Abstract
When yeast cells detect external amino acids via their permease-like Ssy1 sensor, the cytosolic precursor forms of Stp1 and Stp2 transcription factors are activated by endoproteolytic removal of their N-terminal domains, a reaction catalyzed by the Ssy5 endoprotease. The processed Stp factors then migrate into the nucleus, where they activate transcription of several amino acid permease genes including AGP1. We report here that the STP1 and STP2 genes most likely derive from the whole genome duplication that occurred in a yeast ancestor. Although Stp1 and Stp2 have been considered redundant, we provide evidence that they functionally diverged during evolution. Stp2 is the only factor processed when amino acids are present at low concentration, and the transcriptional activation of AGP1 promoted by Stp2 is moderate. Furthermore, only Stp2 can sustain Agp1-dependent utilization of amino acids at low concentration. In contrast, Stp1 is only processed when amino acids are present at high concentration, and it promotes higher level transcriptional activation of AGP1. Domain swapping experiments show that the N-terminal domains of Stp1 and Stp2 are responsible for these proteins being cleaved at different amino acid concentrations. Last, induction of the DIP5 permease gene by amino acids depends on Stp2 but not Stp1. We propose that post-whole genome duplication co-conservation of the STP1 and STP2 genes was favored by functional divergence of their products, likely conferring to cells an increased ability to adapt to various amino acid supply conditions.
Keywords: Evolution/Protein, Genetics/Yeast, Organisms/Fungi, Signal Transduction, Transcription, Transcription/Yeast, Transport/Amino Acid
Introduction
Yeast cells possess a signaling pathway activated by external amino acids. This activation leads to transcriptional induction of a set of amino acid permease genes, including AGP1, GNP1, BAP2, BAP3, and DIP5 (1, 2). At the start of this pathway is Ssy1, a protein member of the amino acid permease family (3). Ssy1 is apparently devoid of transport activity and also differs from classical amino acid permeases by a much larger N-terminal cytosolic domain and two larger external loops between transmembrane domains (4–6). According to a recent model, Ssy1 would bind directly to external amino acids, thus stabilizing a signaling, outward-facing conformation (7). In response to this binding, the cytosolic precursor forms of the Stp1 and Stp2 transcription factors are cleaved by the Ssy5 endoprotease (8, 9). The released C-terminal domains of the Stp factors are then translocated into the nucleus (8), where they activate transcription of amino acid permease genes via an upstream UASAA sequence (10, 11). Ssy1-dependent activation of the Ssy5 protease in response to amino acids requires several intermediary factors, i.e. Ptr3, casein kinase I, and the SCFGrr1 ubiquitin ligase complex (1, 2). Ptr3 is a peripheral membrane protein associated with Ssy1 (3, 6, 12). It is hyperphosphorylated in response to amino acids (13). This phosphorylation depends on casein kinase I (13), known to play a crucial role in the amino acid signaling pathway (9, 13). The positive action of casein kinase I appears to be counteracted by a CKI phosphatase complex containing Rts1 as a regulatory subunit (14). The SCFGrr1 complex also plays an essential role in the pathway, but its exact role remains unclear (15). The mechanism by which the Ssy1-Ptr3-casein kinase I-SCFGrr1 factors activate the Ssy5 endoprotease remains unknown. Experiments suggest that Ssy5 activation involves relief from a negative control exerted by the N terminus of the enzyme on its C-terminal catalytic domain (16).
Here we report that the Stp1 and Stp2 factors most likely originate from the whole genome duplication (WGD)4 event that occurred in the ancestor of the Saccharomyces and related yeast lineages (17, 18). We also show that Stp1 and Stp2 have diverged functionally, and we discuss possible scenarios and advantages of this functional divergence.
EXPERIMENTAL PROCEDURES
Strains and Media
The yeast strains used in this study are all isogenic with wild-type Σ1278b except for the mutations mentioned (Table 1). Cells were grown at 29 °C in minimal buffered medium (pH 6.1) (19) with 3% glucose as the carbon source. To this medium proline (10 mm) was added as the sole nitrogen source. Where indicated, other amino acids were also added. The DNA primers used to generate PCR fragments used in yeast strain construction (20) are listed in supplemental Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference |
|---|---|---|
| Strain | ||
| 23344c | ura3 | Laboratory collection |
| KW018 | stp1Δ ura3 | This study |
| KW021 | stp2Δ ura3 | This study |
| KW023 | stp1Δ stp2Δ ura3 | This study |
| KW097 | uga35Δ ura3 | This study |
| KW098 | uga35Δ stp1Δ ura3 | This study |
| KW052 | uga35Δ stp2Δ ura3 | This study |
| 30629c | gap1Δ ura3 | 5 |
| 37053a | gap1Δ stp1Δ ura3 | 11 |
| KW034 | gap1Δ stp2Δ ura3 | This study |
| 34034b | gap1Δ uga35Δ ura3 | 11 |
| 38009a | gap1Δ uga35Δ stp1Δ ura3 | 11 |
| KW068 | gap1Δ uga35Δ stp2Δ ura3 | This study |
| KW037 | gap1Δ stp1Δ stp2Δ ura3 | This study |
| 30633c | gap1Δ agp1Δ ura3 | 5 |
| CA045 | STP1-HA3 ura3 | This study |
| KW057 | STP1-HA3 stp2Δ ura3 | This study |
| JA827 | STP2-(GA)5-HA3 ura3 | This study |
| KW059 | stp1Δ STP2-(GA)5-HA3 ura3 | This study |
| AA01 | AGP1-lacZ ura3 | Laboratory collection |
| KW067 | AGP1-lacZ ptr3Δ ura3 | This study |
| KW070 | AGP1-lacZ ptr3Δ stp1Δ ura3 | This study |
| KW071 | AGP1-lacZ ptr3Δ stp2Δ ura3 | This study |
| CA006 | rts1Δ ura3 | This study |
| CA099 | rts1Δ stp1Δ ura3 | This study |
| CA117 | rts1Δ stp2Δ ura3 | This study |
| CA119 | rts1Δ stp1Δ stp2Δ ura3 | This study |
| KW109 | AGP1-lacZ stp1Δ stp2Δ ura3 | This study |
| FA064 | stp2 ura3 | 11 |
| Plasmid | ||
| YCpAGP1-lacZ | CEN-ARS URA3 AGP1-lacZ | 5 |
| YCpBAP2-lacZ | CEN-ARS URA3 BAP2-lacZ | This study |
| YCpDIP5-lacZ | CEN-ARS URA3 DIP5-lacZ | This study |
| YEp-ptr3–35 | 2μ URA3 ptr3-35 | 9 |
| pKW010 | CEN-ARS URA3 STP1-HA3 | This study |
| pKW012 | CEN-ARS URA3 STP1-HA3ΔNt | This study |
| pKW014 | CEN-ARS URA3 STP1-HA3STP2prom | This study |
| pKW016 | CEN-ARS URA3 STP1-HA3STP1Nt | This study |
| pKW018 | CEN-ARS URA3 STP2-(GA)5-HA3 | This study |
| pKW021 | CEN-ARS URA3 STP2-(GA)5-HA3ΔNt | This study |
| pKW022 | CEN-ARS URA3 STP2-(GA)5-HA3STP1prom | This study |
| pKW024 | CEN-ARS URA3 STP2-(GA)5-HA3STP1Nt | This study |
Plasmids
The plasmids used in this study are listed in Table 1. STP plasmids were constructed by in vivo recombination between the CEN-based pFL38 vector and one, two, or three DNA fragments amplified by PCR using the genomic DNA of strains CA045 (STP1-HA3 ura3) or JA827 (STP2-(GA)5-HA3 ura3) as a template. The oligonucleotides used to generate the PCR fragments are listed in supplemental Table 1. All plasmids were verified by sequencing.
Yeast Cell Extracts and Immunoblotting
Total protein extracts were prepared as described previously (21). Proteins were resuspended in 200 μl of sample buffer and incubated for 10 min at 95 °C. For immunoblot analysis, equal quantities of proteins were loaded onto an 8% SDS-polyacrylamide gel in a Tricine system. After transfer to a nitrocellulose membrane (PerkinElmer Life Sciences), the Stp1-HA3 and Stp2-(GA)5-HA3 proteins were detected with monoclonal antibodies raised against HA (1:10000; Roche Diagnostics). Phosphoglycerate kinase, used as a loading control, was detected with monoclonal antibodies (1:5000; Invitrogen). Primary antibodies were detected with horseradish peroxidase-conjugated anti-mouse IgG secondary antibody (GE Healthcare) followed by enhanced chemiluminescence (Roche Diagnostics).
β-Galactosidase Assays
β-Galactosidase activities were measured as described previously (22) and are expressed in nmol of ο-nitrophenol/min/mg of protein. Protein concentrations were measured with the Folin reagent, and the standard used was bovine serum albumin. For each β-galactosidase assay, the indicated values correspond to the means of at least two independent sets of cultures and experiments, and the error bars correspond to S.D.
Growth Yield Comparison Assays
Flasks of minimal medium containing increasing concentrations of phenylalanine as the sole nitrogen source were inoculated with identical amounts of cells and incubated for 2 days. The optical density at 660 nm reached by each culture was measured, and cultures without phenylalanine served as blanks.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed essentially as described previously (23). All experiments were performed in duplicate using independent cultures. Stp1-HA3 and Stp2-(GA)5-HA3 were immunoprecipitated with Pan-Mouse IgG Dynabeads (Invitrogen) using an anti-HA antibody (12CA5). Immunoprecipitated DNA was analyzed by quantitative real-time PCR on a StepOne Plus machine (Applied Biosystems) with the Platinium SYBR Green qPCR SuperMix-UDG with Rox (Invitrogen). The occupancy level for a specific fragment was defined as the ratio of immunoprecipitated to total DNA. The GAL1 ORF region was used as a non-transcribed control. The value 1.0 was arbitrarily given to the reference signal provided by amplifying the GAL1 gene. The lengths of the qPCR products were 127 bp (AGP1-0), 120 bp (AGP1-4), and 170 bp (AGP1-8). The positions of the qPCR products are depicted in Fig. 7. The oligonucleotide pairs used are listed in supplemental Table 1. The specificity of the qPCR amplification was checked for each oligonucleotide pair by melting curve analysis at the end of each qPCR run. Negative controls where the template DNA was replaced with sterile water revealed that the primers used did not form dimers.
FIGURE 7.
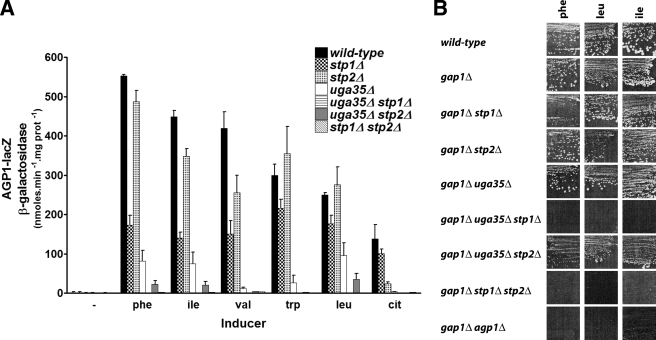
Stp2 is the only factor able to sustain Agp1-dependent utilization of amino acids at low concentration. A, strains 23344c (ura3), 30629C (gap1Δ ura3), 37053a (gap1Δ stp1Δ ura3), KW034 (gap1Δ stp2Δ ura3), and KW037 (gap1Δ stp1Δ stp2Δ ura3) were spread on minimal medium with Phe, Leu, or Ile (100, 200, or 500 μm) as sole nitrogen source and incubated for 5 days at 29 °C. B, strains 30629C (gap1Δ ura3), 37053a (gap1Δ stp1Δ ura3), KW037 (gap1Δ stp2Δ ura3), and KW037 (gap1Δ stp1Δ stp2Δ ura3) were pre-grown to log phase on minimal urea medium. Equal quantities of cells were then added to a minimal medium with phenylalanine at 0, 50, 150, 200, 300, or 400 μm as the sole nitrogen source. The optical density (O.D.) at 660 nm reached by each culture after 2 days was then measured.
Bioinformatic Analysis of STP Orthologues
Orthologues of STP genes were retrieved from the Genolevures data base (24) and used in Blastp analyses (E threshold: 1e-40) against all proteins of several pre-WGD species (Zygosaccharomyces rouxii, Saccharomyces kluyveri, Kluyveromyces thermotolerans, Kluyveromyces lactis, Eremothecium (Ashbya) gossypii) and post-WGD species (Candida glabrata, Kluyveromyces polysporus) (supplemental Table 2) (24). The syntenic context of the STP genes was analyzed with the Yeast Gene Browser online tool (25).
RESULTS
STP1 and STP2 Likely Originate from the Whole Genome Duplication Event
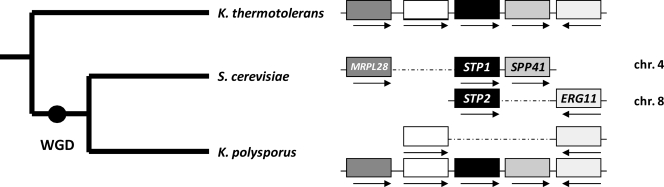
It is established that an ancestor of Saccharomyces cerevisiae underwent a WGD after diverging from yeast lineages like K. lactis, K. thermotolerans, or Kluyveromyces waltii (17, 26). One member of most pairs of “twin” genes was subsequently lost from one of the chromosomes, and a minor fraction of these genes, about 10% in S. cerevisiae, subsisted as duplicates. Analysis of the STP gene orthologues in yeast lineages having diverged before or after the WGD (see “Experimental Procedures”) strongly suggests that the STP1 and STP2 genes derive from the WGD. In support of this view, pre-WGD yeast lineages typically contain a single STP gene, whereas C. glabrata and Saccharomyces castellii, two post-WGD yeast species, contain two. Furthermore, the neighboring genes of the unique STP gene present in pre-WGD yeast species like K. thermotolerans display syntenic conservation with genes close to either STP1 or STP2 of S. cerevisiae (Fig. 1). K. polysporus, a post-WGD species distantly related to S. cerevisiae (27), contains only one STP gene, suggesting that the other gene copy was lost during evolution. In contrast, some of the neighboring genes of this unique STP gene have been conserved as gene duplicates (Fig. 1).
FIGURE 1.
STP1 and STP2 likely derive from the ancestral whole genome duplication event. Left, shown are phylogenetic relationships between three yeast species whose lineages diverged before or after the WGD. Right, shown is a schematic representation of five consecutive genes of K. thermotolerans (chromosome (chr.) 7, genes G05060g, G05082g, G05104g, G05126g, and G05148g) and of their homologues present on chromosomes 4 and 8 of S. cerevisiae and on two chromosomes of K. polysporus. Dotted lines correspond to enlarged spacing between adjacent genes so as to arrange homologous genes in columns. Gene names are indicated only for S. cerevisiae.
Conservation of both STP gene copies in S. cerevisiae and other post-WGD lineages raises the interesting possibility that Stp1 and Stp2 may have diverged functionally during evolution. We have accordingly shown that induction of AGP1 transcription by phenylalanine is reduced by more than 80% in a stp1Δ mutant but remains unaltered in a stp2Δ mutant (11). Yet other papers (8, 28, 29) suggest that Stp1 and Stp2 might be functionally redundant, at least when AGP1 expression was induced by leucine; the stp1Δ and stp2Δ single mutations did not significantly alter this induction, whereas none was observed with the stp1Δ stp2Δ double mutant. We, thus, sought to determine whether the relative contributions of Stp1 and Stp2 to induction of AGP1 expression vary according to the amino acid inducer. For this we isolated a set of mutant strains containing a stp1Δ mutation, a stp2Δ mutation, or both and used them to monitor induction of AGP1 expression by various amino acids. We also analyzed the influence of a deletion mutation altering Uga35/Dal81, a pleiotropic transcription factor contributing importantly to induction of AGP1 transcription (5, 11, 30).
The Relative Contributions of Stp1 and Stp2 to Induction of AGP1 Expression Depend on the Amino Acid Inducer
AGP1 expression was measured 2 h after the addition of phenylalanine, isoleucine, valine, tryptophan, or citrulline at high concentration (5 mm) to cells transformed with a low copy number plasmid bearing the AGP1-lacZ reporter construct (Fig. 2). In the wild type, the induction level varied according to the amino acid, Phe being the strongest and citrulline the weakest inducer. In the stp1Δ stp2Δ double-mutant strain, we observed no induction by any amino acid. Hence, the expression level observed in either single mutant should reflect the ability of the corresponding Stp protein to mediate induction. When Stp1 alone was present, the induction level varied strongly according to the amino acid and was often the same as, or close to that observed in the wild type. As a notable exception, citrulline promoted only weak Stp1-mediated induction. Stp2-mediated induction, on the other hand, varied much less according to the amino acid inducer, its level always remaining lower than the highest Stp1-mediated induction levels. Furthermore, Stp2 appeared largely responsible for induction of AGP1 expression by citrulline, the weakest inducer. Thus, Stp1 and Stp2 do not respond similarly to external amino acids. Interestingly, induction by Leu was about the same in the wild-type, stp1Δ, and stp2Δ strains, indicating that, as previously observed (8, 28, 29), Stp1 and Stp2 are largely redundant under these conditions. With an inducer such as Val, in contrast, the contributions of Stp1 and Stp2 appeared largely additive.
FIGURE 2.
Stp1 and Stp2 are not functionally redundant. A, strains 23344c (ura3), KW018 (stp1Δ ura3), KW021 (stp2Δ ura3), KW097 (uga35Δ ura3), KW098 (uga35Δ stp1Δ ura3), KW052 (uga35Δ stp2Δ ura3), and KW023 (stp1Δ stp2Δ ura3) transformed with the CEN-based plasmid YCpAGP1-lacZ were grown on proline medium. Phe, Ile, Val, Trp, Leu, or citrulline (cit) (5 mm) was then added to the medium for 2 h before harvesting. The reported β-galactosidase activities are the means of at least two experiments. B, strains 23344c (ura3), 30629C (gap1Δ ura3), 37053a (gap1Δ stp1Δ ura3), KW034 (gap1Δ stp2Δ ura3), 34034b (gap1Δ uga35Δ ura3), 38009a (gap1Δ uga35Δ stp1Δ ura3), KW068 (gap1Δ uga35Δ stp2Δ ura3), KW037 (gap1Δ stp1Δ stp2Δ ura3), and 30633c (gap1Δ agp1Δ ura3) were spread on minimal medium with Phe, Leu, or Ile (1 mm) as the sole nitrogen source and incubated for 4 days at 29 °C.
In a previous work (11) we presented results suggesting that the residual induction of AGP1 expression by Phe observed in the stp1Δ mutant was not mediated by Stp2. Our present results obtained with the stp2Δ and stp1Δ stp2Δ strains isolated in this study (Fig. 2) contradict this finding. We have, thus, further analyzed the stp2Δ strain used here and the one (strain FA064) in that earlier study. In the stp2Δ strains used previously, it appears that the antibiotic resistance gene used to delete the STP2 coding region by gene replacement was integrated at the STP2 locus but without deleting the STP2 gene (supplemental Fig. S1). In the newly isolated stp2Δ mutants, STP2 is appropriately deleted. The data obtained with these mutants (Fig. 2) are also fully consistent with those of previous studies showing complete loss of induction in the stp1Δ stp2Δ mutant (8, 28, 29).
Uga35/Dal81 Behaves as a Co-activator of Stp1- and Stp2-mediated Induction
Uga35/Dal81 is a pleiotropic transcription factor containing a Zn(II)2-Cys6 cluster-type DNA binding domain (31, 32). It is essential to full induction of AGP1 expression (5, 11) and to expression of other nitrogen compound-inducible genes, such as those involved in γ-aminobutyric acid, allophanate, and allantoin utilization (31, 32). In each induction mechanism, Uga35/Dal81 acts in conjunction with an inducer-specific transcription factor, e.g. Uga3 for γ-aminobutyric acid (33) and Dal82/DurM for allophanate (34, 35). The results shown in Fig. 2A reveal that Uga35/Dal81 strongly potentiates Stp1-dependent induction, in keeping with previous results (11). A weak though significant induction nevertheless subsists in the absence of Stp2 and Uga35, suggesting that Stp1 alone can activate AGP1 expression to some degree. In contrast, Uga35/Dal81 is essential to Stp2-dependent induction, indicating that Stp2 alone cannot activate AGP1 expression. These results were confirmed by growth tests on media containing a low concentration of Phe, Ile, or Leu as the sole nitrogen source (Fig. 2B). We previously reported that Agp1 is essential to growth on these media when the general amino acid permease, Gap1, is not functional (5). The results presented in Fig. 2B confirm that Agp1 is not functional when both Stp factors or Stp1 and Uga35 are defective, whereas it remains functional if Stp1 alone is functional.
The Stp2 Transcription Factor Responds with Higher Sensitivity to the Amino Acid Sensing System
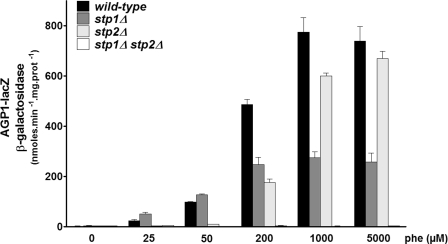
The above results clearly show that Stp1 and Stp2 diverged functionally during evolution. The activating power of Stp1 appears higher, as revealed when induction of AGP1 expression was assayed in response to the strongest inducers like Phe or Ile and by the ability of Stp1 to activate AGP1 expression in the absence of the Uga35/Dal81 co-activator. The activation power of Stp2 is lower, and this might explain why Stp2 absolutely needs Uga35/Dal81 for significant transcriptional activation of AGP1. On the other hand, the stronger response of Stp2 to citrulline, the weakest inducer, raises the possibility that Stp2 might respond more efficiently than Stp1 to limited activation of the amino acid signaling pathway. To test this hypothesis, we examined the induction of AGP1-lacZ expression in response to increasing concentrations of Phe (Fig. 3). In response to low Phe concentrations (25 and 50 μm), AGP1-lacZ induction was weak and principally Stp2-dependent, but at the highest Phe concentrations (1 and 5 mm), the much higher induction of AGP1-lacZ appeared to be largely mediated by Stp1, as shown also in Fig. 2A. Interestingly, in the presence of intermediate Phe concentrations (200 μm), the contributions of Stp1 and Stp2 appeared similar and additive.
FIGURE 3.
The relative contributions of Stp1 and Stp2 to induction of AGP1 expression differ according to the amino acid concentration. Strains 23344c (ura3), KW018 (stp1Δ ura3), KW021 (stp2Δ ura3), and KW023 (stp1Δ stp2Δ ura3) transformed with the CEN-based plasmid YCpAGP1-lacZ were grown on proline medium. Phe was then added to the medium at various concentrations for 2 h.
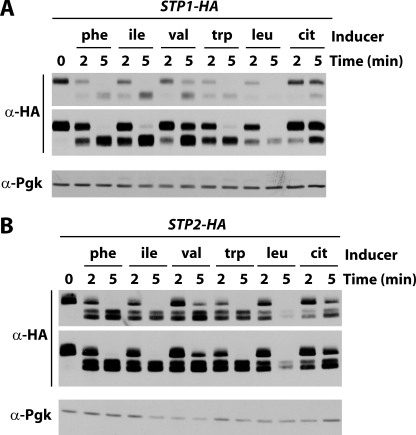
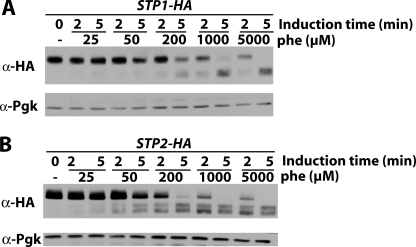
We next tested whether the apparent higher sensitivity of Stp2 to activation by the Ssy1 sensor correlates with a greater propensity to be cleaved by the Ssy5 endoprotease. Each Stp factor was tagged at its C terminus with three repeats of the HA epitope and tested for functionality (supplemental Fig. S2). In the case of Stp2, a linker consisting of glycine-alanine dipeptide repeats had to be inserted between the protein and the 3HA epitope to keep the protein active. In the absence of any amino acid inducer, Stp1 and Stp2 migrated in their uncleaved forms (Fig. 4). Two minutes after the addition of an amino acid, Stp1 and Stp2 appeared to have undergone partial processing. When Phe, Ile, Trp, or Leu was added to the medium, processing of both Stp factors was complete within 5 min. With Val and citrulline, cleavage of Stp1 and Stp2 remained partial even 5 min after the addition of the amino acid to the medium. Stp2 was more efficiently processed than Stp1 in the presence of citrulline, in keeping with the view that Stp2 contributes more importantly than Stp1 to activation of AGP1 expression in response to this poor inducer. We next compared processing of Stp1 and Stp2 in response to increasing concentrations of Phe (Fig. 5). At low Phe concentrations (25, 50, and 200 μm), Stp2 was more efficiently cleaved than Stp1, whereas both Stp factors were processed with similar efficiency in response to higher Phe concentrations (1 and 5 mm).
FIGURE 4.
Efficiencies of Stp1 and Stp2 processing vary according to the amino acid inducer. Strains CA045 (STP1-HA3 ura3) and JA827 (STP2-(GA)5-HA3 ura3) were grown on minimal medium with proline as the sole nitrogen source. Phe, Ile, Val, Trp, Leu, or citrulline (cit) (each at 5 mm) was then added or not to the medium for 2 or 5 min. Total cell extracts were prepared, and immunoblotting was carried out with anti-HA and anti-phosphoglycerate kinase antibodies. Two exposure times are presented for the anti-HA antibody.
FIGURE 5.
Stp2 is more readily processed than Stp1 when amino acids are present at low concentration. Strains CA045 (STP1-HA3 ura3) and JA827 (STP2-(GA)5-HA3 ura3) were grown on minimal medium with proline as sole nitrogen source. Phenylalanine (25, 50, 200, 1000, or 5000 μm) was then added or not (-) to the medium for 2 or 5 min. Total cell extracts were then prepared, and immunoblotting was carried out with anti-HA and anti-phosphoglycerate kinase (PgK) antibodies.
These results indicate that Stp2 has a greater propensity than Stp1 to be processed by the Ssy5 protease when the external amino acid-sensing system is weakly stimulated, i.e. when a good amino acid inducer (like Phe) is present at low concentration or when the sole amino acid present is a weak inducer (like citrulline). Under such conditions, AGP1 induction depends mainly on Stp2. Stp1, in contrast, is efficiently processed only when the amino acid-sensing system is strongly stimulated, i.e. when a good amino acid inducer is present at sufficiently high concentrations. This suggests that Stp1 is less sensitive than Stp2 to cleavage by the Ssy5 endoprotease.
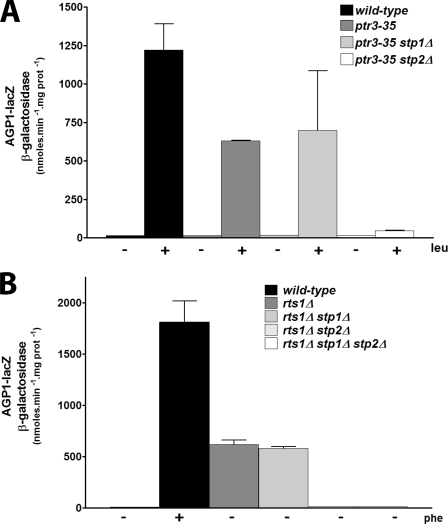
Limited Activation of AGP1 Transcription in the ptr3-35 and rts1Δ Mutants Depends Mainly on Stp2
The Ptr3 factor is associated with Ssy1 and is essential to activation of the Ssy5 endoprotease, with which it interacts (6, 13, 36). In the ptr3-35 mutant, transmission of the amino acid signal is largely impaired, as AGP1 transcription is activable only by leucine (9), the amino acid toward which Ssy1 displays the highest affinity (37). Furthermore, this induction is lower than in the wild type. The limited induction of AGP1 by leucine in the ptr3-35 mutant is, thus, equivalent to weak stimulation of the amino acid sensing system in the wild type and should, thus, depend largely on Stp2. To test this we combined the ptr3-35 mutation with the stp1Δ or stp2Δ mutation. Induction of AGP1 in the ptr3-35 mutant was reduced 2-fold as compared with the wild type, and this residual induction was indeed largely dependent on Stp2 (Fig. 6A). This contrasts with the situation in the wild type, where both Stp1 and Stp2 can mediate maximal induction of AGP1 in response to leucine (Fig. 2).
FIGURE 6.
Limited expression of AGP1 in rts1Δ and ptr3-35 mutants is mediated mainly by Stp2. A, strain AA01 (AGP1-lacZ ura3) and strains KW067 (AGP1-lacZ ptr3Δ ura3), KW070 (AGP1-lacZ ptr3Δ stp1Δ ura3), and KW071 (AGP1-lacZ ptr3Δ stp2Δ ura3) transformed with the YEp-ptr3-35 plasmid were grown on proline minimal medium, and leucine was added (+) or not (-) at 5 mm concentration to the medium for 2 h before cell harvesting. B, strains 23344c (ura3), CA006 (rts1Δ ura3), CA099 (rts1Δ stp1Δ ura3), CA117 (rts1Δ stp2Δ ura3), and CA119 (rts1Δ stp1Δ stp2Δ ura3) transformed with the YCpAGP1-lacZ plasmid were grown on proline minimal medium, and phenylalanine was added (+) or not (-) at 5 mm concentration to the medium for 2 h before cell harvesting.
We also analyzed the respective contributions of Stp1 and Stp2 to induction of AGP1 expression in the rts1Δ mutant. Rts1 is a regulatory subunit of protein phosphatase 2A, a phosphatase complex that probably counteracts the positive action of casein kinase I in the amino acid signaling pathway (14). In the rts1Δ mutant, AGP1 is strongly expressed in the absence of any external amino acid (14). As this constitutive expression does not reach the maximal induction level observed in the wild type, we reasoned that the signaling pathway is only partially activated in the rts1Δ mutant and that the constitutive expression of AGP1 observed in this strain might depend mainly on Stp2. We, thus, assayed AGP1 expression in the rts1Δ strain deleted of the STP1 or STP2 gene (Fig. 6B). The results clearly show that constitutive expression of AGP1 in the rts1Δ mutant depends mainly on Stp2.
In conclusion, limited stimulation of the amino acid signaling pathway in the ptr3-35 and rst1Δ mutants leads to specific activation of Stp2, i.e. the contribution of Stp1 to AGP1 expression is negligible in these mutants. This further shows that Stp2 is more sensitive than Stp1 to activation by the amino acid sensing system.
Stp2 Is Essential to the Utilization of Amino Acids at Low Concentration
As Stp2 is more readily activated than Stp1 when the amino acid sensing system is moderately stimulated, we examined whether this factor might also play a greater role in the utilization of external amino acids present at low concentration. We first compared the growth of the stp1Δ and stp2Δ mutant strains on solid media containing increasing concentrations of Phe, Ile, or Leu as the sole nitrogen source (Fig. 7A). The strains were also deleted of the GAP1 gene, so that amino acid uptake by the mutants was mainly dependent on Agp1 (5). The results clearly show that Stp2 is essential to growth on the lowest concentrations of amino acids. At higher concentrations, both Stp1 and Stp2 can sustain growth. This means that Stp2, but not Stp1, has the ability to activate Agp1 synthesis when amino acids are present at low concentration in the medium. These results were confirmed by comparing growth yields in liquid medium. Cultures containing various initial concentrations of Phe (from 0 to 400 μm) as the sole nitrogen source were inoculated with equal amounts of cells and incubated until stabilization of the optical density (Fig. 7B). At low Phe concentrations (100–250 μm), cell growth appeared to depend mostly on Stp2.
Dissimilar Occupancy of the AGP1 Upstream Region by Stp1 and Stp2
At high Phe concentrations, Stp1 and Stp2 are cleaved with similar efficiency (Fig. 5), but transcriptional induction of AGP1 is largely Stp1-dependent under these conditions (Fig. 3). We, thus, used a chromatin immunoprecipitation assay to compare binding of Stp1 and Stp2 to the AGP1 promoter. Five minutes after Phe addition, both Stp1 and Stp2 were found associated with the AGP1 upstream region but not with the two DNA segments (the gene coding region and the GAL1 gene) used as controls (Fig. 8B). One hour after Phe addition, Stp1-HA was the main factor associated with the AGP1 regulatory region, Stp2 binding being less pronounced. The greater role of Stp1 in induction of AGP1 in response to high concentrations of Phe, thus, correlates with a greater capacity of Stp1 to occupy the AGP1 gene upstream region.
FIGURE 8.
Dissimilar occupancy of the AGP1 upstream region by Stp1 and Stp2. A, schematic representation of the AGP1 gene is shown. The positions of the amplicons (AGP1-0, AGP1-8, and AGP1-4) used as probes in the chromatin immunoprecipitation experiments are indicated. The white box represents the coding region of AGP1. The position of UASAA is also indicated. B, strains CA045 (STP1-HA3 ura3) and JA827 (STP2-(GA)5-HA3 ura3) were grown on proline as the sole nitrogen source, after which Phe was included (+) or not (-) (at 5 mm concentration) for 5 or 60 min. Chromatin extracts were prepared, HA-tagged forms of the Stp factors were immunoprecipitated with anti-HA antibodies, and copurifying DNA was analyzed by qPCR using oligonucleotides hybridizing to the AGP1 upstream region (AGP1-0 and AGP1-8), the AGP1 coding sequence (AGP1-4), or the GAL1 gene (the latter two being used as negative controls). The occupancy level for each specific fragment was defined as the ratio of immunoprecipitated to input DNA. The value 1 was arbitrarily given to the reference signal provided by amplifying the GAL1 gene. C, strains CA045 (STP1-HA3 ura3) and JA827 (STP2-(GA)5-HA3 ura3) were grown on proline as the sole nitrogen source, after which Phe was included (at 5 mm concentration) for 5–120 min. The same strains were also grown on Pro and Phe as sole nitrogen sources (α). Total cell extracts were then prepared, and immunoblotting was carried out with anti-HA and anti-phosphoglycerate kinase (PgK) antibodies.
Why Stp2 binding to the AGP1 upstream region is less pronounced 60 min after the addition of Phe is not known. Stp1 might bind with higher affinity to the AGP1 upstream sequences and progressively displace Stp2 from common binding sites. The Stp2 protein might also be less stable than Stp1. The latter explanation is in fact supported by the results presented in Fig. 8C. The Stp1 and Stp2 proteins tagged with the same HA epitope were detected in the same immunoblot before and several time intervals after the addition of Phe (5 mm) to the cells. In the absence of any amino acid, the signal corresponding to Stp2 was of higher intensity. After the addition of Phe, Stp1 and Stp2 were efficiently processed. Interestingly, whereas the Stp1 signal remained largely stable, the intensity of the Stp2 signal declined and tended to reach that of Stp1 (Fig. 8C). Further experiments showed that Phe-induced degradation of Stp2 is mediated by the proteasome and does not occur in the ssy5Δ mutant in which Stp2 is not processed (data not shown). Hence, Stp2 is more abundant than Stp1 in the absence of any amino acid, but after processing Stp2 is destabilized, and its cellular level turns similar to that of Stp1. It is likely that this destabilization accounts for the reduced occupancy by Stp2 of the AGP1 promoter observed 60 min after Phe addition. Noteworthy, although Stp2 is destabilized upon processing, it promotes efficient AGP1 induction. This destabilization was indeed also observed after the addition of a high Phe concentration to the stp1Δ mutant (data not shown) in which the role of Stp2 in AGP1 transcription is clearly visible (Fig. 3).
The N-terminal Domains of Stp1 and Stp2 Are Responsible for These Proteins Being Cleaved at Different Amino Acid Concentrations
Stp1 and Stp2 are synthesized as latent cytoplasmic proteins with 10-kDa N-terminal domains crucial for their regulation. A study of Stp1 revealed that this N-terminal domain contains a sequence (amino acids 65–97) required for binding to the Ssy5 protease and for Ssy5-dependent processing. The N-terminal domain contains another sequence (amino acids 16–35) required for cytoplasmic retention (or promoter exclusion) of Stp1 when amino acids are not present in the medium (38).
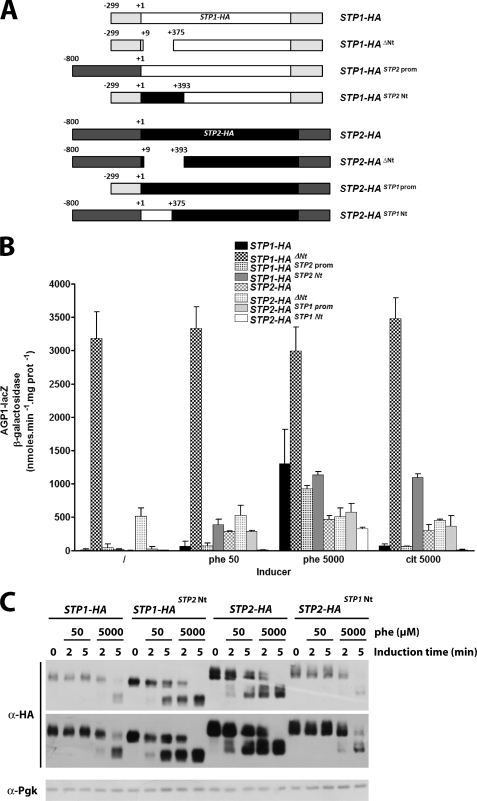
To further study the specific functional properties of Stp1 and Stp2, we constructed genes encoding Stp proteins whose N-terminal domains have been deleted or swapped. We also isolated two gene constructs in which the upstream non-coding sequences of STP1 and STP2 have been swapped (Fig. 9A). These STP genes were first introduced into the stp1Δ stp2Δ mutant, and induction of AGP1 was monitored under the conditions that Stp1 (high Phe concentration) or Stp2 (low Phe concentration or citrulline) normally confers most of the AGP1 induction (Fig. 9B). The results show that the Stp factors lacking their N-terminal domains confer constitutive expression to AGP1, the level of which is much higher in the case of Stp1 versus Stp2. These data are consistent with the proposed role of the N-terminal domains in cytoplasmic retention of Stp factors and confirm that the transcriptional activation power of Stp1 is higher than that of Stp2. The STP genes in which the promoter regions have been swapped behaved like the native STP genes, suggesting that the functional divergence between Stp1 and Stp2 is not due to some differences at gene promoter levels. Finally, the data show that Stp1 containing the N-terminal domain of Stp2 gained the ability to activate AGP1 expression in response to low Phe or citrulline, a typical property of Stp2. Conversely, this property was lost for Stp2 containing the N-terminal domain of Stp1. The latter chimeric protein conserved the ability to respond to a high Phe concentration, but the induction level was limited and close to that conferred by native Stp2. These results show that it is the N-terminal domains of Stp1 and Stp2 that confer to these proteins different abilities to respond to the amino acid concentration. They also confirm that the different transcriptional activation capacities of Stp1 and Stp2 are associated with their C-terminal domains.
FIGURE 9.
The N-terminal domains of Stp1 and Stp2 are responsible for these proteins being cleaved at different amino acid concentrations. A, schematic representation of the plasmid born STP genes used in this experiment is shown. STP1-HA and STP2-HA, STP1 and STP2 genes coding for triple-HA-tagged Stp proteins; STP1-HAΔNt and STP2-HAΔNt, coding for Stp proteins deleted of the N-terminal sequences removed by Ssy5-mediated cleave; STP1-HASTP2prom and STP2-HASTP1prom, in which the upstream non-coding sequences were swapped; STP1-HASTP2Nt and STP2-HASTP2Nt, coding for Stp factors in which the N-terminal sequences were swapped. B, strain KW109 (AGP1-lacZ stp1Δ stp2Δ ura3) transformed with the CEN-based plasmids pKW010 (STP1-HA3), pKW012 (STP1-HA3ΔNt), pKW014 (STP1-HA3STP2 prom), pKW016 (STP1-HA3STP1Nt), pKW018 (STP2-GA5-HA3), pKW021 (STP2-GA5-HA3Nt), pKW022 (STP2-GA5-HA3STP1prom), or pKW024 (STP2-GA5-HA3STP1Nt) was grown on minimal medium with proline as sole nitrogen source. Phe (50 or 5000 μm) or citrulline (5000 μm) was then added or not (/) to the medium for 2 h. C, strain KW022 (stp1Δ stp2Δ ura3) transformed with the CEN-based plasmids pKW010 (STP1-HA3), pKW016 (STP1-HA3switch Nt), pKW018 (STP2-GA5-HA3), or pKW024 (STP2-GA5-HA3switch Nt) was grown on minimal medium with proline as sole nitrogen source. Phenylalanine (50 or 5000 μm) was then added or not to the medium for 2 or 5 min. Total cell extracts were then prepared, and immunoblotting was carried out with anti-HA or anti-phosphoglycerate kinase (Pgk) antibodies. Two exposures times are presented for the anti-HA antibody.
We next compared the processing of the native and chimeric Stp factors in response to low or high Phe concentrations (Fig. 9C). As previously shown, Stp1 is only processed at high Phe concentrations, whereas Stp2 is processed at low or high Phe concentrations. Remarkably, the Stp1 factor with the N-terminal domain of Stp2 was efficiently processed at low Phe concentrations, and the Stp2 factor with the N-terminal domain of Stp1 was only processed at high Phe concentrations. These results correlate with those of the AGP1 induction assay presented above and show that the N-terminal domains of Stp1 and Stp2 are responsible for these proteins being differently cleaved by the Ssy5 endoprotease according to the amino acid concentration.
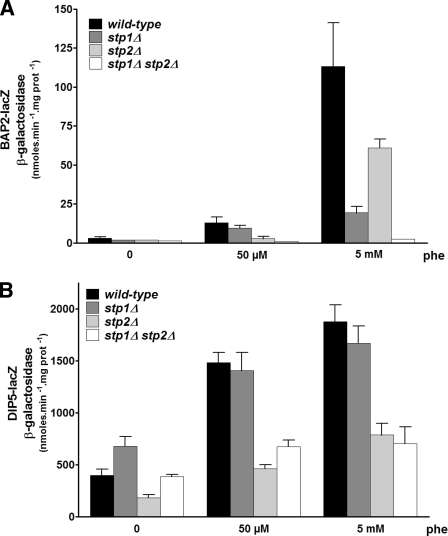
Stp2, but Not Stp1, Mediates Induction of the DIP5 Gene by Amino Acids
The functional divergence of Stp1 and Stp2 during evolution might have led to situations where a single Stp factor mediates amino acid-induced expression of specific amino acid permease genes. To test this we examined the role of each Stp factor in induction by Phe of two additional amino acid permease genes, namely BAP2 and DIP5 (Fig. 10). BAP2 showed an expression profile similar to that of AGP1; that is, moderate, largely Stp2-dependent induction by low Phe and higher, largely Stp1-dependent induction by high Phe. In contrast, the expression of DIP5 was similarly and moderately increased whatever the Phe concentration and in a solely Stp2-dependent manner. Thus, induction of DIP5 by Phe appears mediated specifically by Stp2. It is also entirely dependent on Uga35/Dal81 (data not shown).
FIGURE 10.
Induction of DIP5 expression by amino acids is mediated mainly by Stp2. Strains 23344c (ura3), KW018 (stp1Δ ura3), KW021 (stp2Δ ura3), and KW023 (stp1Δ stp2Δ ura3) transformed with the CEN-based plasmid YCpBAP2-lacZ (A) or YCpDIP5-lacZ (B) were grown on minimal proline medium, and phenylalanine was then added at 50 μm or 5 mm concentration for 2 h before cell harvesting.
DISCUSSION
Traces of WGD events have been found in the genomes of all four eukaryotic kingdoms, including fungi (39). In fungi, the event occurred in the common ancestor of a group of yeast species that includes S. cerevisiae. Only 10% of the resulting duplicated genes survived as duplicates, and a significant fraction of these appear to be transcription factors (39). Here we report that the STP1 and STP2 genes of S. cerevisiae most likely derive from this WGD. This is supported by the presence of only one STP orthologue in pre-WGD yeast species and by the syntenic context conservation of STP genes in post- versus pre-WGD species (Fig. 1). It is worth noting that some pre-WGD species (like S. kluyveri, K. thermotolerans) contain one more distantly related STP gene in addition to the single STP orthologue, suggesting that STP genes have sometimes undergone isolated duplication events. In other pre-WGD species, however, such duplications do not seem to have occurred (e.g. in K. lactis, Z. rouxii). Isolated gene duplication is likely at the origin of the STP1 and STP2 genes existing in Candida albicans. Interestingly, these Stp factors exhibit a clear dichotomy in the genes they transactivate (40).
Conservation of both the STP1 and the STP2 gene in some post-WGD species like S. cerevisiae and C. glabrata suggests that the proteins they encode are not fully redundant. Our results indeed show that Stp1 and Stp2 diverged functionally during evolution; unlike Stp1, Stp2 is efficiently processed by the Ssy5 endoprotease when the Ssy1 sensor system is weakly stimulated, e.g. when good amino acid inducers like Phe are present only at low concentration or when the sole external amino acid is a poor inducer like citrulline. Stp2 is also the main factor responsible for limited AGP1 expression in mutant strains such as ptr3-35 or rts1Δ, where the signaling pathway is only weakly stimulated. The ability of Stp2 to be efficiently activated when amino acids are not abundant is also reflected in its specific ability to sustain Agp1-dependent growth on amino acids at low concentration as the sole nitrogen source. Stp2 also differs from Stp1 in that it exhibits a lesser capacity to activate transcription of the AGP1 gene and probably of other amino acid permease genes as well. Furthermore, this activation is fully dependent on Uga35/Dal81, a protein acting as a transcriptional coactivator of multiple nitrogen source-inducible genes. The lesser transcription-activating capacity of Stp2 as compared with Stp1 is probably related to its more important role when external amino acids are not abundant; under such conditions, high level synthesis of an amino acid permease like Agp1 seems unnecessary.
Unlike Stp2, Stp1 is efficiently processed by the Ssy5 protease only when the amino acid sensing system is highly stimulated, i.e. when a good amino acid inducer is present at a sufficiently high concentration. Furthermore, Stp1 promotes higher level transcription of the AGP1 gene. Another difference between Stp1 and Stp2 is that Stp2 is more abundant that Stp1 in the absence of any amino acid in the medium but is destabilized upon processing so that its cellular level tends to reach that of Stp1.
We could further show by domain swapping experiments that the N-terminal domains of Stp1 and Stp2 are responsible for these proteins being cleaved at different amino acid concentrations. In contrast, their different transcriptional activation capacities, as expected, are associated with the C-terminal parts released after processing. Further experiments will be needed to determine the precise mechanism allowing the N-terminal domains of Stp1 and Stp2 to promote differential processing according to the amino acid concentration. For instance, these domains might interact with the Ssy5 endoprotease with different affinities.
The functional divergence of Stp1 and Stp2 raises interesting questions about the evolutionary scenario that led to this situation. We could envisage that the divergence between STP1 and STP2 has been accompanied by neofunctionalization (39). For instance, the ancestral Stp factor might have been more similar to Stp1, activating, in the presence of amino acids at relatively high concentration, transcription of several genes encoding moderate-affinity amino acid permeases. After WGD, one of the Stp factors (Stp2) would have diverged, acquiring the specific ability to be activated by the Ssy5 protease in response to lower amino acid concentrations along with the ability to activate moderately the transcription of some amino acid permease genes. In parallel to STP gene divergence, other genes might have evolved. For instance, some genes encoding amino acid permeases may have become specifically or more efficiently activable by a single Stp factor. This would notably apply to DIP5, which is regulated only by Stp2. This scenario seems supported by the observation that the unique Stp factor present in pre-WGD species is more closely related to Stp1, as is the unique Stp factor having subsisted in K. polysporus, a post-WGD species.
Our results show that the system enabling yeasts to activate amino acid permease genes in response to external amino acids provides an interesting model for addressing the question of how a signaling pathway can evolve for improved fitness or for adaptation to different environments. This is also true for the glucose signaling pathway activated by the transporter-like Snf3 and Rgt2 glucose sensors. The latter proteins also derive from the WGD and display high and low affinity for glucose, respectively. Furthermore, a recent study reported some functional divergence between Mth1 and Std1, two other factors deriving from WGD and playing a key role in transducing the glucose signal from the cell surface to the nucleus (41). Investigating these nutrient signaling pathways in other pre- and post-WGD yeast species certainly appears as a promising direction of future investigation.
Supplementary Material
Acknowledgments
We are grateful to Catherine Jauniaux and Lydia Spedale for excellent technical assistance and to Maxime Wéry and Denis Lafontaine for help in chromatin immunoprecipitation experiments. We also thank members of the laboratory for fruitful discussions.
This work was supported by Fonds de la Recherche Scientifique Médicale Grant 3.4.592.08.F and Actions de Recherche Concertée Grant 04/09-307 of the Communauté Française de Belgique and by CIBLES Grant 716760 of the Région Wallonne de Belgique.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- WGD
- whole genome duplication
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- HA
- hemagglutinin
- qPCR
- quantitative PCR.
REFERENCES
- 1.Boles E., André B. (2004) in Molecular Mechanisms Controlling Transmembrane Transport (Boles E., Krämer R., eds) pp. 121–153, Springer-Verlag, Berlin [Google Scholar]
- 2.Ljungdahl P. O. (2009) Biochem. Soc. Trans. 37, 242–247 [DOI] [PubMed] [Google Scholar]
- 3.Jørgensen M. U., Bruun M. B., Didion T., Kielland-Brandt M. C. (1998) Yeast 14, 103–114 [DOI] [PubMed] [Google Scholar]
- 4.Didion T., Regenberg B., Jørgensen M. U., Kielland-Brandt M. C., Andersen H. A. (1998) Mol. Microbiol. 27, 643–650 [DOI] [PubMed] [Google Scholar]
- 5.Iraqui I., Vissers S., Bernard F., de Craene J. O., Boles E., Urrestarazu A., André B. (1999) Mol. Cell. Biol. 19, 989–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klasson H., Fink G. R., Ljungdahl P. O. (1999) Mol. Cell. Biol. 19, 5405–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu B., Ottow K., Poulsen P., Gaber R. F., Albers E., Kielland-Brandt M. C. (2006) J. Cell Biol. 173, 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andréasson C., Ljungdahl P. O. (2002) Genes Dev. 16, 3158–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdel-Sater F., El Bakkoury M., Urrestarazu A., Vissers S., André B. (2004) Mol. Cell. Biol. 24, 9771–9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Boer M., Bebelman J. P., Gonçalves P. M., Maat J., Van Heerikhuizen H., Planta R. J. (1998) Mol. Microbiol. 30, 603–613 [DOI] [PubMed] [Google Scholar]
- 11.Abdel-Sater F., Iraqui I., Urrestarazu A., André B. (2004) Genetics 166, 1727–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes D., Lai W., Breslav M., Naider F., Becker J. M. (1998) Mol. Microbiol. 29, 297–310 [DOI] [PubMed] [Google Scholar]
- 13.Liu Z., Thornton J., Spírek M., Butow R. A. (2008) Mol. Cell. Biol. 28, 551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckert-Boulet N., Larsson K., Wu B., Poulsen P., Regenberg B., Nielsen J., Kielland-Brandt M. C. (2006) Eukaryot. Cell 5, 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernard F., André B. (2001) FEBS Lett. 496, 81–85 [DOI] [PubMed] [Google Scholar]
- 16.Andréasson C., Heessen S., Ljungdahl P. O. (2006) Genes Dev. 20, 1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfe K. H., Shields D. C. (1997) Nature 387, 708–713 [DOI] [PubMed] [Google Scholar]
- 18.Scannell D. R., Butler G., Wolfe K. H. (2007) Yeast 24, 929–942 [DOI] [PubMed] [Google Scholar]
- 19.Jacobs P., Jauniaux J. C., Grenson M. (1980) J. Mol. Biol. 139, 691–704 [DOI] [PubMed] [Google Scholar]
- 20.Wach A. (1996) Yeast 12, 259–265 [DOI] [PubMed] [Google Scholar]
- 21.Hein C., Springael J. Y., Volland C., Haguenauer-Tsapis R., André B. (1995) Mol. Microbiol. 18, 77–87 [DOI] [PubMed] [Google Scholar]
- 22.André B., Hein C., Grenson M., Jauniaux J. C. (1993) Mol. Gen. Genet. 237, 17–25 [DOI] [PubMed] [Google Scholar]
- 23.Wery M., Ruidant S., Schillewaert S., Leporé N., Lafontaine D. L. (2009) RNA. 15, 406–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherman D. J., Martin T., Nikolski M., Cayla C., Souciet J. L., Durrens P. (2009) Nucleic Acids Res. 37, D550–D554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrne K. P., Wolfe K. H. (2006) Nucleic Acids Res. 34, D452–D455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellis M., Birren B. W., Lander E. S. (2004) Nature 428, 617–624 [DOI] [PubMed] [Google Scholar]
- 27.Scannell D. R., Frank A. C., Conant G. C., Byrne K. P., Woolfit M., Wolfe K. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8397–8402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Boer M., Nielsen P. S., Bebelman J. P., Heerikhuizen H., Andersen H. A., Planta R. J. (2000) Nucleic Acids Res. 28, 974–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen P. S., van den Hazel B., Didion T., de Boer M., Jørgensen M., Planta R. J., Kielland-Brandt M. C., Andersen H. A. (2001) Mol. Gen. Genet. 264, 613–622 [DOI] [PubMed] [Google Scholar]
- 30.Boban M., Ljungdahl P. O. (2007) Genetics 176, 2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bricmont P. A., Daugherty J. R., Cooper T. G. (1991) Mol. Cell. Biol. 11, 1161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coornaert D., Vissers S., André B. (1991) Gene 97, 163–171 [DOI] [PubMed] [Google Scholar]
- 33.André B. (1990) Mol. Gen. Genet. 220, 269–276 [DOI] [PubMed] [Google Scholar]
- 34.Dorrington R. A., Cooper T. G. (1993) Nucleic Acids Res. 21, 3777–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.André B., Jauniaux J. C. (1990) Nucleic Acids Res. 18, 7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernard F., André B. (2001) Mol. Microbiol. 41, 489–502 [DOI] [PubMed] [Google Scholar]
- 37.Gaber R. F., Ottow K., Andersen H. A., Kielland-Brandt M. C. (2003) Eukaryot. Cell 2, 922–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andréasson C., Ljungdahl P. O. (2004) Mol. Cell. Biol. 24, 7503–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conant G. C., Wolfe K. H. (2008) Nat. Rev. Genet. 9, 938–950 [DOI] [PubMed] [Google Scholar]
- 40.Martínez P., Ljungdahl P. O. (2005) Mol. Cell. Biol. 25, 9435–9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabina J., Johnston M. (2009) J. Biol. Chem. 284, 29635–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.