Abstract
Elevated levels of the calcium-binding protein S100A4 promote metastasis and in carcinoma cells are associated with reduced survival of cancer patients. S100A4 interacts with target proteins that affect a number of activities associated with metastatic cells. However, it is not known how many of these interactions are required for S100A4-promoted metastasis, thus hampering the design of specific inhibitors of S100A4-induced metastasis. Intracellular S100A4 exists as a homodimer through previously identified, well conserved, predominantly hydrophobic key contacts between the subunits. Here it is shown that mutating just one key residue, phenylalanine 72, to alanine is sufficient to reduce the metastasis-promoting activity of S100A4 to 50% that of the wild type protein, and just 2 or 3 specific mutations reduces the metastasis-promoting activity of S100A4 to less than 20% that of the wild type protein. These mutations inhibit the self-association of S100A4 in vivo and reduce markedly the affinity of S100A4 for at least two of its protein targets, a recombinant fragment of non-muscle myosin heavy chain isoform A, and p53. Inhibition of the self-association of S100 proteins might be a novel means of inhibiting their metastasis-promoting activities.
Keywords: Calcium/Binding Proteins, Methods/Site-directed Mutagenesis, Methods/Surface Plasmon Resonance, Protein/Protein-protein interactions, Tumor/Metastases, myosin, p53
Introduction
S100A4 protein belongs to the S100 family of EF-hand containing calcium-binding proteins. The presence of S100A4 protein in human carcinoma cells has been associated with reduced survival in patients suffering from malignant melanoma (1), non-small cell lung carcinoma (2, 3), and cancers of the breast (4, 5), colorectum (6, 7), urinary bladder (8, 9), gall bladder (10), esophagus (11), mouth (12), and stomach (13). S100A4 was the first S100 protein to be shown to promote metastases in a model system of breast cancer (14).
S100A4 protein interacts with a number of intracellular targets, including the heavy chain of non-muscle myosin (15, 16) isoform A (NMMIIHCA)5 in vitro (17) and in vivo (18), by which it might affect cell motility (19, 20). S100A4 also interacts with the C-terminal region of the tumor suppressor protein, p53 (21, 22), affecting p53 oligomerization (22) and cell apoptosis (21). S100A4 also exhibits less well characterized interactions with other intracellular proteins, β-liprin (23) and tropomyosin (24), and with extracellular proteins, annexin II and plasminogen (25).
S100A4 exists as a self-assembling homodimer, and the dimerization arises from hydrophobic residues, particularly Ile-12, Val-13, Phe-16 in helix I and Phe-72, Tyr-75, Phe-78, and Leu-79 in helix 4 (26) in human S100A4. In the yeast two-hybrid system, interaction between newly synthesized S100A4 mutant molecules and wild type S100A4 was not detectable for proteins containing single point mutations, F72A, Y75K, or F78A (26). However, F78A behaved like wild type protein for dimerization in separate immunoprecipitation experiments and for interaction with NMMIIHCA (27). Single mutations of either F72Q or Y75K prevented dimerization of S100A4 and also interaction with non-muscle myosin (27). However, the effect of such dimerization-disrupting mutations on the biological activity of S100A4 is presently unknown.
EXPERIMENTAL PROCEDURES
Expression and Purification of Human S100A4
Wild type S100A4 cDNA was cloned into the NdeI/BamHI sites of bacterial expression vector pET15b (Novagen) and recombinant protein produced in Escherichia coli C41 (28). The fraction containing the S100A4 recombinant protein was dialyzed against phosphate-buffered saline (PBS), and the purity of the S100A4 was checked by SDS-15% (w/v) polyacrylamide gel electrophoresis. To remove the His tag, the purified protein was incubated with 100 units/ml thrombin and immobilized on Affi-10 resin (Bio-Rad) for >12 h with shaking. After centrifugation at 5000 × g for 5 min to remove the immobilized thrombin, the supernatant containing the cleaved S100A4 protein was collected and loaded onto a His-select column to remove any undigested His-tagged proteins.
Site-directed Mutagenesis
Mutations at amino acids 72, 72.78, and 16.72.78 encoded by the S100A4 cDNA cloned into pET15b were performed using a QuikChange site-directed mutagenesis kit (Stratagene). Pairs of complementary oligonucleotides (supplemental Data 1) containing the desired mutation(s) were used to change phenylalanine residues at positions 16, 72, and 78 to alanine, glutamine, and alanine, respectively. PCR reactions were set up as described previously (28) with an initial denaturing step at 95 °C for 3 min, 16 cycles of 95 °C for 1 min, 55 °C for 1 min, and 68 °C for 10 min with a final extension step of 68 °C for 15 min, and the desired mutations were confirmed by DNA sequencing. The mutagenized constructs were transformed into E. coli C41 for propagation and recombinant protein production.
Transfection of Mammary Cell Lines
The S100A4 wild type and mutated cDNAs in pET-15b were amplified by PCR using a pair of primers bearing (underlined) BamHI and HindIII restriction enzyme sites (forward human S100A4 primer, 5′- CCGGATCC70ATGGCGTGCCCTCTGG85; reverse-human S100A4 primer, 5′-CGAAGCTTT375CATTTCTTCCTGGGCTG358, numbering is from GenBank accession number NM_002961). The PCR products were cloned into pCDNA3.1(−) vector that had been double-digested with BamHI and HindIII. 2–3 μg of recombinant constructs were used to transfect the rat mammary tumor-derived Rama 37 cells (29) using Lipofectamine 2000 reagent (Invitrogen), and transfected cells were selected in medium containing 0.8 mg/ml Geneticin, replaced every 2–3 days. Surviving colonies of cells were picked and expanded, and the remaining survivors were expanded as a pool. The resulting cloned and pooled transfected cells were tested for the presence of S100A4 proteins and actin by Western blotting (28, 30, 31). Cell lines and pools with similar levels of S100A4 proteins were selected.
Growth rates of transfected cell clones were determined by plating 12,000 cells/ml in triplicate and counting the number of cells after 48 h of incubation. The growth rates in cells/h for the various transfected cell lines were compared using Student's t tests on Stats Direct software Version 2.6.2 (Stats Direct Ltd., Altringham, UK).
In Vitro Caspase Activity Assays
In vitro caspase-3 activity was measured using Caspase-Glo 3/7 assay kit (Promega, Southampton, UK) according to the manufacturer's instructions. Briefly, 4 × 104 cells of the indicated cell lines were plated in white 96-multiwell microplates (Greiner, Stonehouse, UK). 24 h after plating, basal caspase-3 activity was assessed by luminescence measurement using a 5-s measurement time in a PerkinElmer Life Sciences EnVision plate reader.
Immunofluorescent Staining
Rama 37 cell lines were cultured as described above and plated onto fibronectin-coated (2.5 mg/cm2) glass coverslips in 24-well plates and grown for 48 h. Cells were washed once in cytoskeleton buffer (CB: 150 mm NaCl, 5 mm MgCl2, 5 mm EGTA, 5 mm glucose, 10 mm mes, pH 6.1) before fixation with methanol at −18 °C for 20 min. Cells were then further washed in CB before being permeabilized with 0.2% (v/v) Triton X-100 in CB for 10 min, rinsed with CB, and blocked with blocking solution (3% (w/v) bovine serum albumin in CB) for 60 min. Samples were incubated with primary antibodies against non-muscle myosin IIA (Covance, Princeton, NJ) and S100A4 (DAKO, Ely, UK) in blocking solution for 45 min at 37 °C. After washing three times with blocking solution, cells were incubated with anti-rabbit and anti-goat antibodies labeled with fluorescein isothiocyanate and Cy3, respectively, in blocking solution for 45 min at 37 °C. After washing with blocking solution, coverslips were rinsed with water, mounted in Vectashield mounting medium (Vector, Peterborough, UK), and viewed using a Zeiss LSM510 confocal laser scanning microscope.
In Vivo Metastasis Assay
Transfected cell clones and pools were subjected to metastasis assays as described previously (14, 31). Lung tissue for detection of metastases was fixed in formalin, embedded in paraffin wax, sectioned, and stained with hematoxylin and eosin (29). Invasion of the primary tumor cells in vivo was determined as described previously (32). Animals were maintained according to United Kingdom Coordinating Committee for Cancer Research guidelines under UK Home Office Project License 40/2395 to Professor P. S. Rudland.
Transwell Cell Migration Assays
Twelve-well (wells 1.0 cm2) Transwell permeable devices (Corning Costar) with 8.0-μm pore size polycarbonate membranes were used to measure chemotactic migration of 1 × 105 cells induced by a 1–5% (v/v) fetal calf serum gradient over 24 h. The wells and the inserts were washed three times with PBS, and the filters were fixed and stained using a Diffquik histochemical kit (Reagena, Toivala, Finland) following the manufacturer's instructions. The stained cells on the lower side of the membrane were counted using a light microscope with a ×20 objective lens. Mean values ± S.D. were of 4 replicate wells.
Binding of rS100A4 to Calcium, rNMMIIHCA, and p53
The binding of Ca2+ to apoS100A4 proteins was determined using isothermal titration calorimetry (ITC). Traces of Ca2+ were stripped from the recombinant protein samples by adding 1 mm EGTA before dialysis against 20 mm Tris-Cl, pH 7.4, 140 mm NaCl overnight. The dialysate buffer was saved for diluting the protein samples, making CaCl2 solution and blank ITC controls. An iTC200 Microcalorimeter (GE Healthcare) was used for all measurements. The sample cell was filled with 250 μl of 0.25 mm mutant 72 protein, 0.5 mm mutant 72.78, or 0.6 mm mutant 16.72.78. 1 mm CaCl2 titrant dissolved in the dialysate mentioned above was injected in 2-μl aliquots with injection durations of 4 s and a 150-s interval between injections, except the first injection, for which the injection volume and duration were 0.4 μl and 2s, respectively, to remove an irregularity due to capillary action. Cell temperature was set at 30 °C for all experiments. A blank control, acquired by adding the titrant into the buffer without protein, was subtracted from the titration curve. The results were analyzed using the “Origin for ITC” software supplied with iTC200 Microcalorimeter.
Binding of S100A4 to a recombinant protein consisting of the 149 C-terminal amino acids of NMMIIHCA or recombinant wild type p53 (GenWay Biotech Inc., San Diego, Ca; catalog no. 10-663-45596) was tested using a dual-Channel IAsys resonant mirror biosensor (Neosensors, Sedgefield, UK) as described previously (33, 34). rNMMIIHCA rods or recombinant p53 were covalently bound to aminosilane sensor surfaces (Neosensors) using bissuccinimidyl suberate (Perbio, Chester, UK) as the cross-linker according to supplier's instructions. Different concentrations of S100A4 proteins were injected into the rNMMIIHCA- or recombinant p53-derivatized sensor cuvette that contained 45 μl of 1× PBS buffer containing 0.5 mm CaCl2, 0.02% (v/v) Tween 20. The immobilized protein was regenerated by washing with 20 mm HCl, which removed all detectable S100A4 proteins. No detectable binding of S100A4 was observed to non-derivatized aminosilane surfaces (not shown). The kinetics of the binding of S100A4 to rNMMIIHCA or p53 proteins were calculated using non-linear curve fitting Fast Fit software (Neosensors) (34). The association and dissociation curves fit a single-site binding model at least as well as a two-site binding model.
Protein Interactions in Living Cells
Fluorescence resonance energy transfer (FRET) by fluorescence lifetime imaging (FLIM) was used to test the dimerization of wild type and mutant S100A4 proteins and their binding to rNMMIIHCA in vivo. The S100A4 wild type and mutant cDNAs were amplified by PCR using a pair of oligonucleotides having HindIII and BamHI restriction enzyme sites (forward primer (restriction sites are underlined), 5′-CCGGATCC70ATGGCGTGCCCTCTGG85; reverse primer, 5′-CGAAGCTT375TCATTTCTTCCTGGGCTG358, numbering is from GenBankTM accession number NM_002961) and inserted into pYFP and pAmCyan vectors such that the fluorescent protein resided at the N terminus of the S100A4 protein (18). For the insertion of non-muscle myosin rNMMIIHCA DNA (GenBankTM accession number M31013) into pYFP and pAmCyan vectors, a pair of oligonucleotides was used as described previously (33). For FLIM experiments, HeLa cells, grown in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal calf serum at 37 °C in 10% (v/v) CO2, were transfected with 3 μg of plasmid DNAs using FuGENE 6 (Roche Applied Science). Each pAmCyan construct was transfected alone or with a pYFP construct. Empty vector pYFP or pAmCyan were also used as a negative control. For FRET-FLIM experiments, 100,000 living cells/well of each transfection were plated out for 12–24 h onto IWAKI glass base dishes (Asahi Techno Glass Corp., Japan). An in-house-built confocal laser-scanning microscope system was used to detect the interaction of AmCyan and YFP at a single cell level. A frequency-doubled laser (Coherent, Santa Clara, CA; λ = 435 nm, 76 MHz repetition rate) was delivered to the sample via a dichroic mirror (51017, Chroma Technology, Rockingham, VT) focused through the 1.3-NA Zeiss oil immersion lens (Carl Zeiss). The FLIM images were analyzed for FRET valuation using SPCImage (Becker-Hickl GmbH) FLIM analysis software (18).
Analytical Gel Filtration Chromatography
60–100 μg of purified protein samples dialyzed overnight against PBS buffer were applied to Superdex 75 10/300 GL columns (GE Healthcare) equilibrated with PBS. In some experiments proteins dialyzed against TBS buffer (20 mm Tris-HCl, pH 7.4, 140 mm NaCl, pH 7.4) were applied to Superdex 200 10/300 GL columns equilibrated with TBS buffer either in the presence or absence of calcium. Columns were operated at a flow rate of 0.5 ml/min, and elution was monitored at 215 nm. Separations on Superdex 200 were calibrated using β-amylase (molecular mass 200 kDa), bovine γ-globulin (molecular mass monomer, 160 kDa; dimer, 320 kDa), bovine serum albumin (molecular mass 66 kDa), cytochrome c (molecular mass 12.4 kDa).
RESULTS
cDNAs encoding three mutant S100A4 proteins were produced: mutant 72, in which phenylalanine 72 was converted to glutamine, Mutant 72.78, in which phenylalanines 72 and 78 were converted to glutamine and alanine, respectively, and Mutant 16.72.78, in which phenylalanines 16, 72, and 78 were converted to alanine, glutamine, and alanine, respectively. Expression plasmid constructs bearing wild type S100A4 or these mutant cDNAs were transfected into Rama 37 cells. The benign non-metastatic rat mammary tumor-derived cell line, Rama 37 (29), was used because expression of wild type rat (14) or human (35) S100A4 in these cells induces a metastatic phenotype. After selection in Geneticin, permanent cell clones and pools of the transfected Rama 37 cells were isolated.
Immunofluorescent staining of cells for non-muscle myosin IIA showed the presence of fibers in the mock-transfected cells and a more punctate staining pattern in the wild type S100A4-transfected cells. Mutant 72 cells resembled the S100A4-transfected cells in this regard. Generally, mutants 72.78 and 16.72.78 resembled the mock-transfected cells, with less punctate staining and more evidence of fibers. The fibers, however, were not as well developed as in the mock-transfected cells and seemed to be collapsed toward the periphery of the cells. The distribution of S100A4 generally reflected filament structures apart from being present at a low level in the mock-transfected cell line (supplemental Data 2), and the transfected pools exhibited similar characteristics (not shown). S100A4 was also found between myosin fibers and particularly at the tips of fibers at the cell periphery.
The transfected Rama 37 cloned cell lines did not exhibit significantly different growth rates from one another (cell lines transfected with empty vector, mutant 72, mutant 72.78, and mutant 16.72.78 compared with wild type, p = 0.37, 0.24, 0.21, 0.61, respectively, Student's t test). This result suggests that expression of wild type S100A4 does not increase the growth rate of cells, supporting previous reports in mouse (19) and rat (35) cells and that the expression of the mutant S100A4 proteins per se does not adversely affect the growth rate of the cells.
Determination of caspase 3/7 activity in two experiments (supplemental Data 3) as a marker of apoptosis showed that there were no consistent differences in caspase 3/7 activities between cells expressing wild type or mutant S100A4 proteins, a result that is entirely consistent with the similarity in growth rates of these cells in culture. However, there were minor variations between the various mutants in the two experiments, with mutants 72 and 16.72.78 showing no change or significantly less caspase 3/7 activity than wild type S100A4 (mutant 72, p = 0.6 or 0.002; mutant 16.72.78, p < 0.001 or < 0.001, Student's t test) and mutant 72.78 showing significantly increased caspase 3/7 activities compared with wild type (p = 0.05 or < 0.0001, Student's t test) that were not associated with observed cell death. It was noted that the cells expressing wild type S100A4 or mutant 72, 72.78, or 16.72.78 exhibited significantly, 2–5-fold higher, caspase 3/7 activity in the two separate experiments than untransfected Rama 37 cells (wild type S100A4, p = 0.0002 and p < 0.0001; mutant 72, p = 0.0035 and p = 0.05; mutant 72.78, p < 0.0001 and p < 0.0001; mutant 16.72.78, p = 0.0009 and p = 0.006), whereas cells transfected with empty PBKCMV or pCDNA3 exhibited significantly decreased or not significantly increased caspase activity relative to Rama 37 cells (pBKCMV p = 0.46 and p = 0.017; pCDNA3, p = 0.29 and p = 0.86). It is possible that cells expressing wild type and mutant S100A4 proteins exhibit higher rates of growth but also higher rates of apoptosis than mock-transfected/untransfected cells. However, it is probably more likely that the S100A4-expressing cells exhibit a higher basal level of caspase 3/7 activity, an idea that is supported by the fact that caspase 3/7 activity in all cell lines tested could be further triggered by the treatment of the cells with pro-caspase activator 1 (not shown).
Clones and pools that expressed the corresponding mutant proteins at similar levels to one another were selected (Fig. 1), as determined by measuring the densities of the chemiluminescent Western blot bands and corrected for variations in actin levels (mutant clones, 91% ± S.D. 3.6 wild type clone; mutant pools, 100% ± S.D. 9.3 wild type pool).
FIGURE 1.
The level of wild type and mutant S100A4 proteins in transfected Rama 37 cell clones and cell pools. Cells were transfected with expression vectors containing S100A4 wild type or mutant 72, 72.78, or 16.72.78 cDNAs as described under “Experimental Procedures.” Extracts of cloned cell lines (A and B) or pooled transfectants (C and D) were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membranes. Membranes were incubated with either a polyclonal S100A4 antibody (A and C) or anti-actin serum (B and D) as a loading control. Bound antibodies were detected by chemiluminescence with horseradish peroxidase-conjugated secondary antibody as described previously (28, 30, 31). The intensities of the bands of S100A4 protein corrected for actin levels in cloned cell lines, Rama 37, Rama 37 + vector, Rama 37 + S100A4 mutant 72, Rama 37 + S100A4 mutant 72.78, and Rama 37 + S100A4 mutant 16.72.78 were 16, 10, 95, 90, and 88% that of wild type and, for cell pools, 10, 5, 99, 110, and 91% that of wild type.
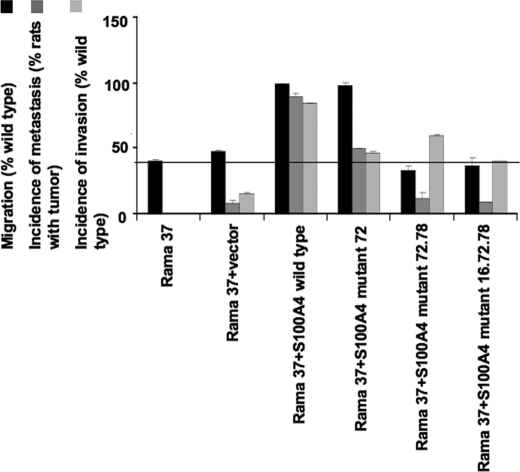
The transfected Rama 37 cloned cell lines and pools expressing S100A4 proteins bearing mutant 72, mutant 72.78, or mutant 16.72.78 all exhibited a significantly reduced ability to promote metastases in the lungs compared with wild type protein when injected into mammary fat pads of syngeneic rats (Table 1). However, the reduction in metastasis-promoting potential for mutant 72 (cell clone, 43% of rats against 90% of rats for wild type S100A4, p = 0.0029, Fisher's Exact Test; transfected cell pool, 57% of rats against 89% for wild type S100A4, p = 0.0625, Fisher's Exact Test) was less than for mutant 72.78 (cell clone, 15% of rats against 90% of rats for wild type S100A4, p < 0.0001, Fisher's Exact Test; transfected cell pool, 8% of rats against 89% for wild type S100A4, p < 0.0001, Fisher's Exact Test) or mutant 16.72.78 (cell clone, 0% of rats against 90% of rats for wild type S100A4, p < 0.0001, Fisher's Exact Test; transfected cell pool, 19% of rats against 89% for wild type S100A4, p < 0.0001, Fisher's Exact Test). These results show that the mutations 72.78 and 16.72.78 had reduced the ability of S100A4 protein to promote metastases by more than 85%.
TABLE 1.
Incidences of mammary tumors, metastases, and invasion in rats injected with various transfected cell lines
| Transfected DNA (designation of cell line)a | Incidence of mammary tumorsb | p valuec | Incidence of metastasisd | p valuee | Incidence of invasionf | p valueg |
|---|---|---|---|---|---|---|
| % | % | % | ||||
| None; untransfected Rama 37h | 18/20 (90) | p = 0.99 | 0/20 (0) | p < 0.0001 | 0/20 (0) | p < 0.0001 |
| Vector onlyh | ||||||
| Clone | 19/20 (95) | p = 1.00 | 0/20 (0) | p < 0.0001 | ||
| Pool | 23/23 (100) | p = 1.00 | 2/23 (9) | p < 0.0001 | 3/18 (16) | p < 0.0001 |
| S100A4 wild typeh | ||||||
| Clone | 22/22 (100) | 20/22 (90) | ||||
| Pool | 19/19 (100) | 17/19 (89) | 17/20 (85) | |||
| S100A4 mutant 72 | ||||||
| Clone | 16/16 (100) | p = 1.00 | 7/16 (43) | p = 0.0029 | ||
| Pool | 19/19 (100) | p = 1.00 | 11/19 (57) | p = 0.063 | 9/19 (47) | p = 0.017 |
| S100A4 mutant 72.78 | ||||||
| Clone | 18/19 (94) | p = 1.00 | 3/19 (15) | p < 0.0001 | ||
| Pool | 21/23 (91) | p = 1.00 | 2/23 (8) | p < 0.0001 | 3/5 (60) | p = 0.289 |
| S100A4 mutant 16.72.78 | ||||||
| Clone | 16/16 (100) | p = 1.00 | 0/16 (0) | p < 0.0001 | ||
| Pool | 14/15 (93) | p = 1.00 | 6/31 (19) | p < 0.0001 | 7/17 (41) | p = 0.007 |
a S100A4 mutant 72, cells expressing mutant S100A4 protein in which residue 72 has been changed from phenylalanine to glutamine; S100A4 mutant 72.78, cells expressing mutant S100A4 protein in which residues 72 and 78 have been changed from phenylalanine to glutamine and alanine respectively; S100A4 mutant 16.72.78, cells expressing mutant S100A4 protein in which residues 16, 72, and 78 have been changed from phenylalanine to alanine, glutamine, and alanine, respectively.
b Number of tumors/number of animals inoculated.
c p values for tumor incidences in cloned or in pooled transfectant cells of mutant 72, mutant 72.78, and mutant 16.72.78 are compared with cloned or pooled transfectant cells of the wild type S100A4 transfections (Fisher's Exact test).
d Numbers of animals (%) with lung metastases/numbers of animals with tumors.
e p values for incidences of metastasis in cloned cell lines or in cell pools transfected with S100A4 mutant 72, mutant 72.78, mutant 16.72.78, empty pBKCMV vector, or cloned untransfected Rama 37 cells compared with cloned or pooled transfectant cells of the wild type S100A4 transfections (Fisher's Exact test).
f Numbers of animals (%) with muscle invasion/numbers of animals with muscle around tumors. No peripheral muscle tissue was recovered in sections of primary tumors from 10 rats with clones and 11 rats with pools of wild type S100A4, from 7 rats with clones and 9 rats with pools of mutant 72, in 16 rats with clones and 21 rats with pools of mutant 72.78, in 8 rats with clones and 22 rats with pools of mutant 16.72.78, and in 11 rats with clones and 14 rats with pools of pBKCMV vector transfected cells. In view of the low proportion of primary tumors with muscle in the available histological sections of wild type, mutants, and vector control, the results for clones and pools were combined for statistical analysis.
g p values for muscle invasion in tumors from combined cell lines and pooled transfectant cells of mutant 72, mutant 72.78, and mutant 16.72.78, pBKCMV vector or cloned untransfected Rama 37 cells compared with combined cloned and pooled transfectant cells of the wild type S100A4 transfectants (Fisher's Exact test).
The effect of the mutations on the ability of S100A4 to enhance the invasion of tumor cells into surrounding muscle on histological sections of primary tumors was examined. However, because of the absence of muscle in the sections of some of the primary tumors, the results for invasion of cell clones and pools were combined for each mutant (Table 1). S100A4 mutants 72 and 16.72.78 exhibited significantly reduced invasion relative to wild type S100A4 (p = 0.017 and p = 0.007, respectively, Fisher's Exact Test). However, mutant 72.78 was not significantly different from wild type protein (p = 0.29), probably reflecting the very small number of 72.78 tumors with muscle blocks in the sections.
Using the Boyden chamber assay, the migration of the cells expressing S100A4 mutant 72-transfected clone/pooled cells was not significantly different from the wild type S100A4 clone/pooled cells (p = 0.99 test, Student's t test; Fig. 2). However, transfected clone/pool cells expressing S100A4 mutant 72.78 or S100A4 mutant 16.72.78 showed significantly reduced migration relative to clone/pool cells expressing wild type S100A4 (mutant 72.78 clone, p = 0.0001 and pools, p = 0.0009; mutant 16.72.78 clone, p = 0.0003 and pool, p = 0.0005 Student's t test; Fig. 2).
FIGURE 2.
The effects of interface mutations of S100A4 on metastasis and invasion in vivo and S100A4-induced migration of Rama 37 cells in vitro. Cell pools or clones of Rama 37 cells expressing wild type S100A4 or mutants 72, 72.78, or 16.72.78 were subjected to migration assays in quadruplicate Transwell chambers as described under “Experimental Procedures.” Parental Rama 37 cell line and cells transfected with empty vector were used as negative controls. For statistical analysis, at least four independent assays were conducted on clones and four on pools of cells. The results were converted to a percentage of wild type migration. The horizontal line denotes the basal level of migration of the Rama 37 cells in the Transwell assay. Incidences of metastasis and invasion are expressed as the percentages of animals with primary tumors using data from Table 1.
The abilities of the mutant 72, 72.78, and 16.72.78 proteins to bind calcium were determined using isothermal titration calorimetry. The results showed that the mutant 72 protein bound calcium with a Kd value of 31 ± 10 mm, a value that is consistent with those obtained previously for S100 proteins (30, 36, 37) albeit with reduced stoichiometry (supplemental Data 4). In contrast, the binding of calcium ions for mutants 72.78 and 16.72.78 was too low to analyze.
The kinetics of interaction between rNMMIIHCA and mutant proteins 72, 72.78, and 16.72.78 were determined in vitro using an optical biosensor and compared with previously obtained data for wild type S100A4 protein (28). For each mutant the observed level of binding of S100A4 increased as the concentration of S100A4 protein increased. A plot of the observed on-rate (kon) against the concentration of S100A4 yielded a straight line. A plot of the slope of initial rate against concentration of S100A4 yielded a straight line. Results obtained from the binding curves with a one-site model resulted in a near random distribution of the data about the model. The data obtained covered at least 90% of the curve described by the single-site model. The interaction of S100A4 with rNMMIIHCA was monophasic and was not limited by diffusion (supplemental Data 5).
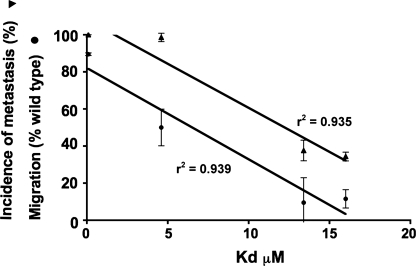
The kinetics of interaction of wild type S100A4 and three interface mutant proteins with rNMMIIHCA showed a dramatic reduction in the kass in all the mutants compared with the wild type protein and a consequent increase in the equilibrium dissociation constant Kd from the nm into the μm range (Table 2), with mutant S100A4 proteins 72.78 and 16.72.78 exhibiting lower affinities (14–16 and 13–15 μm, respectively) than mutant S100A4 protein 72 (4.6–4.8 μm) for rNMMIIHCA. The value of the Kd for the interaction between the mutant S100A4 proteins and rNMMHCIIA correlated significantly with both migration in the Transwell assay (r2 = 0.935, p = 0.033, linear regression) and metastasis (r2 = 0.939, p = 0.031, linear regression) (Fig. 3).
TABLE 2.
Kinetics of binding of mutant recombinant S100A4 proteins to immobilized recombinant C-terminal fragment of non-muscle myosin II heavy chain
| Proteina | Kass ± S.E.b | Correlation coefficientc | Kdiss ± S.E.d | Kd (kinetic)e | Kdf (equilibrium) |
|---|---|---|---|---|---|
| m−1s−1 | s−1 | nm | nm | ||
| Wild type S100A4g | 173,000 ± 63,000 | 0.91 | 0.015 ± 0.002 | 91 ± 36 | 55 ± 8 |
| Mutant 72 | 5,000 ± 1300 | 0.92 | 0.0237 ± 0.0014 | 4,600 ± 1270 | 4,800 ± 440 |
| Mutant 72.78 | 1,150 ± 360 | 0.93 | 0.02 ± 0.0023 | 16,000 ± 5,400 | 14,600 ± 8,700 |
| Mutant 16.72.78 | 730 ± 215 | 0.96 | 0.01 ± 0.003 | 13,400 ± 3,900 | 15,000 ± 3,500 |
a Mutants 72, 72.78, and 16.72.78 refer to S100A4 proteins in which phenylalanine residues at positions 16, 72, and 78 have been substituted with alanine, glutamine and alanine, respectively.
b kass and S.E. from three experiments were derived as described previously (33).
c The correlation coefficient of the linear regression through kon values used for obtaining kass..
d kdiss of the mean ± S.E. of at least 5 values at high concentrations of S100A4. No evidence was found for a two-site model of dissociation, suggesting that the S100A4 binding sites on NMMIIHCA were homogeneous.
e Kd kinetics were calculated from the ratio of kdiss/kass.
f Kd equilibrium was calculated from the extent of binding near equilibrium as described previously (33).
FIGURE 3.
Relationship between motility, metastasis, and the interaction of rNMMIIHCA with wild type and mutant S100A4 proteins. The equilibrium dissociation constants for the interaction of mutant S100A4 proteins and immobilized rNMMIIHCA were determined using an optical biosensor and plotted against the motility and metastasis-promoting properties of Rama 37 cells expressing wild type or mutant S100A4 proteins.
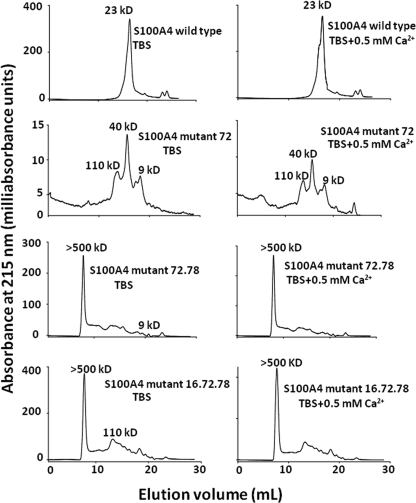
To find out the effect of the mutations of S100A4 on its self-association, purified wild type and mutant proteins were analyzed by Superdex 75 gel filtration chromatography in the absence of calcium ions, employing the buffer used for the biosensor experiments. Wild type S100A4 protein chromatographed as a single peak with a relative molecular mass of 28 kDa, consistent with it being a dimer in solution (not shown). Re-running the main peak after 17 h at 4 °C showed an absence of higher molecular weight material (not shown). In contrast to the wild type protein, the majority of the mutant proteins were present in higher molecular weight associations. Analysis of wild type and mutant proteins by Superdex 200 chromatography in a TBS buffer showed a single peak for wild type S100A4 protein of molecular mass ∼23 kDa, similar to that obtained with Superdex 75, but the mutant 72 protein chromatographed heterogeneously with molecular masses in the range 9–110 kDa. Mutants 72.78 and 16.72.78, although heterogeneous in size, chromatographed with a prominent peak of greater than 500 kDa (Fig. 4), thus, showing a higher degree of aggregation than the mutant 72 protein. The results suggest that the point mutations have seriously disrupted the normal dimerization of the S100A4 protein. The addition of 0.5 mm Ca2+ did not alter the pattern of elution of the proteins, thus, showing that the aggregation state of the mutants was not dependent upon the presence of Ca2+ ions (Fig. 4).
FIGURE 4.
Elution profile of S100A4 wild type and mutant proteins on gel filtration chromatography. Samples were applied to a Superdex 200 10/300 GL column equilibrated with TBS in the absence of (left-hand column) or in the presence of 0.5 mm Ca2+ (right-hand column). Proteins were separated at a flow rate of 0.5 ml/min, and elution was monitored at 215 nm. The retention times were related to those of standard proteins (“Experimental Procedures”) to determine the molecular masses of the S100A4 wild type and mutant proteins.
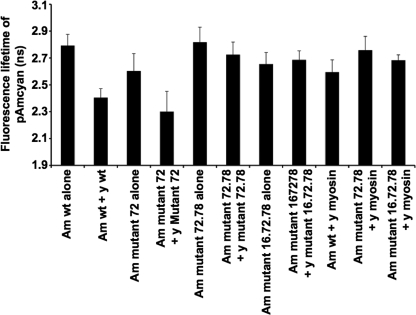
To find out whether these mutations to S100A4 protein affected its ability to self-associate and to associate with rNMMIIHCA inside living cells, FRET experiments using FLIM were carried out in HeLa cells (“Experimental Procedures”). The results (Fig. 5) showed a significant reduction in the fluorescence lifetime of pAmCyan in cells transfected with pAmCyan-wild type S100A4 and pYFP-wild type S100A4 constructs (7 cells were measured) compared with cells transfected with pAmCyan-wild type S100A4 alone (9 cells were measured; p < 0.0001, Student's t test; supplemental Data 6A), indicating self-association of the S100A4 monomers, as described previously (18). Similarly, there was a significant reduction in the fluorescence lifetime of pAmCyan in cells transfected with pAmCyan-S100A4 mutant 72 and pYFP-S100A4 mutant 72 constructs (4 cells were measured) compared with cells transfected with pAmCyan-S100A4 mutant 72 alone (5 cells were measured; p = 0.015, Student's t test; supplemental Data 6A); the results were not significantly different from pAmCyan-wild type S100A4 and pYFP-wild type S100A4 (p = 0.15, Student's t test; supplemental Data 6A), suggesting that mutant 72 is able to self-associate in vivo. In contrast, there was a borderline significant reduction in the fluorescence lifetime of pAmCyan in cells transfected with pAmCyan-S100A4 mutant 72.78 and pYFP-S100A4 mutant 72.78 constructs (4 cells were measured) compared with cells transfected with pAmCyan-S100A4 mutant 72.78 alone (5 cells were measured; p = 0.053, Student's t test; supplemental Data 6A); fluorescence lifetime was significantly longer than for pAmCyan-wild type S100A4 and pYFP-wild type S100A4 (p < 0.0001, Student's t test; supplemental Data 6A), suggesting that mutant 72.78 was not able to self-associate efficiently in vivo. For mutant 16.72.78 there was no significant reduction in fluorescence lifetime of pAmCyan in cells transfected with pAmCyan-S100A4 mutant 16.72.78 and pYFP-S100A4 mutant 16.72.78 constructs (5 cells were measured) compared with cells transfected with pAmCyan-S100A4 mutant 16.72.78 alone (9 cells were measured; p = 0.52, Student's t test; supplemental Data 6A), but the fluorescence lifetime was significantly different from pAmCyan-wild type S100A4 and pYFP-wild type S100A4 (p < 0.0001, Student's t test; supplemental Data 6A), showing that mutant 16.72.78 was not able to self-associate in vivo. These results suggest that the aggregation observed in vitro with the purified protein is not evident in vivo, at least as detected by FLIM, and that mutants 72.78 and 16.72.78 fail to self-associate efficiently in vivo.
FIGURE 5.
FLIM-FRET analysis. Fluorescence lifetime imaging of cells expressing AmCyan-S100A4, YFP-S100A4, and myosin-YFP proteins are shown. The interaction at the level of one living cell was measured using laser scanning microscopy as described under “Experimental Procedures.” Cells expressing AmCyan-wild type S100A4 and YFP-wild type S100A4 (7 cells were measured) and cells expressing AmCyan-S100A4 mutant 72 protein and YFP-S100A4 mutant 72 protein (4 cells were measured) exhibited significantly reduced fluorescence lifetime compared with control cells expressing AmCyan-wild type S100A4 alone (9 cells were measured) or AmCyan mutant 72 protein alone (5 cells were measured). In contrast, cells expressing AmCyan-S100A4 mutant 72.78 and YFP-S100A4 mutant 72.78 (11 cells were measured) or AmCyan-S100A4 mutant 16.72.78 and YFP-S100A4 mutant 16.72.78 (5 cells were measured) did not show reduced fluorescence lifetimes relative to controls expressing AmCyan-S100A4 mutant 72.78 (11 cells were measured) or AmCyan-S100A4 mutant 16.72.78 alone (9 cells were measured). Am, AmCyan fluorescent protein; y, yellow fluorescent protein; wt, wild type.
The wild type and mutant pAmCyan-S100A4 constructs 72.78 and 16.72.78 were tested for their ability to interact with rNMMIIHCA-YFP in vivo. The results (Fig. 5) showed that for cells transfected with pAmCyan-wild type S100A4 and pYFP-rNMMIIHCA constructs (10 cells were measured), there was a significant reduction in fluorescence lifetime compared with cells transfected with pAmCyan-wild type S100A4 alone (p = 0.00016, Student's t test; supplemental Data 6A) as reported before (18). For cells transfected with pAmCyan-S100A4 mutant 72.78 and pYFP-rNMMIIHCA (7 cells were measured), the fluorescence lifetime was not significantly different from that of cells transfected with pAmCyan-S100A4 mutant 72.78 alone (p = 0.285, Student's t test; supplemental Data 6A) but was significantly longer than from cells transfected with pAmCyan-wild type S100A4 and pYFP-NMMIIHCA constructs (p = 0.004, Student's t test; supplemental Data 6A), suggesting that mutant 72.78 fails to associate with rNMMIIHCA in vivo. A similar result was obtained with pAmCyan-S100A4 mutant 16.72.78 and pYFP-NMMIIHCA (6 cells were measured), which exhibited a fluorescence lifetime of pAmCyan that was not significantly different from that of pAmCyan-S100A4 mutant 16.72.78 alone (p = 0.479, Student's t test; supplemental Data 6A) but significantly longer than pAmCyan-wild type S100A4 and pYFP-NMMIIHCA constructs (p = 0.048, Student's t test; supplemental Data 6A), suggesting that this mutant also fails to interact with rNMMIIHCA in vivo.
In separate experiments (supplemental Data 6B) it was confirmed that cells transfected with pAmCyan-mutant 72.78 or pAmCyan-mutant 16.72.78 together with pYFP-rNMMIIHCA exhibited significantly longer fluorescence lifetimes than pAmCyan-wild type S100A4 with pYFP-rNMMIIHCA (p = 0.0007 and p < 0.0001, respectively, Student's t test). It was further shown that pAmCyan-mutant 72 also exhibited significantly longer fluorescence lifetimes than pAmCyan-wild type S100A4 with pYFP-rNMMIIHCA (p = 0.0012), suggesting that all three mutants of S100A4 had an impaired ability to interact with rNMMIIHCA in living cells.
Mutations to S100A4 affected its calcium-dependent interaction with another potential target, p53, using the optical biosensor (Table 3 and supplemental Data 7). Wild type S100A4 exhibited Kd values in the range 200–300 nm with immobilized p53 protein, whereas the effect of all the interface mutants was to increase the Kd ∼10-fold to μm concentrations, mainly by a decrease in the association rate constant (Table 3). The results suggest that the mutations which affect dimerization of S100A4 also reduce the affinity of its interaction with more than one of its intracellular targets.
TABLE 3.
Kinetics of binding of mutant recombinant S100A4 proteins to immobilized wild type p53
| Proteina | kass ± S.E.b | Correlation coefficientc | kdiss ± S.E.d | Kd (kinetic)e | Kd (equilibrium)f |
|---|---|---|---|---|---|
| m−1s−1 | s−1 | nm | nm | ||
| Wild type S100A4 | 65,000 ± 1 000 | 0.93 | 0.016 ± 0.003 | 243 ± 72 | 190 ± 10 |
| Mutant 72 | 4,200 ± 920 | 0.97 | 0.014 ± 0.003 | 3,400 ± 1000 | 1,800 ± 720 |
| Mutant 72.78 | 3,600 ± 770 | 0.97 | 0.015 ± 0.002 | 4,200 ± 1110 | 1,400 ± 450 |
| Mutant 16.72.78 | 6,500 ± 1,700 | 0.93 | 0.013 ± 0.0005 | 2,000 ± 540 | 1,400 ± 230 |
a Mutants 72, 72.78, and 16.72.78 refer to S100A4 proteins in which phenylalanine residues at positions 16, 72, and 78 have been substituted with alanine, glutamine, and alanine, respectively.
b kass and S.E. from 3–5 experiments are derived as described previously (33).
c The correlation coefficient of the linear regression through kon values used for obtaining kass..
d kdiss of the mean ± S.E. of three values at high concentrations of S100A4. No evidence was found for a two-site model of dissociation, suggesting that the S100A4 binding sites on p53 were homogeneous.
e Kd kinetics were calculated from the ratio of kdiss/kass.
f Kd equilibrium was calculated from the extent of binding near equilibrium as described previously (33).
DISCUSSION
The effects of mutations in the dimer interface of S100A4 on its target binding and metastasis-promoting ability have been tested. Transfected cell lines and cell pools derived from the rat mammary tumor-derived Rama 37 cells expressed the mutant proteins at levels that were similar to those of cells expressing wild type S100A4, suggesting that the mutations did not affect the stability of the mutant proteins in the Rama 37 cells, in contrast to human embryonic kidney cells, 293 (27). In vitro, the purified mutant S100A4 proteins associated to produce large aggregates, mutants 72.78 and 16.72.78 having a greater propensity to form higher molecular weight associations than the mutant 72 protein. In living cells the results of the FRET-FLIM experiments showed evidence for mutant 72-self-associating similar to the wild type protein; however, mutants 72.78 and 16.72.78 were not significantly different from non-associating controls, suggesting that these mutant proteins do not show the aggregate formation in vivo as they do in vitro. However, it is possible, but considered unlikely, that the FRET assay fails to detect the aggregates in living cells. It is also possible that the mutant proteins fused to the cyan and yellow fluorescent proteins do not aggregate to the same extent as the non-fused proteins.
It is shown for the first time that the double mutant, 72.78, and the triple mutant, 16.72.78, almost completely abolished the metastasis-promoting ability of S100A4 protein in vivo and, in the case of the triple mutant, reduced its invasion-inducing ability. However, the small number of primary tumors that contained muscle blocks on the histological sections of primary tumors might have affected the results for the double mutant, 72.78.
There was a linear relationship between metastasis-promoting ability and rate of cell migration (p = 0.011, r2 = 0.837, 95% confidence interval 0.4–0.99, linear regression) for mutants 72, 72.78, and 16.72.78, as observed previously in mouse mammary tumor cells bearing S100A4 transgenes (19), further establishing the link between S100A4-dependent cell migration and metastasis.
In the dual immunofluorescence staining, S100A4 was found in between and at the ends of myosin fibers, raising the possibility that S100A4 is associated with the dynamics of assembly of the myosin fibers. Furthermore, in the cells expressing mutant proteins 72.78 and 16.72.78, the myosin fibers were collapsed around the periphery of the cell, further raising the possibility that changes in the supramolecular structure of the myosin filaments are associated with the loss of migration, invasion, and metastasis in cells expressing mutant 72.78 or 16.72.78.
Measurement of the binding parameters of the interaction between mutant S100A4 proteins and the S100A4 target, rNMMIIHCA, in vitro showed that the three mutants, 72, 72.78, and 16.72.78 exhibited equilibrium dissociation constants with rNMMIIHCA in the 5–15 μm range compared with the low nm value for wild type S100A4 (28). There was an inverse relationship between the value of the equilibrium dissociation constant and the ability of the mutant proteins to promote metastasis/invasion. Although in vitro the increased equilibrium dissociation constant was associated with an increasingly aggregated state of the protein, the FLIM FRET results showed that mutants 72.78 and 16.72.78 did not self-associate in vivo nor, importantly, did they interact with rNMMIIHCA in vivo. These results suggest that proper dimerization of S100A4 is necessary for its interaction with myosin heavy chains and for S100A4-induced migration and metastasis.
The results for S100A4 mutant 72 protein were less clear cut. The 72 mutant protein resembled wild type protein in its abilities to bind calcium ions, to self-associate, and to induce migration. However, it exhibited defective interaction with rNMMIIHCA in vivo, partially reduced affinity for rNMMIIHCA (Kd 4–5 μm) in vitro, and partially reduced metastasis induction relative to the 72.78 and 16.72.78 mutants. We have previously shown (28) that deletion of 14 amino acids from the C terminus of S100A4 (Δ14) yields a protein that exhibits near wild type levels of migration but a Kd for myosin interaction in vitro of 3–4 μm, an ∼50% of wild type level of metastasis induction, and near wild type levels of Ca2+ interaction, properties that are similar to those of the current mutant 72 protein. In the case of the Δ14 mutant, it was suggested that previously described interdimer interactions (38) had been disrupted. It is possible that the mutation residue 72 causes changes in the structure of the protein that only become evident in the longer term assays, but the 72.78 and 16.72.78 mutations have more wide-ranging effects on the S100A4 protein which abrogate Ca2+ binding, reduce myosin affinity further, and reduce migration and metastasis closer to basal levels.
S100A4 interacts with a number of proposed, mainly intracellular (16, 17, 21, 23, 24, 39, 40) but also extracellular (25), protein targets. Of particular relevance in cancer is the interaction of S100A4 with the tumor suppressor protein, p53 (21, 22, 33). It is now shown that disruption of normal S100A4 self-association also disrupts the interaction of S100A4 with p53, at least in vitro, using immobilized recombinant p53 protein in an optical biosensor. In these experiments, the Kd values of the interaction between the wild type S100A4 protein and p53 were of the order of 200 nm, somewhat lower than obtained previously using immobilized S100A4, where it adopts an altered, calcium-independent conformation (33). In contrast, in the present experiments recombinant p53 was immobilized on the surface. The Kd values were lower than those in the μm range obtained using fluorescence anisotropy for a recombinant fragment consisting of 100 amino acids from the C-terminal region of p53 protein (22). In the present experiments the entire p53 protein was used, and the exact state of the p53 protein on the biosensor surface after the acid regenerative wash is not known. However, it has been proposed that S100A4 binds to the tetramerization domain in the C-terminal regions of p53 when exposed to lower oligomerization states of p53 (22). For interaction with p53, there was a large, 8–17-fold increase in the Kd for the mutant proteins relative to the wild type S100A4 protein, with a 14-fold increase in Kd evident with just the single mutant, 72. The observation that it is possible to disrupt the interaction between S100A4 and two proposed cancer-related intracellular protein targets, rNMMIIHCA and p53, with just two point mutations of amino acids conserved as phenylalanine/hydrophobic amino acids in 54 S100 proteins from fish to humans (26), opens up the possibility of inhibiting the metastatic action of S100A4 and other prometastatic S100 proteins, such as S100P (41), by targeting the self-association of these proteins.
Supplementary Material
Acknowledgment
We acknowledge the excellent technical assistance of Joe Carroll.
This work was supported by North West Cancer Research Fund CR655 and the Cancer and Polio Research Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Data 1–7.
- NMMIIHCA
- non-muscle myosin II heavy chain, isoform A
- FLIM
- fluorescence lifetime imaging
- FRET
- fluorescence resonance energy transfer
- PBS
- phosphate-buffered saline
- r
- recombinant
- TBS
- Tris-buffered saline
- MES
- 2-(N-morpholino)ethanesulfonic acid
- ITC
- isothermal titration calorimetry
- YFP
- yellow fluorescent protein.
REFERENCES
- 1.Andersen K., Nesland J. M., Holm R., Flørenes V. A., Fodstad Ø., Maelandsmo G. M. (2004) Mod. Pathol. 17, 990–997 [DOI] [PubMed] [Google Scholar]
- 2.Kimura K., Endo Y., Yonemura Y., Heizmann C. W., Schafer B. W., Watanabe Y., Sasaki T. (2000) Int. J. Oncol. 16, 1125–1131 [DOI] [PubMed] [Google Scholar]
- 3.Matsubara D., Niki T., Ishikawa S., Goto A., Ohara E., Yokomizo T., Heizmann C. W., Aburatani H., Moriyama S., Moriyama H., Nishimura Y., Funata N., Fukayama M. (2005) Cancer Sci. 96, 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudland P. S., Platt-Higgins A., Renshaw C., West C. R., Winstanley J. H., Robertson L., Barraclough R. (2000) Cancer Res. 60, 1595–1603 [PubMed] [Google Scholar]
- 5.Lee W. Y., Su W. C., Lin P. W., Guo H. R., Chang T. W., Chen H. H. (2004) Oncology 66, 429–438 [DOI] [PubMed] [Google Scholar]
- 6.Gongoll S., Peters G., Mengel M., Piso P., Klempnauer J., Kreipe H., von Wasielewski R. (2002) Gastroenterology 123, 1478–1484 [DOI] [PubMed] [Google Scholar]
- 7.Hemandas A. K., Salto-Tellez M., Maricar S. H., Leong A. F., Leow C. K. (2006) J. Surg. Oncol. 93, 498–503 [DOI] [PubMed] [Google Scholar]
- 8.Davies B. R., O'Donnell M., Durkan G. C., Rudland P. S., Barraclough R., Neal D. E., Mellon J. K. (2002) J. Pathol. 196, 292–299 [DOI] [PubMed] [Google Scholar]
- 9.Agerbaek M., Alsner J., Marcussen N., Lundbeck F., Von der Maase H. (2006) Eur. Urol. 50, 777–785 [DOI] [PubMed] [Google Scholar]
- 10.Nakamura T., Ajiki T., Murao S., Kamigaki T., Maeda S., Ku Y., Kuroda Y. (2002) Int. J. Oncol. 20, 937–941 [PubMed] [Google Scholar]
- 11.Ninomiya I., Ohta T., Fushida S., Endo Y., Hashimoto T., Yagi M., Fujimura T., Nishimura G., Tani T., Shimizu K., Yonemura Y., Heizmann C. W., Schäfer B. W., Sasaki T., Miwa K. (2001) Int. J. Oncol. 18, 715–720 [DOI] [PubMed] [Google Scholar]
- 12.Moriyama-Kita M., Endo Y., Yonemura Y., Heizmann C. W., Schäfer B. W., Sasaki T., Yamamoto E. (2004) Oral Oncol. 40, 496–500 [DOI] [PubMed] [Google Scholar]
- 13.Yonemura Y., Endou Y., Kimura K., Fushida S., Bandou E., Taniguchi K., Kinoshita K., Ninomiya I., Sugiyama K., Heizmann C. W., Schafer B. W., Sasaki T. (2000) Clin. Cancer Res. 6, 4234–4242 [PubMed] [Google Scholar]
- 14.Davies B. R., Davies M. P., Gibbs F. E., Barraclough R., Rudland P. S. (1993) Oncogene 8, 999–1008 [PubMed] [Google Scholar]
- 15.Ford H. L., Salim M. M., Chakravarty R., Aluiddin V., Zain S. B. (1995) Oncogene 11, 2067–2075 [PubMed] [Google Scholar]
- 16.Ford H. L., Silver D. L., Kachar B., Sellers J. R., Zain S. B. (1997) Biochemistry 36, 16321–16327 [DOI] [PubMed] [Google Scholar]
- 17.Kriajevska M. V., Cardenas M. N., Grigorian M. S., Ambartsumian N. S., Georgiev G. P., Lukanidin E. M. (1994) J. Biol. Chem. 269, 19679–19682 [PubMed] [Google Scholar]
- 18.Zhang S., Wang G., Fernig D. G., Rudland P. S., Webb S. E., Barraclough R., Martin-Fernandez M. (2005) Eur. Biophys. J. 34, 19–27 [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson S. R., Barraclough R., West C. R., Rudland P. S. (2004) Br. J. Cancer 90, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z. H., Bresnick A. R. (2006) Cancer Res. 66, 5173–5180 [DOI] [PubMed] [Google Scholar]
- 21.Grigorian M., Andresen S., Tulchinsky E., Kriajevska M., Carlberg C., Kruse C., Cohn M., Ambartsumian N., Christensen A., Selivanova G., Lukanidin E. (2001) J. Biol. Chem. 276, 22699–22708 [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Fernandez M. R., Veprintsev D. B., Fersht A. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4735–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kriajevska M., Fischer-Larsen M., Moertz E., Vorm O., Tulchinsky E., Grigorian M., Ambartsumian N., Lukanidin E. (2002) J. Biol. Chem. 277, 5229–5235 [DOI] [PubMed] [Google Scholar]
- 24.Takenaga K., Nakamura Y., Sakiyama S., Hasegawa Y., Sato K., Endo H. (1994) J. Cell Biol. 124, 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semov A., Moreno M. J., Onichtchenko A., Abulrob A., Ball M., Ekiel I., Pietrzynski G., Stanimirovic D., Alakhov V. (2005) J. Biol. Chem. 280, 20833–20841 [DOI] [PubMed] [Google Scholar]
- 26.Tarabykina S., Scott D. J., Herzyk P., Hill T. J., Tame J. R., Kriajevska M., Lafitte D., Derrick P. J., Dodson G. G., Maitland N. J., Lukanidin E. M., Bronstein I. B. (2001) J. Biol. Chem. 276, 24212–24222 [DOI] [PubMed] [Google Scholar]
- 27.Kim E. J., Helfman D. M. (2003) J. Biol. Chem. 278, 30063–30073 [DOI] [PubMed] [Google Scholar]
- 28.Ismail T. M., Fernig D. G., Rudland P. S., Terry C. J., Wang G., Barraclough R. (2008) Carcinogenesis 29, 2259–2266 [DOI] [PubMed] [Google Scholar]
- 29.Dunnington D. J., Hughes C. M., Monaghan P., Rudland P. S. (1983) J. Natl. Cancer Inst. 71, 1227–1240 [PubMed] [Google Scholar]
- 30.Gibbs F. E., Wilkinson M. C., Rudland P. S., Barraclough R. (1994) J. Biol. Chem. 269, 18992–18999 [PubMed] [Google Scholar]
- 31.Zhang S., Wang G., Liu D., Bao Z., Fernig D. G., Rudland P. S., Barraclough R. (2005) Oncogene 24, 4401–4411 [DOI] [PubMed] [Google Scholar]
- 32.Wang G., Zhang S., Fernig D. G., Martin-Fernandez M., Rudland P. S., Barraclough R. (2005) Oncogene 24, 1445–1454 [DOI] [PubMed] [Google Scholar]
- 33.Chen H., Fernig D. G., Rudland P. S., Sparks A., Wilkinson M. C., Barraclough R. (2001) Biochem. Biophys. Res. Commun. 286, 1212–1217 [DOI] [PubMed] [Google Scholar]
- 34.Rahmoune H., Chen H. L., Gallagher J. T., Rudland P. S., Fernig D. G. (1998) J. Biol. Chem. 273, 7303–7310 [DOI] [PubMed] [Google Scholar]
- 35.Lloyd B. H., Platt-Higgins A., Rudland P. S., Barraclough R. (1998) Oncogene 17, 465–473 [DOI] [PubMed] [Google Scholar]
- 36.Pedrocchi M., Schäfer B. W., Durussel I., Cox J. A., Heizmann C. W. (1994) Biochemistry 33, 6732–6738 [DOI] [PubMed] [Google Scholar]
- 37.Baudier J., Glasser N., Gerard D. (1986) J. Biol. Chem. 261, 8192–8203 [PubMed] [Google Scholar]
- 38.Malashkevich V. N., Varney K. M., Garrett S. C., Wilder P. T., Knight D., Charpentier T. H., Ramagopal U. A., Almo S. C., Weber D. J., Bresnick A. R. (2008) Biochemistry 47, 5111–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kriajevska M., Tarabykina S., Bronstein I., Maitland N., Lomonosov M., Hansen K., Georgiev G., Lukanidin E. (1998) J. Biol. Chem. 273, 9852–9856 [DOI] [PubMed] [Google Scholar]
- 40.Ford H. L., Zain S. B. (1995) Oncogene 10, 1597–1605 [PubMed] [Google Scholar]
- 41.Wang G., Platt-Higgins A., Carroll J., de Silva Rudland S., Winstanley J., Barraclough R., Rudland P. S. (2006) Cancer Res. 66, 1199–1207 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.