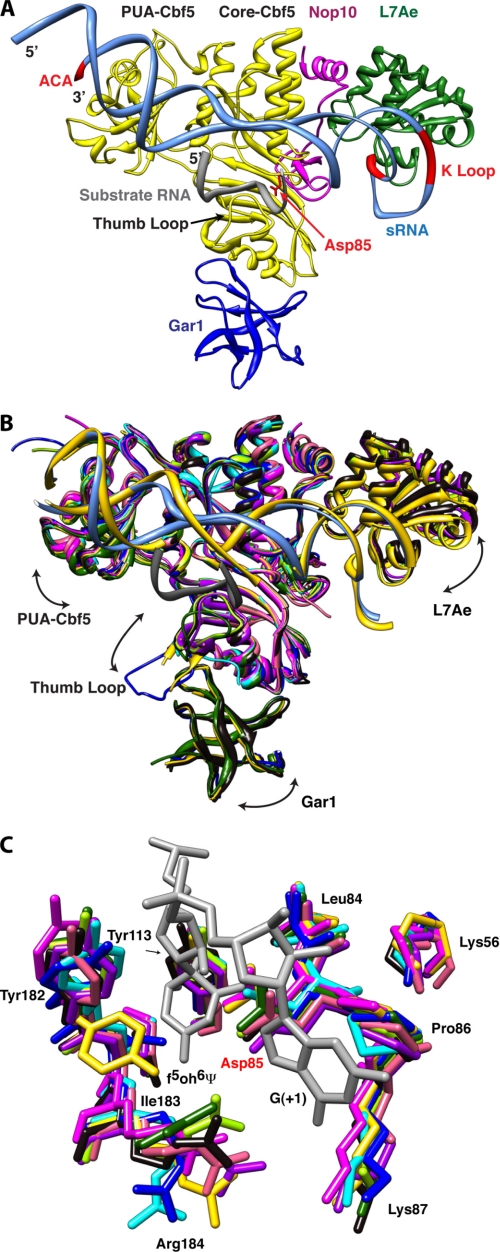
The box H/ACA RNP is composed of four proteins whose structural cores are highly conserved between Archaea and Eukarya (the eukaryotic proteins often have low sequence complexity extensions that hinder structural analyses). Cbf5 (dyskerin in human) is a protein with high sequence identity and structural similarity to members of the TruB family of single-polypeptide Ψ synthases. Like TruB, Cbf5 is composed of a core catalytic domain, which is structurally conserved among all Ψ synthases (4, 36), and a peripheral “PUA” domain (8, 28, 37, 38). The other three conserved proteins are Nop10, L7Ae (called Nhp2 in Eukarya), and Gar1. Nop10 is an elongated protein that associates tightly with Cbf5 and stabilizes its active site structure (27, 28). L7Ae is an RNA-binding protein that specifically recognizes the K-turn (39) and K-loop (40) structural motifs of RNA. The stem-loop structures of box H/ACA RNAs typically contain one of these motifs distal to their Ψ pocket (41). Gar1 is a small basic protein that binds to Cbf5. Fig. 1A shows the overall structure of the RNP.
FIGURE 1.
Structural comparison of box H/ACA RNPs. A, overall structure of the box H/ACA RNP. The conserved ACA sequence and K-turn motif of the guide RNA and the catalytic aspartate residue of Cbf5 are shown in red. Substrate RNA (gray), guide RNA (aqua), Cbf5 (yellow), Gar1 (blue), L7Ae (green), and Nop10 (magenta) are shown. In this structure, the thumb loop is in its closed (or RNA-bound) conformation. B, structural comparison of box H/ACA RNP complexes and subcomplexes. Arrows indicate the directions of molecular motion suggested by variability between structures. Ribbon representations of the superimposed structures of the Cbf5-Nop10 complexes (pink, Protein Data Bank code 2APO (28); and salmon, code 2AUS (27)), the Cbf5-Gar1-Nop10 complex (green, code 2EY4 (26)), the Cbf5-Gar1-L7Ae-Nop10-guide RNA complex (yellow, code 2HVY (24)), the Cbf5-Nop10-guide RNA-substrate RNA complex (sky blue, code 3HJY (21)), the Cbf5-Gar1-Nop10-guide RNA-substrate RNA complex (blue, code 2RFK (25)), the Cbf5-L7Ae-Nop10-guide RNA-substrate RNA complex (light green, code 3HAX (20); and purple, code 3HJW (21)), and the full complex with substrate RNA (black, code 3HAY (20)) are shown. A single guide RNA from substrate-bound (aqua) and substrate-free (yellow) structures and the docked substrate RNA (gray) are shown. C, conserved active site residues of Cbf5. The color scheme is as described for B. Only two nucleotides from the substrate RNA, including the isomerized 5-fluorouridine (5-fluoro-6-hydroxypseudouridine (f5oh6Ψ)), are shown in gray. Structures were superimposed and displayed using UCSF Chimera (48, 49) employing the Cbf5-Nop10 complex (code 2APO (28)) as the reference.
Comparison of the available structures of the H/ACA RNP reveals a combination of rigid-body docking and induced fit in the sequential assembly of the RNP (Fig. 1B). By analogy with the human body, the core of Cbf5 would correspond to the torso, and the PUA domain and L7Ae would be the two arms. The active site cleft runs horizontally across the torso. Gar1 binds lower down. The guide RNA is held between the two arms, with the 3′-ACA sequence at one end of the RNA bound by the PUA domain and with the K-loop at the other end bound by L7Ae. Deletion of the PUA domain of Cbf5 or mutation of the ACA sequence disrupts association of the guide RNA with Cbf5 and results in drastic reduction of pseudouridylation activity (19, 27). The same is true when interaction between L7Ae and the K-loop is disrupted (19). Superposition of the structures of the free Cbf5-Nop10 heterodimer (27, 28) or the free Cbf5-Nop10-Gar heterotrimer (26) with their RNA-bound forms (20, 21, 24) shows that RNA binding results in both L7Ae and the PUA domain moving away from the active site cleft as if stretching the chest (Fig. 1B). L7Ae moves by as much as 4 Å between structures. Gar1 moves up (toward the “belly”) when the proteins bind to guide RNA. Nop10 does not move relative to the core of Cbf5. The catalytic and PUA domains of Cbf5, L7Ae, and Gar1 appear to move essentially as rigid bodies relative to each other. The lower lip of the active site cleft of TruB-type Ψ synthases, including Cbf5, is flanked by a loop called the “thumb loop” (3). This loop shows evidence of induced fit. The loop moves as much as 65° vertically and twists by as much as 50° as it travels between Gar1 and the active site. The thumb loop is up (closed conformation) in the active H/ACA RNP, where it interacts with the guide RNA-substrate RNA complex (Fig. 1B). In the absence of the guide-substrate complex, the thumb can explore different conformations and associates with Gar1 when in its fully open conformation.
Role of Proteins in RNA Remodeling
Structures have been determined of box H/ACA guide RNAs in complex with substrate RNAs in solution (22, 23). Comparison of these structures with those of the RNAs as part of the RNP (20, 21) shows that, although the overall topology of the guide RNA-substrate RNA complex is similar in protein-free and protein-bound states, the structures do not superimpose on each other without substantial distortion (both bending of the helical axis and twisting around it). The conformation of the Ψ pocket of the guide RNA in particular is strikingly altered by protein binding, which appears to unwind and bend the guide RNA-substrate RNA complex to increase accessibility of the target uridine to the active site of Cbf5. Comparison of the structures suggests that movement of the K-loop-bound L7Ae relative to the rest of the complex (Fig. 1B) is an important aspect of the RNA distortion leading to the active RNP.
Liang et al. (25, 42) employed fluorescence spectroscopy to monitor productive docking of the substrate RNA into the active RNP and found that L7Ae is a key player. Comparison of the crystal structures of the guide RNA-substrate RNA complex bound to Cbf5-Nop10 with (20, 21) and without (25) L7Ae reveals markedly different RNA conformations. In addition to structural differences around the K-loop itself, the pseudouridylation site of the RNA undergoes large changes concomitant with L7Ae binding. The U-shaped substrate RNA in the L7Ae-free RNP approaches the active site of Cbf5 from the same direction and orientation as in the full RNP. However, in the complex lacking L7Ae, the uridine that is to be isomerized to Ψ is ∼10 Å away from the catalytic aspartate residue of Cbf5 (21, 25). In addition, the guide RNA near the site of pseudouridylation is poorly ordered. In the presence of L7Ae, the substrate RNA is twisted and widened, the Ψ pocket becomes ordered, and the uridine that is to be isomerized is docked in the active site of Cbf5 (Fig. 1C) (20, 21).
Conserved Histidines, Base Flipping, and a Proline Spine
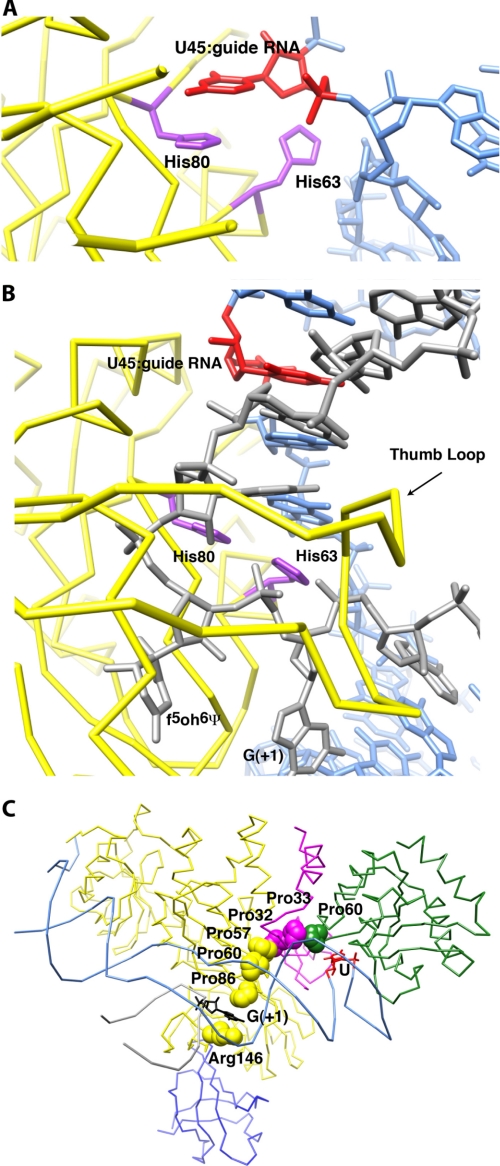
Structure determination of Escherichia coli TruB bound to the TΨC stem-loop of a tRNA revealed that this single-polypeptide Ψ synthase positions its substrate uridine in its active site by flipping the base out of the helical context where it resides in free tRNA. A histidine residue inserts its imidazole ring into the RNA helix, occupying the space vacated by the flipped-out uridine (8). This histidine is conserved in Cbf5 (His80 in Pyrococcus furiosus Cbf5 numbering). Our comparison of box H/ACA structures indicates that both this histidine and His63 (which is conserved in Cbf5 orthologs but not in TruB orthologs) play important roles in positioning the uridine of the substrate RNA in the active site. In the structure of the RNP without substrate RNA (Fig. 2A), the side chain of His80 stacks under a nucleotide from the guide RNA, and the side chain of His63 faces away from the protein. When the substrate RNA binds (Fig. 2B), His80 and His63 interact with the substrate RNA backbone on either side of the site of modification (the crystal structures have the unnatural base 5-fluoro-6-hydroxypseudouridine at this position (4, 8)). The guide RNA nucleobase that stacked on His80 in the substrate-free state is now rotated into the interior of the guide RNA-substrate RNA duplex. His80 does not change conformation between the substrate-free and substrate-bound RNPs. In contrast, His63 rotates over 90°. Mutational analysis has demonstrated the functional importance of His80 (43). Although His63 has not been subjected to site-directed mutagenesis, structural comparison suggests that it plays a key role in coordinating substrate docking in the active site with structural accommodation between the guide RNA and the protein components of the RNP during assembly (and possibly disassembly; see below) of the RNP. The orientation of the side chain of His63 correlates with the status of substrate docking. In structures containing partially docked substrate RNA (21, 25), this side chain occupies orientations that are in between those it adopts in the substrate-free and substrate-bound RNPs.
FIGURE 2.
Putative protein motifs that respond to substrate RNA binding and the isomerization reaction. A, structure of the two histidines that appear to sense the substrate RNA in the substrate-free RNP structure (Protein Data Bank code 2EVY (23)). A nucleotide that stacks on top of one of the histidines is shown in red. The guide RNA is shown in aqua, and the Cbf5 backbone is shown in yellow. B, structure of the same histidines in the presence of substrate RNA (gray). This panel is in the same orientation as A. Substantial changes in the orientation of His63 and guide RNA upon substrate RNA binding are seen. Intercalation of His63 flips out G(+1). The thumb loop hovers over this area. C, the flipped-out G(+1) is stabilized by the conserved Arg146 and a series of prolines that span from Cbf5 to L7Ae. The last proline (Pro60) is also in proximity to the flipped-out U in the K-turn motif of the guide RNA (recognized by L7Ae). f5oh6Ψ, 5-fluoro-6-hydroxypseudouridine.
For His63 to stack against the backbone of the substrate RNA in the substrate-bound state of the box H/ACA RNP, the side chain of the base of the nucleotide immediately 3′ to the site of modification (position +1) needs to be flipped out (Fig. 2B). The active site of Cbf5 makes numerous non-sequence-specific interactions with this nucleotide, whose identity varies from substrate RNA to substrate RNA (Fig. 1C). In particular, the flipped-out base of residue +1 of the substrate stacks between the side chain of the conserved Arg146 and Pro86 residues of Cbf5 (Figs. 1C and 2C). Arg146 is located at the end of the thumb loop. The stacking interaction between the guanidinium group of the side chain of Arg146 and the flipped-out nucleotide +1 is not sequence-specific. Pro86 is immediately adjacent to the catalytic Asp85 in the active site (Fig. 1C). Our comparative structural analysis reveals that Pro86 is part of a conserved “proline spine” that traverses the RNP from the active site of Cbf5 through Nop10 to L7Ae (Fig. 2C). Pro86 is in van der Waals contact with Pro57 and Pro60, which are conserved residues of Motif I of Cbf5. Motif I is a sequence element conserved throughout Ψ synthases that is important for Cbf5 stability and function (44, 45); specifically, these two prolines position Lys56 that hydrogen bonds with the backbone of the catalytic aspartate (Fig. 1C). Pro57 further interacts with the absolutely conserved Pro32 and the less conserved Pro33 of Nop10. Pro33 interacts in turn with the absolutely conserved Pro60 of L7Ae. Finally, Pro60 of L7Ae is in contact with the characteristic (29) flipped-out U residue of the K-loop of the guide RNA. The spectroscopic studies mentioned above underlined the importance of L7Ae binding to the guide RNA for function of the box H/ACA RNP. Our structural analysis of the substrate-bound and substrate-free RNPs indicates that flipping of nucleotide +1 is correlated with productive positioning of the uridine to be modified in the active site of Cbf5. In addition to distortion of the guide RNA that results from binding to the protein components of the RNP, the proline spine may provide a second communication path between the flipped-out nucleotides of the active site and the functionally important L7Ae (Fig. 2C).
Active Site and Enzymatic Turnover
Three active site residues (Asp85, Arg184, and Tyr113 in P. furiosus Cbf5 numbering) are absolutely conserved among all Ψ synthases, including TruB and Cbf5. Their conformations are almost identical among all the H/ACA RNP structures compared herein (Fig. 1C). How is completion of the pseudouridylation reaction detected by Cbf5, and how is this signaled to the other components of the RNP to enable enzymatic turnover? Structural analysis of E. coli TruB led to the suggestion that the conserved active site arginine (which forms a salt bridge with the catalytic aspartate and hydrogen bonds to the isomerized nucleotide) detects the conversion of uridine to Ψ and signals the change in the chemical status of the active site to the thumb loop for substrate release (46). In one of the fully assembled H/ACA RNP structures (21), the equivalent arginine (Arg184), along with neighboring residues Ile183, Thr181, and Gly180, interacts with the isomerized nucleotide through the polypeptide backbone. This segment of Cbf5 is in a β strand-like conformation and appears to be sensitive to the presence of substrate because Tyr182 (which points in the same direction as Arg184) adopts completely different conformations in the structures of the RNP free of substrate (24) and the RNP bound to substrate (20, 21) (Fig. 2C). This segment of Cbf5 is therefore a candidate to mediate communication between the thumb loop and the isomerized nucleotide. It is noteworthy that Tyr182 interacts with the absolutely conserved residues Arg154 and Pro144 of the thumb loop through hydrogen bonding and stacking (also only in the fully assembled RNP structures). These two residues further interact with the substrate RNA through water-mediated hydrogen bonding, and site-directed mutagenesis of Arg154 has been shown to abolish pseudouridylation (20).
The web of interactions outlined above could functionally link the thumb loop conformation to the active site status. As indicated above (Fig. 1B), the thumb loop is quite conformationally variable, binding to the guide RNA-substrate RNA complex in the fully assembled RNA and interacting with Gar1 in other complexes. The thumb loop may need to adopt the open conformation for product release and enzymatic turnover by associating with Gar1, and the hydrogen bonding network connecting it to the active site may bias its conformation. A mutational analysis of Gar1 by Duan et al. (20) suggested effects on the steady-state production of Ψ by the box H/ACA RNP. These data tentatively implicate Gar1 in product release. Fluorescence spectroscopic analysis of the RNP showed that whereas L7Ae promotes formation of the active structure of the RNP, Gar1 does the opposite (42). Gar1 may serve to stabilize the undocked inactive conformation of the RNP by biasing the thumb loop to its open conformation. We therefore speculate that L7Ae and Gar1 sit on opposite ends of a chain of communication that extends from L7Ae through the proline spine, to the active site, to the thumb loop, and finally to Gar1 and that this communication chain allows the RNP sequentially to adopt the different conformations needed for substrate binding, catalysis, and product release.
Perspectives
Although the wealth of crystallographic, NMR, and biochemical characterizations of the H/ACA RNP in many functional states now provides a detailed picture of the conformational transitions undergone by the complex in the course of its enzymatic function, many questions remain. An obvious limitation of the studies so far is that they do not shed light into the role of protein and RNA dynamics in the function of the particle. The catalytic mechanism of Ψ synthases remains shrouded in mystery (4). Little is known about turnover of the H/ACA RNP. For instance, it is not known if a guide RNA remains associated with Cbf5 during turnover and therefore functions processively or if Cbf5 (and Nop10 and Gar1) dissociates from the guide RNA after one round of catalysis. Differences in assembly and function of archaeal and eukaryal box H/ACA RNPs will need to be explored. Crystallographic studies so far have employed guide RNAs that contain a single stem-loop. Many natural guide RNAs have multiple such stem-loops in tandem (47), and it is unknown if each stem-loop assembles an independent catalytic complement of proteins or if the stem-loops interact with each other, for instance, displaying cooperativity. The structures now available provide a solid foundation for future in-depth analyses of the evolutionarily conserved box H/ACA RNP.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM063576 (to A. R. F.). This work was also supported by a special fellowship from the Leukemia and Lymphoma Society (to T. H.). This is the third article in the thematic minireview series on RNA Molecular Machines. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- Ψ
- pseudouridine
- RNP
- ribonucleoprotein.
REFERENCES
- 1.Rozenski J., Crain P. F., McCloskey J. A. (1999) Nucleic Acids Res. 27, 196–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czerwoniec A., Dunin-Horkawicz S., Purta E., Kaminska K. H., Kasprzak J. M., Bujnicki J. M., Grosjean H., Rother K. (2009) Nucleic Acids Res. 37, D118–D121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamma T., Ferré-D'Amaré A. R. (2006) Chem. Biol. 13, 1125–1135 [DOI] [PubMed] [Google Scholar]
- 4.Mueller E. G., Ferré-D'Amaré A. R. (2009) in DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution (Grosjean H. ed.) pp. 363–376, Landes Bioscience, Austin, TX [Google Scholar]
- 5.Reichow S. L., Hamma T., Ferré-D'Amaré A. R., Varani G. (2007) Nucleic Acids Res. 35, 1452–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terns M., Terns R. (2006) Cold Spring Harbor Symp. Quant. Biol. 71, 395–405 [DOI] [PubMed] [Google Scholar]
- 7.Ni J., Tien A. L., Fournier M. J. (1997) Cell 89, 565–573 [DOI] [PubMed] [Google Scholar]
- 8.Hoang C., Ferré-D'Amaré A. R. (2001) Cell 107, 929–939 [DOI] [PubMed] [Google Scholar]
- 9.Hoang C., Chen J., Vizthum C. A., Kandel J. M., Hamilton C. S., Mueller E. G., Ferré-D'Amaré A. R. (2006) Mol. Cell 24, 535–545 [DOI] [PubMed] [Google Scholar]
- 10.Pan H., Ho J. D., Stroud R. M., Finer-Moore J. (2007) J. Mol. Biol. 367, 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charette M., Gray M. W. (2000) IUBMB Life 49, 341–351 [DOI] [PubMed] [Google Scholar]
- 12.Offengand J., Fournier M. J. (1998) in Modification and Editing of RNA (Grosjean H., Benne R. eds.) pp. 229–253, ASM Press, Washington, D. C. [Google Scholar]
- 13.Auffinger P., Westhof E. (1998) in Modification and Editing of RNA (Grosjean H., Benne R. eds) pp. 103–112, ASM Press, Washington, D. C. [Google Scholar]
- 14.Eliceiri G. L. (2006) J. Cell. Biochem. 98, 486–495 [DOI] [PubMed] [Google Scholar]
- 15.Meier U. T. (2005) Chromosoma 114, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vulliamy T. J., Dokal I. (2008) Biochimie 90, 122–130 [DOI] [PubMed] [Google Scholar]
- 17.Kirwan M., Dokal I. (2009) Biochim. Biophys. Acta 1792, 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charpentier B., Muller S., Branlant C. (2005) Nucleic Acids Res. 33, 3133–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker D. L., Youssef O. A., Chastkofsky M. I., Dy D. A., Terns R. M., Terns M. P. (2005) Genes Dev. 19, 1238–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan J., Li L., Lu J., Wang W., Ye K. (2009) Mol. Cell 34, 427–439 [DOI] [PubMed] [Google Scholar]
- 21.Liang B., Zhou J., Kahen E., Terns R. M., Terns M. P., Li H. (2009) Nat. Struct. Mol. Biol. 16, 740–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin H., Loria J. P., Moore P. B. (2007) Mol. Cell 26, 205–215 [DOI] [PubMed] [Google Scholar]
- 23.Wu H., Feigon J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6655–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Ye K. (2006) Nature 443, 302–307 [DOI] [PubMed] [Google Scholar]
- 25.Liang B., Xue S., Terns R. M., Terns M. P., Li H. (2007) Nat. Struct. Mol. Biol. 14, 1189–1195 [DOI] [PubMed] [Google Scholar]
- 26.Rashid R., Liang B., Baker D. L., Youssef O. A., He Y., Phipps K., Terns R. M., Terns M. P., Li H. (2006) Mol. Cell 21, 249–260 [DOI] [PubMed] [Google Scholar]
- 27.Manival X., Charron C., Fourmann J. B., Godard F., Charpentier B., Branlant C. (2006) Nucleic Acids Res. 34, 826–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamma T., Reichow S. L., Varani G., Ferré-D'Amaré A. R. (2005) Nat. Struct. Mol. Biol. 12, 1101–1107 [DOI] [PubMed] [Google Scholar]
- 29.Hamma T., Ferré-D'Amaré A. R. (2004) Structure 12, 893–903 [DOI] [PubMed] [Google Scholar]
- 30.Khanna M., Wu H., Johansson C., Caizergues-Ferrer M., Feigon J. (2006) RNA 12, 40–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reichow S. L., Varani G. (2008) Biochemistry 47, 6148–6156 [DOI] [PubMed] [Google Scholar]
- 32.Kramer G., Boehringer D., Ban N., Bukau B. (2009) Nat. Struct. Mol. Biol. 16, 589–597 [DOI] [PubMed] [Google Scholar]
- 33.Patel S. B., Bellini M. (2008) Nucleic Acids Res. 36, 6482–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Hir H., Andersen G. R. (2008) Curr. Opin. Struct. Biol. 18, 112–119 [DOI] [PubMed] [Google Scholar]
- 35.Wild K., Weichenrieder O., Strub K., Sinning I., Cusack S. (2002) Curr. Opin. Struct. Biol. 12, 72–81 [DOI] [PubMed] [Google Scholar]
- 36.Hoang C., Ferré-D'Amaré A. R. (2004) RNA 10, 1026–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferré-D'Amaré A. R. (2003) Curr. Opin. Struct. Biol. 13, 49–55 [DOI] [PubMed] [Google Scholar]
- 38.Pérez-Arellano I., Gallego J., Cervera J. (2007) FEBS J. 274, 4972–4984 [DOI] [PubMed] [Google Scholar]
- 39.Klein D. J., Schmeing T. M., Moore P. B., Steitz T. A. (2001) EMBO J. 20, 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nolivos S., Carpousis A. J., Clouet-d'Orval B. (2005) Nucleic Acids Res. 33, 6507–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozhdestvensky T. S., Tang T. H., Tchirkova I. V., Brosius J., Bachellerie J. P., Hüttenhofer A. (2003) Nucleic Acids Res. 31, 869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang B., Kahen E. J., Calvin K., Zhou J., Blanco M., Li H. (2008) RNA 14, 2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller S., Fourmann J. B., Loegler C., Charpentier B., Branlant C. (2007) Nucleic Acids Res. 35, 5610–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koonin E. V. (1996) Nucleic Acids Res. 24, 2411–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spedaliere C. J., Hamilton C. S., Mueller E. G. (2000) Biochemistry 39, 9459–9465 [DOI] [PubMed] [Google Scholar]
- 46.Hoang C., Hamilton C. S., Mueller E. G., Ferré-D'Amaré A. R. (2005) Protein Sci. 14, 2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lübben B., Fabrizio P., Kastner B., Lührmann R. (1995) J. Biol. Chem. 270, 11549–11554 [DOI] [PubMed] [Google Scholar]
- 48.Meng E. C., Pettersen E. F., Couch G. S., Huang C. C., Ferrin T. E. (2006) BMC Bioinformatics 7, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.