Abstract
Mutations in N-RAS and B-RAF, which commonly occur in melanomas, result in constitutive activation of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated protein kinase (ERK) signaling. Active ERK increases expression and activity of the c-Jun transcription factor, linking ERK and Jun N-terminal kinase (JNK) cascades. Here, we show that c-Jun regulates transcription of phosphoinositide-dependent kinase 1 (PDK1) with a concomitant impact on Akt and protein kinase C (PKC) activity and related substrates. Inhibition of c-Jun reduces PDK1 expression and attenuates Akt and PKC activity, which can be restored by exogenous PDK1. c-Jun regulation of PDK1 in melanoma contributes to growth rate and the ability to form tumors in mice. Correspondingly, increased levels of c-Jun in melanoma cell lines coincide with up-regulation of PDK1 and phosphorylation of PKC and Akt. The identification of c-Jun as a transcriptional regulator of PDK1 expression highlights key mechanisms underlying c-Jun oncogenic activity, and provides new insight into the nature of up-regulated Akt and PKC in melanoma.
Keywords: Spleen, Tissue Plasminogen Activator, AKT, ERK, PDK1, PKC, c-Jun, Melanoma
Introduction
c-Jun is a primary component of the AP-1 transcription complex regulated by the JNK4 signaling pathway, and contributes to cell proliferation, cell death, and inflammation in response to a plethora of extracellular stimuli (1). Mounting evidence indicates that cellular Jun is involved in human cancer. It is frequently overexpressed, and the full oncogenic properties of some cancer cells might require elevated Jun function. No activating Jun mutations have been found in human cancers, and there have been no reports of C-JUN amplification or recurring chromosomal translocations that involve C-JUN. Instead, c-Jun can be constitutively up-regulated by activation of upstream kinases. We recently found that constitutively active ERK, which is commonly found in melanoma due to activating mutations in B-RAF or N-RAS (2, 3), increases C-JUN transcription and stability via cAMP response element-binding activation and glycogen synthase kinase 3 (GSK3) inactivation, respectively (4). Consistent with its regulation by ERK, c-Jun phosphorylation is required for Ras-induced transformation of fibroblasts in vitro and Ras-induced skin tumorigenesis in vivo (5). Along these lines, c-Jun has been shown to be oncogenic in several tumor types, including human sarcomas (6), breast cancer (7), and lung cancer (8). Moreover, a genetic mouse model revealed c-Jun oncogenic activity in liver tumors, in part through antagonizing the tumor suppressor protein p53 (9). Studies from our laboratory indicate an important role of c-Jun together with its heterodimeric partner activating transcription factor 2 in acquisition of resistance to apoptosis by melanoma cells (10). This observation, combined with up-regulation of c-Jun in melanoma due to re-wired ERK signaling (4), underscores its role in melanoma.
Although several c-Jun target genes have been identified, the mechanism by which c-Jun affects tumorigenesis is not completely understood (11). In the present work we show that c-Jun regulates cell growth and tumorigenesis by regulating phosphoinositide-dependent kinase 1 (PDK1) and their targets. PDK1 is a Ser/Thr kinase required for activation of protein kinases in the AGC kinase superfamily that play important roles in cancer progression, such as protein kinase B (also known as Akt), protein kinase C (PKC), and S6K (12, 13). To be active, these kinases require phosphorylation at both the activation loop and the hydrophobic domain (e.g. Thr308 and Ser473 for Akt1) by PDK1 and mammalian target of rapamycin complex 2, respectively (14, 15). Because PDK1 is activated upon autophosphorylation, its kinase activity is thought to be regulated by protein-protein interaction, changes in subcellular localization, or other phosphorylation-independent mechanisms (16). c-Jun-dependent regulation of PDK1 and subsequent activation of downstream substrates Akt and PKC in melanoma offers important new insight into the mechanism underlying c-Jun oncogenic activity.
EXPERIMENTAL PROCEDURES
Melanoma Samples and Cell Lines
Melanoma samples were obtained from regional dermal metastases, nodal metastases, or distant sites of metastases as described (4). Melanoma lines were kindly provided by Dr. M. Herlyn. c-Jun mutant fibroblasts were kindly provided by R. Wisdom. Cells were transfected with calcium phosphate or by Lipofectamine PLUS reagent (Invitrogen) following the manufacturer's protocol.
Constructs
Constructs encoding glutathione S-transferase-Jun-(1–89), MEKK, and FLAG-TAM67 were previously described (10, 17). FLAG-TAM67 was cloned into pBABE-Puro. TAM67 inhibits AP-1 activity by dimerizing with wild-type AP-1 proteins to yield low activity dimers containing only one transactivation domain. A plasmid encoding ΔMEKK was kindly provided by M. Karin. pBABE myr-Akt was provided by Dr. P. Tsichlis. Plasmids encoding WT PDK1 and the A280V mutant were kindly provided by Dr. Liu (18). cDNAs from these plasmids were cloned into pBABE-Hygro. Viral particles were obtained from supernatants of transfected human embryonic kidney 293T cells.
RNA Interference
siCONTROL (scrambled) and PDK1-specific SMARTpool reagents were used (Dharmacon). c-Jun-specific siRNA oligonucleotide was obtained from Qiagen, and Sp1-specific and Ets-1-specific siRNA oligonucleotides from Ambion. c-Jun-specific shRNA clones were obtained from Open Biosystems. Transfection of 50 nm duplexes into melanoma cells was performed using Lipofectamine 2000 or nucleofection using Amaxa reagent V. Lentiviral shRNA clones packaged in human embryonic kidney 293T cells were used for corresponding infections. Protein extracts were obtained 48–60 h after transfection.
Antibodies and Immunoblotting
Antibodies and reagents were purchased as follows: antibodies to JNK1, JNK2, P-c-Jun, c-Jun, P-RSK2 (Ser227), GSK, SP1, and Ets-1 were from Santa Cruz Biotechnologies. Antibodies to PDK1, P-Akt (Thr308), P-Akt (Ser473), Akt, pan-P-PKC (activation loop site, Thr514 of PKCγ), pan-P-PKC (hydrophobic site, Ser660 of PKCβII), pan-PKC, P-JNK (Thr183/Tyr185), P-Foxo3a, Foxo3a, P-GSK3β (S9), RSK2, S6K1, proliferating cell nuclear antigen, and c-Jun (L70B11) were from Cell Signaling. Because PKC autophosphorylation at the hydrophobic site in a given melanoma cell line depends on the efficiency of activation loop phosphorylation by PDK1, we used the P-PKC (Ser660) antibody to monitor activation loop phosphorylation. Antibodies to PRAS40 and P-PRAS40 (Thr246) were from Invitrogen. Antibodies to P-S6K1 (Thr229) were from Abcam. Antibodies against PKC isoforms were from BD Biosciences. Antibodies to hemagglutinin and FLAG were from Babco and Sigma, respectively. Final concentrations of reagents were as follows: LY294002 (Calbiochem), 10 μm; MG132 (Calbiochem), 10 μm; actinomycin D (5 μg/ml). Protein extract preparation and Western blot analysis were as described (4). β-Actin or α-tubulin antibodies were used to monitor loading. Where multiple bands are present (i.e. in PDK1 and c-Jun blots) the relevant band (see Fig. 1, arrow) was confirmed by siRNA and/or by using different antibodies.
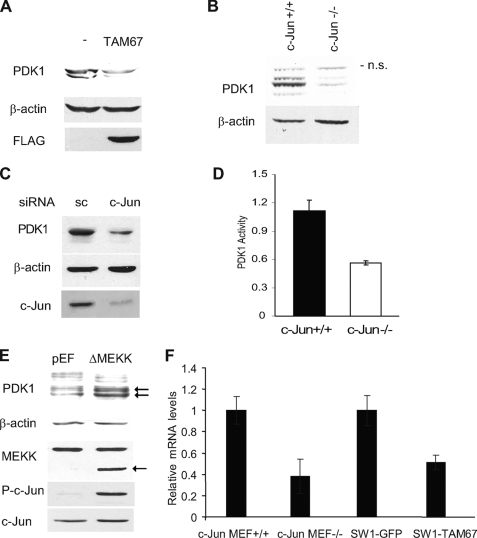
FIGURE 1.
c-Jun regulates PDK1 transcription. A, c-Jun regulates PDK1 expression. Protein extracts (40 μg) from SW1 cells stably transfected with FLAG-TAM67 and control SW1 cells were blotted with PDK1 and FLAG antibodies. B, PDK1 levels are regulated by c-Jun expression. Protein extracts (40 μg) from c-Jun+/+ fibroblasts and c-Jun−/− fibroblasts were blotted with the indicated antibodies. C, c-Jun siRNA decreases PDK1 protein levels in melanoma. Protein extracts (40 μg) from Lu1205 cells transfected with scrambled (sc) oligonucleotides or a siRNA specific for c-Jun were analyzed by Western blots using the indicated antibodies. D, PDK1 activity is reduced in c-Jun−/− fibroblasts. Protein extracts from c-Jun+/+ or c-Jun−/− fibroblasts were immunoprecipitated with PDK1 antibody, and PDK1 kinase activity was measured by an in vitro kinase assay as indicated under “Experimental Procedures.” Results shown are mean ± S.D. of three independent experiments. E, activation of the JNK/c-Jun pathway increases PDK1 expression. Human embryonic kidney 293T cells were transfected with 0.2 μg of ΔMEKK1 or empty pEF plasmid. Protein extracts were obtained 48 h post-transfection and blotted with the indicated antibodies. The arrow indicates bands corresponding to PDK1 and ΔMEKK1. F, inhibition of c-Jun decreases PDK1 mRNA levels. Relative levels of PDK1 mRNA were determined by real time PCR in c-Jun−/− and control fibroblasts and in control SW1 and SW1-TAM67 cells. Results shown are mean ± S.D. of the respective relative concentrations. A representative experiment (of three performed) is shown.
Real Time PCR
Real Time PCR was performed as described (4). Specific primers used for PCR were as follows: PDK1 forward, tgactgcaaagatggaaacg; PDK1 reverse, tgagaaggtccgagttcttg. Human β-actin served as an endogenous control. Reactions were run in triplicate. The target mRNA concentration of control cells, normalized to the level of β-actin mRNA, was set to 1.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed using the Magna-Chip (Upstate) according to the manufacturer's instructions. Cells were fixed with 37% formaldehyde and sheared chromatin from Lu1205 melanoma cells was immunoprecipitated and subjected to PCR. The following primers corresponding to the proximal region of the PDK1 promoter were used: forward, cggcgcgacggaagtcctgc; reverse, tcctcctccccgaagcggag. Antibodies against Sp1, c-Fos, and c-Jun (sc-44 and sc-45) were from Santa Cruz Biotechnologies. IgG control, anti-AcH3 antibody, and glyceraldehyde-3-phosphate dehydrogenase oligonucleotides were provided by the kit. All antibodies were polyclonal.
Avidin-Biotin Complex DNA Binding Assay
Nuclear extracts obtained by using NE-PER extraction reagents (Thermo Scientific) were supplemented with 0.5% Nonidet P-40 and incubated with 3 μg of biotin-labeled box 3 oligo (5′-GGGCGACGGGGCGGGCGCAGGATG-3′) in the presence of poly-(dI-dC) (20 μg/ml) for 2 h. Box 3 oligo-bound proteins were captured using streptavidin-agarose (Invitrogen) (1 h incubation), followed by extensive washes with washing buffer (20 mm HEPES, 150 mm NaCl, 20% glycerol, 0.5 mm EDTA, and 0.5% Nonidet P-40) and analysis using SDS-PAGE and Western blots.
Luciferase Assays
The PDK1 promoter was amplified from genomic DNA from HeLa cells. Fragments of different lengths from the PDK1 promoter were amplified using the GC-rich PCR kit (Roche) and cloned into pGL2-basic vector using KpnI and HindIII. Site-directed mutagenesis of AP-1 and Sp1 sites were performed using the QuikChange II kit (Stratagene) following the manufacturer's protocol. HeLa, MeWo, SW1, or Lu1205 cells were transfected with reporter plasmid along with control vector, c-Jun, or TAM67. Cell lysates were prepared from cells after 24 or 48 h. Luciferase activity was normalized with β-galactosidase activity. Results are shown as the mean (bar) ± S.D.
PDK1 and Akt Kinase Activity
PDK1 kinase activity was measured in PDK1 immunoprecipitates obtained from 600 μg of protein extracts from Jun+/+ and Jun−/− fibroblasts. PDK1 kinase activities were measured using the PDK1 Immunoprecipitation Kinase Assay Kit (Upstate). The activity of Akt was measured in immunoprecipitates obtained from 600 μg of protein extracts by an Akt kinase assay using histone 2B as a substrate as described by Chan et al. (19).
Xenograft Assay and Cell Growth in Culture
For measuring in vitro growth, Lu1205, Lu1205 expressing TAM67, and Lu1205 cells expressing both TAM67 and the PDK1 constitutive active form (A280V) were plated (104 cells/well) at day 0. At days 3, 5, 7, and 9, cells were trypsinized and counted. The medium was changed every 3 days. For the assessment of in vivo tumor growth, the cells described above were trypsinized, resuspended in phosphate-buffered saline, and injected subcutaneously (1.5 × 106) into 8-week-old female nude mice in the lower left flank. For tumor growth of c-Jun knockdown cells, Lu1205 and Lu1205 cells stably expressing PDK1(A280V) were transduced with c-Jun shRNA lentiviral clones. After selection using puromycin, each group of cells (5 × 105) was injected subcutaneously into a group of 5 mice as described above. Tumor size was measured every week.
RESULTS
c-Jun Regulates PDK1 Transcription
We recently showed that ERK activation of c-Jun contributes to increased JNK activity, through transcription of receptor for activated C kinase-1 (RACK1), a PKC adaptor protein that enhances JNK activity (17, 20). In pursuing these studies we observed a decrease in total and P-PKC levels in cells with impaired c-Jun function (see below). Because PKC phosphorylation within its activation loop by PDK1 affect its stability and activity (12, 21), we have explored the possible impact of c-Jun on PDK1 activity. Analysis of mouse fibroblasts deficient in c-Jun (22) and melanoma cells in which c-Jun transcriptional activities are effectively inhibited by a dominant-negative c-Jun construct (TAM67) or c-Jun siRNA revealed a marked reduction of PDK1 protein levels (Fig. 1, A–C). In line with diminished PDK1 expression, c-Jun-deficient fibroblasts also exhibited reduced PDK1 kinase activity (Fig. 1D). Consistent with these findings, expression in human embryonic kidney 293T cells of constitutively active MEKK1 (ΔMEKK1), which elicits strong activation of JNK and c-Jun, increased activation of c-Jun and elevated PDK1 expression levels (Fig. 1E). These data suggest that c-Jun positively regulates PDK1 expression. To assess a possible role of c-Jun in regulation of PDK1 mRNA, we performed real time PCR. A marked inhibition of PDK1 transcription was observed in cells in which c-Jun expression was either absent or inhibited by TAM67 (Fig. 1F), suggesting that changes seen at the protein level may be attributed to regulation of PDK1 mRNA levels by c-Jun.
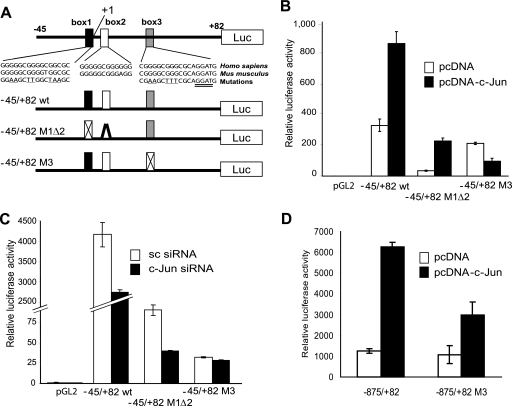
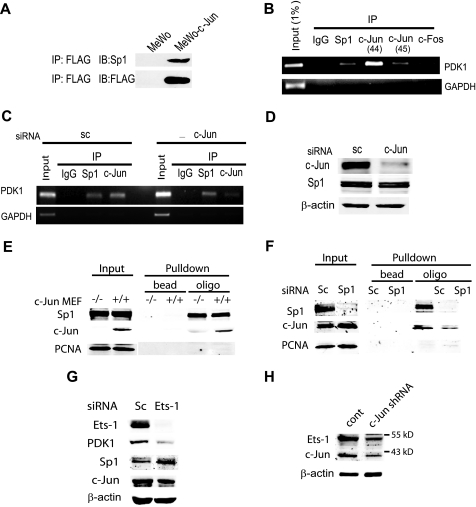
To determine whether c-Jun also affects PDK1 protein levels by affecting its degradation, we monitored changes in PDK1 steady state levels in the presence of the proteasome inhibitor MG132. PDK1 protein levels were slightly increased following MG132 treatment (supplemental Fig. S1); however, we attribute this to increased PDK1 mRNA levels (supplemental Fig. S2) resulting from MG132-induced c-Jun stabilization. Similarly, neither PDK1 mRNA nor protein levels increased following MG132 treatment in TAM67-expressing cells (supplemental Figs. S2 and S3). These results suggest that PDK1 degradation is not altered by c-Jun and that changes in PDK1 levels observed in c-Jun-deficient cells are due to decreased PDK1 mRNA levels. Because c-Jun did not affect PDK1 mRNA stability either (supplemental Fig. S4), we assessed whether c-Jun regulates PDK1 mRNA levels by increasing its transcription. Consensus AP-1 sites were identified at position −924 and between −900 and −300 of the PDK1 promoter (supplemental Fig. S5). Although transfection of c-Jun increased the luciferase activity driven by the −971 to +82 fragment of PDK1 promoter (supplemental Figs. S6–S9) mutating or deleting the AP-1 site did not affect the reporter activity (supplemental Fig. S6–S8). Similarly, the decrease on reporter activity induced by TAM67 persisted after mutation or deletions were performed on the PDK1 promoter (supplemental Fig. S6–S8). Intriguingly, 3 GC-rich boxes between −45 and +82 were found to mediate c-Jun-dependent trans-activation of the PDK1 promoter (supplemental Figs. S8 and S9 and Fig. 2, A–C). These boxes contain 5 Sp1 (5′-GGGCGG-3′) sites that are conserved between human and mouse, but not AP-1 response elements (Fig. 2A). Mutations that alter Sp1 box 1, together with deletion of box 2 (−45/+82 M1Δ2) reduced overall reporter activity (Fig. 2B), but did not abolish responsiveness to c-Jun expression (−45/+82 WT, Fig. 2B). On the other hand, mutations within box 3 (−45/+82 M3) barely affected basal transcription but attenuated (>8-fold) c-Jun-mediated transcription (Fig. 2B). Consistent with these observations, c-Jun siRNA reduced transcription driven by the −45/+82 M1Δ2 fragment but not by the box 3-mutated construct (Fig. 2C). In line with this data, c-Jun mediated transcription was abrogated when box 3 mutations were introduced in a large fragment (−875/+82) of the PDK1 promoter (Fig. 2D). These results suggest that c-Jun enhances PDK1 transcription through Sp1 sites located in box 3. Consistent with our findings, Sp1 was shown capable of recruiting c-Jun for transactivation of a number of genes including 12(S)-lipoxygenase, keratin 16, neuronal nicotinic acetylcholine receptor β4, and p21WAF1/CIP1 (23–26). Further support for the role of c-Jun in the regulation of PDK1 transcription comes from immunoprecipitation experiments and chromatin immunoprecipitation analysis. Sp1 and c-Jun do interact in melanoma cells (Fig. 3A) and chromatin immunoprecipitation of Sp1, c-Jun, or c-Fos enabled amplification of PDK1 promoter sequences bearing the Sp1 box 3 sequences (Fig. 3B), demonstrating in vivo binding of these transcription factors to a 130-bp fragment from the PDK1 promoter containing Sp1 boxes 1 to 3. As expected, binding of c-Jun to the promoter was reduced in cells transfected with c-Jun siRNA (Fig. 3, C and D). To further demonstrate binding of c-Jun to box 3 we performed gel shift assays. Binding of c-Jun to the 21-bp box 3 bearing oligo was inhibited by a competitor oligo but not by a oligo with a mutated box 3 sequence (data not shown). Furthermore, we determined c-Jun binding to box 3 by a pull-down experiment using a biotinylated oligo. As shown in Fig. 3E, the box 3 oligo efficiently pulled down both c-Jun and Sp1 as revealed by Western blot. To test whether Sp1 is a prerequisite for c-Jun binding to the box 3 sequence, we performed an oligo pull-down assay using protein extracts from Sp1 knockdown cells. In line with previous observations, the binding of c-Jun to the box 3 sequence was significantly reduced (Fig. 3F). However, despite efficient inhibition of Sp1 expression, residual binding of c-Jun to box 3 oligonucleotide was observed. Furthermore, inhibition of Sp1 expression did not decrease PDK1 expression (data not shown), indicating that either other members of the Sp1 family, or additional transcription factors mediate c-Jun regulation of PDK1 transcription. Among possible transcription factors that can bind to GC-rich regions of the PDK1 promoter and have been associated with AP-1, we identified the Ets-1 binding sequence in the 3′ end of the box 3 oligonucleotide (Fig. 2A, double underlined sequence, and Refs. 27–29). To test the possible role of Ets-1 in PDK1 expression we have assessed the levels of transcripts upon inhibition of Ets1 expression. Reduced expression of Ets-1 by RNA interference resulted in marked inhibition of PDK1 expression without changes in Sp1 and c-Jun expression (Fig. 3G). Furthermore, Ets-1 expression was affected by c-Jun, consistent with a previous report on regulation of Ets-1 expression by AP-1 (see Ref. 30 and Fig. 3H). Altogether, these results indicate that c-Jun regulates the transcription of PDK1 through cooperation with Sp1 and Ets-1 and their corresponding response elements on the PDK1 promoter. Because c-Jun also controls Ets-1 transcription, regulation of PDK1 involves both direct and indirect function of c-Jun.
FIGURE 2.
c-Jun enhances PDK1 transactivation through a Sp1 site. A, structure of the proximal region of the human PDK1 promoter. The region from −45 to +82 contains 3 boxes highly conserved between human and mouse (box 1 from −14 to +2, box 2 from +4 to +15, and box 3 from +26 to +37 of the human promoter). In addition to the WT sequence, fragments containing mutations in box 1 and deletion of box 2 as well as mutation in box 3 were cloned in pGL2 vector for reporter assays. B, c-Jun enhances PDK1 transactivation through a Sp1 site in box 3. Reporter plasmids described in panel A were transfected into MeWo cells together with empty vector or pcDNA-c-Jun. Results are shown as the mean ± S.D. C, c-Jun siRNA no longer affect transcription driven by a mutant Sp1 site on box 3. Reporter plasmids described in panel A were transfected into Lu1205 cells together with scramble (sc) or c-Jun siRNA. D, mutation on the Sp1 site on box 3 abrogates responsiveness to c-Jun of the PDK1 promoter. A large region of the PDK1 promoter (−875 to +82) was subjected to mutagenesis of the Sp1 site on box 3 and the activity of both the WT and mutant promoter was assessed using reporter assays. Luciferase data are shown as the mean ± S.D.
FIGURE 3.
A, c-Jun enhances PDK1 transactivation through Sp1 and Ets. c-Jun interacts with Sp1 in melanoma cells. Nuclear extracts from MeWo cells and MeWo cells stably transfected with FLAG-c-Jun were immunoprecipitated (IP) with FLAG antibodies and blotted (IB) with the indicated antibodies. B, chromatin immunoprecipitation assay. The products of PDK1- and glyceraldehyde-3-phosphate dehydrogenase-specific PCR amplification on IP material from Lu1205 cells are shown. c-Jun antibodies sc-44 and sc-45 were used. C, c-Jun siRNA decrease binding of c-Jun to the PDK1 promoter. A chromatin immunoprecipitation assay was performed with extracts from Lu1205 cells transfected with scramble (sc) or c-Jun siRNA. D, protein extracts from cells described in panel C were analyzed by Western blot using the indicated antibodies. E and F, c-Jun binds to the box 3 region from the PDK1 promoter. Protein extracts from c-Jun−/− and c-Jun+/+ mouse embryonic fibroblasts (MEFs) and Lu1205 cells were incubated with a biotinylated oligo bearing the box 3 sequence. Complexes were pulled down with streptavidin and blotted with the indicated antibodies. G, Ets-1 is involved regulation of PDK1 expression. Protein extracts obtained from Lu1205 cells transfected with scramble (sc) or Ets-1 siRNA were analyzed by Western blot using the indicated antibodies. H, c-Jun regulates Ets-1 expression. Protein extracts from Lu1205 transduced with c-Jun lentiviral shRNA were analyzed by Western blot using the indicated antibodies. PCNA, proliferating cell nuclear antigen.
c-Jun Regulation of PDK1 Modulates Akt Activity
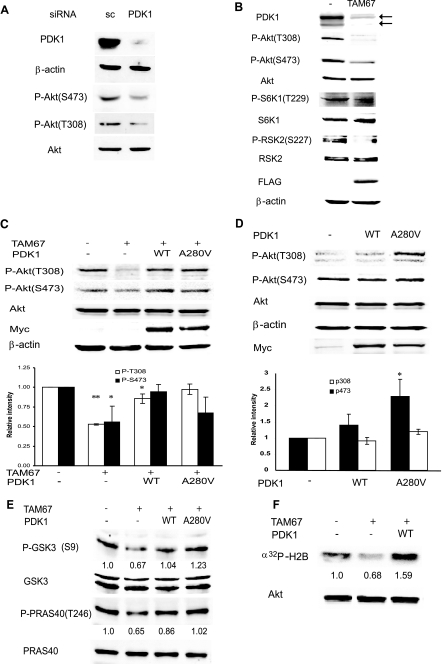
Because the PDK1 level is a major determinant in its activity given its autophosphorylation capacity, we assessed the effect of c-Jun on PDK1 targets Akt and PKC. It is important to note that these experiments were performed in the absence of any stimuli to activate c-Jun, Akt, or PKC because these proteins are constitutively activated in melanoma cell lines (4). Consistent with previous findings (31, 32) inhibition of PDK1 expression by corresponding siRNA in Lu1205 melanoma cells, which harbors mutant PTEN (phosphatase and tensin homolog deleted on chromosome Ten) and exhibit constitutive phosphorylation of Akt, reduced the level of Akt phosphorylation on both Thr308 and Ser473 sites (Fig. 4A). As expected, PDK1 knockdown affected phosphorylation on Thr308, the site directly phosphorylated by PDK1 on Akt, to a greater extent than Ser473 phosphorylation. Given the role of PDK1 in Akt activation and the finding that PDK1 is regulated by c-Jun, we determined whether changes in c-Jun expression alter Akt activity. Expression of TAM67 reduced PDK1 expression and Akt phosphorylation on both Thr308 and Ser473 in melanoma cell lines Lu1205 (Fig. 4B) and A375 (supplemental Fig. S10). Reduction of PDK1 levels by TAM67 expression also decreased the degree of phosphorylation of S6K1, RSK2 (Fig. 4B), and PKC (see below), indicating that multiple PDK1 targets are affected by c-Jun. Expression of either WT PDK1 or PDK1-A280V (a constitutively active form of PDK1 (33)) in TAM67-expressing cells restored Akt phosphorylation to levels observed in Lu1205 control cells (Fig. 4C). Of note, expression of either WT PDK1 or PDK1-A280V increased phosphorylation of Thr308 in Lu1205 control cells (Fig. 4D). Furthermore, phosphorylation levels of Akt substrates PRAS40 and GSK3 were decreased by TAM67 and efficiently rescued by PDK1 expression (Fig. 4E). Consistent with these observations, Akt activity, assessed by an in vitro kinase assay, was also reduced by TAM67 and rescued by PDK1 (Fig. 4F). These results suggest that by controlling PDK1 transcription, c-Jun contributes to the degree of Akt phosphorylation and activation. Given that PTEN mutation in Lu1205 cells promotes maximal translocation of Akt to the membrane, regulation of PDK1 transcription by c-Jun constitutes a PTEN-independent regulation of Akt activity.
FIGURE 4.
c-Jun regulates Akt activity via PDK1. A, PDK1 is required for Akt phosphorylation and activation. Protein extracts (40 μg) from Lu1205 cells transfected with scrambled (sc) oligonucleotides or siRNA specific for PDK1 were analyzed by Western blots using the indicated antibodies. B, TAM67 inhibits constitutive phosphorylation of Akt, S6K1, and RSK2. Protein extracts from Lu1205 or Lu1205 cells stably transfected with FLAG-TAM67 were blotted with the indicated antibodies. C, c-Jun-dependent regulation of Akt activation is mediated by PDK1. Lu1205-FLAG-TAM67 cells were infected with retrovirus expressing either WT PDK1 or the constitutively active PDK1 mutant A280V, and stable lines expressing exogenous PDK1 were established. Both PDK1 proteins are Myc-tagged. Protein extracts (40 μg) were blotted with the indicated antibodies. The intensity of bands corresponding to Thr308 and Ser473 phosphorylation from three independent experiments was determined, normalized to levels of total Akt, and plotted. Results are shown as the mean ± S.D. **, represents p < 0.0005. *, represents p < 0.05. D, expression of exogenous PDK1 causes a minimal increase in Akt phosphorylation. Lu1205 cells, which harbor mutant Pten, were infected with retrovirus expressing either WT PDK1 or the constitutively active mutant A280V, and stable cell lines were established. Protein extracts were analyzed as indicated in panel C. * represents p < 0.05. E, c-Jun-dependent regulation of PDK1 affects phosphorylation of Akt substrates. Protein extracts from cells described in panel C were analyzed with the indicated antibodies. The intensity of bands corresponding to p-GSK3β (S9) and p-PRAS40 (T246) was determined and normalized to total GSK3 and PRAS40, respectively. The degree of phosphorylation in the first lane was set to 1. Similar results were obtained in three separate experiments. F, c-Jun-dependent regulation of PDK1 affects Akt kinase activity. Akt was immunoprecipitated from cells described in the legend for Fig. 3C, and an in vitro kinase assay was performed. The relative intensity of the phosphorylated substrate is indicated. Similar results were obtained in three separate experiments.
c-Jun Modulates PKC and JNK Activities via PDK1
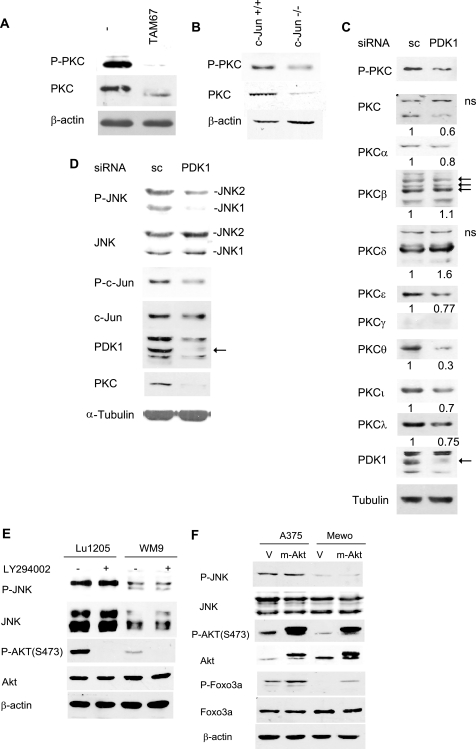
In addition to Akt, PDK1 also phosphorylates PKC kinases. Shortly after its translation and as part of its maturation, PKC is phosphorylated by PDK1 in the activation loop, which is important for its stabilization and subsequent autophosphorylation at residues Thr638 and Ser660 (21). Deficient phosphorylation within the activation loop results in PKC degradation (34). To determine whether c-Jun regulation of PDK1 transcription indirectly regulates PKC levels and phosphorylation, we assessed P-PKC and total PKC levels in melanoma cells expressing TAM67. Inhibition of c-Jun function either via TAM67 expression or c-Jun knockdown reduced both PDK1 and PKC levels, the latter detected by a pan-PKC antibody (Fig. 5, A and B). Also, the prominent protein band detected by the pan-PKC antibody migrated faster in melanoma cells expressing TAM67, suggesting PKC dephosphorylation (Fig. 5A, and data not shown). Among the 9 PKC isoforms tested, the expression levels of α, ϵ, θ, ι, and λ were reduced following inhibition of PDK1 expression (Fig. 5C). These data are in agreement with the notion that PDK1 regulates phosphorylation and stability of select PKC isoforms (12, 21).
FIGURE 5.
c-Jun modulates PKC and JNK via PDK1 in melanoma. A, TAM67 inhibits PKC protein levels. Protein extracts (40 μg) from Lu1205 cells stably transfected with FLAG-TAM67 and control cells were blotted with pan-p-PKC (Ser660) and pan-PKC antibodies. B, PKC levels are reduced in c-Jun−/− fibroblasts. Protein extracts (40 μg) from c-Jun−/− and control fibroblasts were blotted with pan-p-PKC (Ser660) and pan-PKC antibodies. C, PDK1 regulates protein levels of select PKC isoforms. Protein extracts (40 μg) from Lu1205 cells transfected with scrambled (sc) oligonucleotides or a PDK1-specific siRNA were analyzed by Western blots using antibodies specific for the indicated PKC isoforms. D, PDK1 siRNA inhibits JNK and c-Jun activity. Protein extracts (40 μg) from cells described in C were analyzed by Western blots using the indicated antibodies. E, inhibition of the phosphatidylinositol 3-kinase/Akt pathway does not affect p-JNK levels. WM9 and Lu1205 cells were treated with 10 μm LY294002 for 60 min. Protein samples were analyzed by Western blots with the indicated antibodies. F, active Akt does not increase p-JNK levels in melanoma. A375 and MeWo cells were stably transfected with empty vector (V) or pBabe expressing myr-Akt (m-Akt). Protein extracts were blotted with the indicated antibodies.
We recently showed that PKC-dependent phosphorylation enhances JNK activation in concert with activity of upstream MAPKK (17, 20). Data presented here imply that by regulating transcription of PDK1 and, subsequently, PKC levels and activity, c-Jun might also affect PKC-dependent JNK activation. Consistent with this possibility, reduced PDK1 levels by corresponding siRNA resulted in decreased PKC, P-JNK, and P-c-Jun levels (Fig. 5D). Altogether, these data establish that c-Jun regulation of PDK1 expression also contributes to PKC activity with a concomitant effect on JNK activity.
The possibility that PTEN-Akt can also affect JNK signaling was recently demonstrated in epidermoid carcinoma A431 cells and Pten−/− MEFs (35). Given that PTEN regulates inositol 1,4,5-trisphosphate cellular levels that are critical for PDK1 phosphorylation of Akt, we used a constitutively active Akt myristoylated Akt (myr-Akt) to assess whether JNK regulation by PTEN is mediated by Akt. As shown (Fig. 5E), myr-Akt expression did not increase levels of P-JNK in A375 or MeWo cells, which express mutant or WT PTEN, respectively. Treatment of these melanoma cells with the phosphatidylinositol 3-kinase inhibitor LY294002 inhibited Akt phosphorylation but did not alter levels of P-JNK (Fig. 5F). These observations suggest that, in melanoma, the PTEN-Akt axis does not constitute a major pathway regulating JNK activity.
c-Jun-dependent PDK1 Expression Contributes to Melanoma Growth Rate and Tumorigenesis
Because PDK1 controls AGC family kinases, including Akt, PKC, S6K, and RSK, c-Jun-dependent changes in PDK1 expression and activity are likely to affect numerous processes.
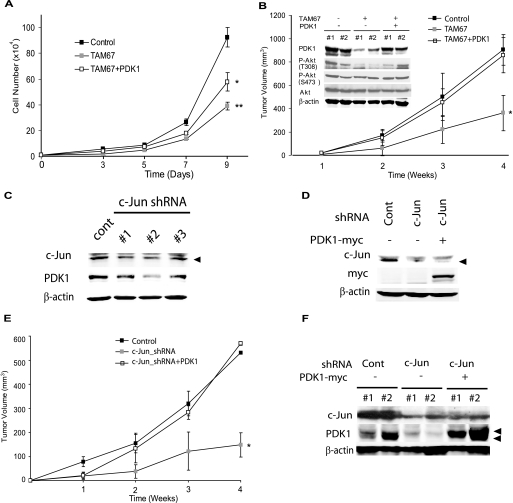
Through its ability to influence cell growth and survival via numerous pathways, c-Jun also promotes tumor development and progression (1). Because our data identified PDK1 as a c-Jun target, we next addressed whether PDK1 mediates effects of c-Jun on cell growth and tumorigenesis. Consistent with c-Jun oncogenic activity, TAM67 expression efficiently inhibited cell growth of Lu1205 melanoma cells (Fig. 6A). Significantly, transfection of PDK1-A280V into Lu1205 cells expressing TAM67 partially restored the growth rate (Fig. 6A), indicating that PDK1 mediates, at least in part, the effect of c-Jun on cell growth. To assess whether changes in growth rate would affect tumorigenesis, WT, TAM67, and TAM67 + PDK1-A280V cultures were assayed in a xenograft model. Notably, the xenograft tumor volume from Lu1205 cells expressing TAM67 was markedly lower compared with Lu1205 control cells. Significantly, reduced tumor volume was not seen in tumor cells co-expressing PDK1 and TAM67 (Fig. 6B). Reduced levels of PDK1 and the corresponding decrease in Akt phosphorylation were also seen in the tumor samples (Fig. 6B).
FIGURE 6.
c-Jun-dependent PDK1 regulation contributes to melanoma growth and tumorigenesis. A, c-Jun-dependent PDK1 expression regulates in vitro cell growth. Cells (104) described in the legend for Fig. 4C were plated and the cell number determined on the indicated days. Results are shown as mean ± S.D. (n = 3). ** represents p < 0.0001. * represents p < 0.001. B, c-Jun-dependent PDK1 expression regulates tumor growth in vivo. Cells described in the legend for Fig. 4C were injected subcutaneously into mice and tumor volume was measured at the indicated times. Results are shown as mean ± S.D. (n = 5). * represents p < 0.0005. Protein extracts (40 μg) obtained from tumors removed from mice were analyzed by Western blots using the indicated antibodies. #1 and #2 indicate samples from different mice. C and D, lentiviral c-Jun-specific shRNA efficiently knockdowns c-Jun expression and reduces PDK1 expression. Protein extracts (30 μg) from cells transduced with c-Jun-specific shRNA were analyzed by Western blotting using the indicated antibodies. E and F, PDK1 expression reverts the defect in tumor growth of c-Jun knockdown cells. Cells described in D were injected subcutaneously into mice. Tumor growth was measured as described in B. Results are shown as mean ± S.D. (n = 4). * represents p < 0.0001. F, protein extracts (40 μg) obtained from tumors removed from mice were analyzed by Western blots using the indicated antibodies. #1 and #2 indicate samples from different mice.
To verify that tumor growth is affected by c-Jun-dependent regulation of PDK1 expression, we repeated the xenograft assay using melanoma cells that were transduced with c-Jun shRNA. The melanoma cells that were inhibited for c-Jun expression were reconstituted with wild type or the constitutively active form of PDK1 (A280V) (Fig. 6, C and D). In line with the previous finding, tumor growth of c-Jun knockdown cells was greatly reduced. Significantly, the inhibition of tumor growth upon knockdown of c-Jun was markedly attenuated upon expression of active PDK1 (Fig. 6, E and F). Taken together, these findings provide direct evidence that PDK1 is required for c-Jun oncogenic capacity.
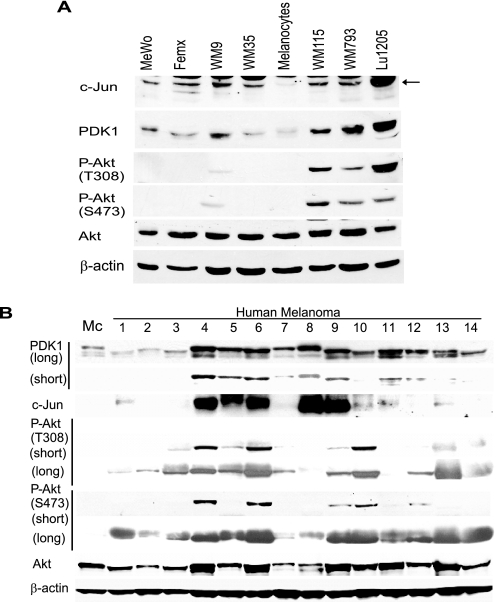
Given the link between c-Jun and PDK1 expression and the observation of increased c-Jun levels in human melanomas, we asked whether PDK1 levels were up-regulated in melanoma. Compared with normal melanocytes, most melanoma cell lines exhibit elevated PDK1 levels (Fig. 7A). Increased levels of PDK1 coincide with elevated expression of c-Jun (5 of 7 cell lines: MeWo, WM9, WM115, WM793, and Lu1205; Fig. 7A). However, as expected, increased PDK1 levels do not suffice to phosphorylate Akt in WT PTEN cells (Mewo, Femx, and WM35) because Akt translocation to the membrane is not expected to take place in these cells. Furthermore, and significantly, analysis of melanoma tumor samples revealed that those that harbor increased c-Jun expression also exhibit elevated PDK1 levels (5 of 5 samples; Fig. 7B). Correlation between elevated c-Jun, PDK1, and Akt phosphorylation was seen in over 50% of these cases, further substantiating the link established between c-Jun and PDK1, whereas suggesting that additional regulatory components contribute to Akt activation in these tumors.
FIGURE 7.
A, c-Jun and PDK1 levels increase in melanoma cell lines. Protein extracts (40 μg) from melanoma cell lines were analyzed by Western blots using the indicated antibodies. Human melanocytes served as controls. The cell lines WM115, WM793, and Lu1205 have deregulated PTEN. B, PDK1 levels are increased in melanoma tumor specimens and correlate with c-Jun levels. Protein samples (80 μg) obtained from melanoma tumors were analyzed by Western blot using the indicated antibodies.
DISCUSSION
Rewired signaling is one mechanism by which tumors overcome barriers that block uncontrolled proliferation, motility, and cell death programs in non-transformed cells. Recently we demonstrated how such re-wiring impacts the c-Jun/JNK pathway. Constitutive ERK activation increases transcription and the stability of c-Jun, with consequent effects on the JNK signaling cascade. The present study shows that the signal conveyed by ERK and JNK is transmitted by c-Jun to PDK1, a master kinase that regulates activation of multiple signaling pathways such as Akt, PKC, S6K, and RSK.
c-Jun, which is frequently up-regulated in melanoma, increases PDK1 transcription, leading to elevated PDK1 levels seen in both cell lines and tumor specimens. Given that PDK1 is self-activated (trans-autophosphorylation), positive regulation of PDK1 levels by c-Jun increases the amount of active PDK1 available to phosphorylate its targets.
Of importance is to note that whereas c-Jun activation of PDK1 is seen in both non-transformed and transformed cells, PDK1 activation of downstream targets, as shown for Akt, seems to differ between normal and cancer cells. Consistent with our findings in melanoma cells, Flynn et al. (31) and Feldmann et al. (32) showed that inhibiting PDK1 results in inhibition of Akt phosphorylation in glioblastoma and prostate cancer cells, respectively, suggesting that PDK1 levels or activity can be a limiting factor for Akt activation. Furthermore, recent studies have demonstrated the importance of PDK1 signaling in breast cancer, where increased PDK1 expression has been associated, in part, with increased copy number for the PDK1 gene (36). Moreover, the PDK1 contribution to human breast tumor development was also shown to be Akt-independent (37). Thus, mounting evidence suggest that in human cancer PDK1 up-regulation can be mediated by multiple pathways, including Jun-dependent transcription (this study), and increased PDK1 gene copy number (36), resulting in activation of downstream pathways, which include Akt, PKC (this study), and SGK3 (37).
Of note, Akt activity was not reduced in mice expressing a low level of PDK1 due to hypomorphic mutation (38). This difference may also be related to the activation of additional signaling pathways by treatment with growth factors used to activate Akt in normal cells (38). Such activation is not required in tumors cells such as the melanoma studied here, because Akt is already activated (this article and see Refs. 31 and 32). Among the possible factors that may explain the different requirements of PDK1 for Akt activation in tumor cells are priming phosphorylation by other activated protein kinases, including Src (39), or other post-translational modifications.
Here we demonstrate that c-Jun-dependent PDK1 regulation has a direct and a major impact on Akt and PKC activation in melanoma. Elevated c-Jun and PDK1 levels result in greater Akt activity and phosphorylation of Thr308 and, to a lesser extent, Ser473, possibly due to additional regulatory components involved in regulation of Ser473 phosphorylation. The up-regulation of c-Jun also result in PDK1-dependent phosphorylation and stabilization of PKC. This, together with c-Jun-dependent regulation of the receptor of the PKC, RACK1 (4), provide a mechanistic explanation for the reported up-regulation of PKC in melanoma (40).
Because c-Jun was identified as an oncogene, extensive efforts have been devoted to understanding the mechanisms underlying that activity. The present study links for the first time c-Jun with activation of two major signal transduction pathways, Akt and PKC, whose role in tumor development and progression is well established in multiple tumor types including melanoma (reviewed by Lopez Bergami et al. (40)). We show that the underlying mechanism for such activation is c-Jun control of PDK1 transcription. The up-regulation of c-Jun in melanoma increases PDK1 transcription in concert with transcription factors Sp1 and Ets-1. Accordingly, inhibiting c-Jun is expected to have major effects on melanoma development and progression. Indeed, expression of TAM67 and knockdown of c-Jun in melanoma cells elicited PDK1-dependent inhibition of cell growth in vitro and reduced in vivo tumor formation. In identifying a role for c-Jun in regulating Akt and PKC signaling, our findings provide important insight into the mechanism underlying c-Jun oncogenic activity.
Supplementary Material
Acknowledgments
We thank Dr. M. Herlyn for providing melanoma cell lines used in this study, and R. Wisdom for the c-Jun null cells. We also thank Drs. M. Karin, P. Tsichlis, and F. Liu for constructs used in the present study. We thank members of the Ronai Lab for advice.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1CA051995 and PO1CA128814 from the NCI (to Z. R.), grants from the Roemmers Foundation, The Harry J. Lloyd Charitable Trust, and Agencia Nacional de Promocion Cientifica y Tecnologica (ANPCyT) Grant PICT-2007-01010 (to P. L. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10.
- JNK
- Jun N-terminal kinase
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated protein kinase
- PDK1
- phosphoinositide-dependent kinase 1
- PKC
- protein kinase C
- GSK3
- glycogen synthase kinase 3
- PTEN
- phosphatase and tensin homolog deleted on chromosome 10
- myr-Akt
- myristoylated Akt
- S6K
- S6 kinase
- MEKK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- WT
- wild-type
- siRNA
- small interfering RNA
- shRNA
- short hairpin RNA
- RSK
- ribosomal S6 kinase.
REFERENCES
- 1.Vogt P. K. (2001) Oncogene 20, 2365–2377 [DOI] [PubMed] [Google Scholar]
- 2.Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B. A., Cooper C., Shipley J., Hargrave D., Pritchard-Jones K., Maitland N., Chenevix-Trench G., Riggins G. J., Bigner D. D., Palmieri G., Cossu A., Flanagan A., Nicholson A., Ho J. W., Leung S. Y., Yuen S. T., Weber B. L., Seigler H. F., Darrow T. L., Paterson H., Marais R., Marshall C. J., Wooster R., Stratton M. R., Futreal P. A. (2002) Nature 417, 949–954 [DOI] [PubMed] [Google Scholar]
- 3.Gorden A., Osman I., Gai W., He D., Huang W., Davidson A., Houghton A. N., Busam K., Polsky D. (2003) Cancer Res. 63, 3955–3957 [PubMed] [Google Scholar]
- 4.Lopez-Bergami P., Huang C., Goydos J. S., Yip D., Bar-Eli M., Herlyn M., Smalley K. S., Mahale A., Eroshkin A., Aaronson S., Ronai Z. (2007) Cancer Cell 11, 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens A., Jochum W., Sibilia M., Wagner E. F. (2000) Oncogene 19, 2657–2663 [DOI] [PubMed] [Google Scholar]
- 6.Mariani O., Brennetot C., Coindre J. M., Gruel N., Ganem C., Delattre O., Stern M. H., Aurias A. (2007) Cancer Cell 11, 361–374 [DOI] [PubMed] [Google Scholar]
- 7.Vleugel M. M., Greijer A. E., Bos R., van der Wall E., van Diest P. J. (2006) Hum. Pathol. 37, 668–674 [DOI] [PubMed] [Google Scholar]
- 8.Maeno K., Masuda A., Yanagisawa K., Konishi H., Osada H., Saito T., Ueda R., Takahashi T. (2006) Oncogene 25, 271–277 [DOI] [PubMed] [Google Scholar]
- 9.Eferl R., Ricci R., Kenner L., Zenz R., David J. P., Rath M., Wagner E. F. (2003) Cell 112, 181–192 [DOI] [PubMed] [Google Scholar]
- 10.Bhoumik A., Jones N., Ronai Z. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4222–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews C. P., Birkholz A. M., Baker A. R., Perella C. M., Beck G. R., Jr., Young M. R., Colburn N. H. (2007) Cancer Res. 67, 2430–2438 [DOI] [PubMed] [Google Scholar]
- 12.Mora A., Komander D., van Aalten D. M., Alessi D. R. (2004) Semin. Cell Dev. Biol. 15, 161–170 [DOI] [PubMed] [Google Scholar]
- 13.Storz P., Toker A. (2002) Front. Biosci. 7, d886–902 [DOI] [PubMed] [Google Scholar]
- 14.Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 15.Huang J., Manning B. D. (2009) Biochem. Soc. Trans. 37, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makris C., Voisin L., Giasson E., Tudan C., Kaplan D. R., Meloche S. (2002) Oncogene 21, 7891–7896 [DOI] [PubMed] [Google Scholar]
- 17.López-Bergami P., Habelhah H., Bhoumik A., Zhang W., Wang L. H., Ronai Z. (2005) Mol. Cell 19, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wick M. J., Ramos F. J., Chen H., Quon M. J., Dong L. Q., Liu F. (2003) J. Biol. Chem. 278, 42913–42919 [DOI] [PubMed] [Google Scholar]
- 19.Chan T. O., Rodeck U., Chan A. M., Kimmelman A. C., Rittenhouse S. E., Panayotou G., Tsichlis P. N. (2002) Cancer Cell 1, 181–191 [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Bergami P., Ronai Z. (2008) Int. J. Biochem. Cell Biol. 40, 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton A. C. (2003) Biochem. J. 370, 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson R., Spiegelman B., Hanahan D., Wisdom R. (1996) Mol. Cell. Biol. 16, 4504–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B. K., Chang W. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10406–10411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kardassis D., Papakosta P., Pardali K., Moustakas A. (1999) J. Biol. Chem. 274, 29572–29581 [DOI] [PubMed] [Google Scholar]
- 25.Melnikova I. N., Gardner P. D. (2001) J. Biol. Chem. 276, 19040–19045 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y. N., Chang W. C. (2003) J. Biol. Chem. 278, 45848–45857 [DOI] [PubMed] [Google Scholar]
- 27.Wasylyk B., Wasylyk C., Flores P., Begue A., Leprince D., Stehelin D. (1990) Nature 346, 191–193 [DOI] [PubMed] [Google Scholar]
- 28.Logan S. K., Garabedian M. J., Campbell C. E., Werb Z. (1996) J. Biol. Chem. 271, 774–782 [DOI] [PubMed] [Google Scholar]
- 29.Bassuk A. G., Leiden J. M. (1995) Immunity 3, 223–237 [DOI] [PubMed] [Google Scholar]
- 30.Majérus M. A., Bibollet-Ruche F., Telliez J. B., Wasylyk B., Bailleul B. (1992) Nucleic Acids Res. 20, 2699–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldman R. I., Wu J. M., Polokoff M. A., Kochanny M. J., Dinter H., Zhu D., Biroc S. L., Alicke B., Bryant J., Yuan S., Buckman B. O., Lentz D., Ferrer M., Whitlow M., Adler M., Finster S., Chang Z., Arnaiz D. O. (2005) J. Biol. Chem. 280, 19867–19874 [DOI] [PubMed] [Google Scholar]
- 32.Flynn P., Mellor H., Casamassima A., Parker P. J. (2000) J. Biol. Chem. 275, 11064–11070 [DOI] [PubMed] [Google Scholar]
- 33.Wick M. J., Dong L. Q., Riojas R. A., Ramos F. J., Liu F. (2000) J. Biol. Chem. 275, 40400–40406 [DOI] [PubMed] [Google Scholar]
- 34.Balendran A., Hare G. R., Kieloch A., Williams M. R., Alessi D. R. (2000) FEBS Lett. 484, 217–223 [DOI] [PubMed] [Google Scholar]
- 35.Vivanco I., Palaskas N., Tran C., Finn S. P., Getz G., Kennedy N. J., Jiao J., Rose J., Xie W., Loda M., Golub T., Mellinghoff I. K., Davis R. J., Wu H., Sawyers C. L. (2007) Cancer Cell 11, 555–569 [DOI] [PubMed] [Google Scholar]
- 36.Maurer M., Su T., Saal L. H., Koujak S., Hopkins B. D., Barkley C. R., Wu J., Nandula S., Dutta B., Xie Y., Chin Y. R., Kim D. I., Ferris J. S., Gruvberger-Saal S. K., Laakso M., Wang X., Memeo L., Rojtman A., Matos T., Yu J. S., Cordon-Cardo C., Isola J., Terry M. B., Toker A., Mills G. B., Zhao J. J., Murty V. V., Hibshoosh H., Parsons R. (2009) Cancer Res. 69, 6299–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasudevan K. M., Barbie D. A., Davies M. A., Rabinovsky R., McNear C. J., Kim J. J., Hennessy B. T., Tseng H., Pochanard P., Kim S. Y., Dunn I. F., Schinzel A. C., Sandy P., Hoersch S., Sheng Q., Gupta P. B., Boehm J. S., Reiling J. H., Silver S., Lu Y., Stemke-Hale K., Dutta B., Joy C., Sahin A. A., Gonzalez-Angulo A. M., Lluch A., Rameh L. E., Jacks T., Root D. E., Lander E. S., Mills G. B., Hahn W. C., Sellers W. R., Garraway L. A. (2009) Cancer Cell 16, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawlor M. A., Mora A., Ashby P. R., Williams M. R., Murray-Tait V., Malone L., Prescott A. R., Lucocq J. M., Alessi D. R. (2002) EMBO J. 21, 3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang T., Qiu Y. (2003) J. Biol. Chem. 278, 15789–15793 [DOI] [PubMed] [Google Scholar]
- 40.Lopez-Bergami P., Fitchman B., Ronai Z. (2008) Photochem. Photobiol. 84, 289–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.