Abstract
Postmenopausal estrogen depletion is a characterized risk factor for Alzheimer disease (AD), a human disorder linked to high levels of β-amyloid peptide (Aβ) in brain tissue. Previous studies suggest that estrogen negatively regulates the level of Aβ in the brain, but the molecular mechanism is unknown. Here, we provide evidence that estrogen promotes Aβ degradation mainly through a principal Aβ degrading enzyme, neprilysin, in neuroblastoma SH-SY5Y cells. We also demonstrate that up-regulation of neprilysin by estrogen is dependent on both estrogen receptor α and β (ERα and ERβ), and ligand-activated ER regulates expression of neprilysin through physical interactions between ER and estrogen response elements (EREs) identified in the neprilysin gene. These results were confirmed by in vitro gel shift and in vivo chromatin immunoprecipitation analyses, which demonstrate specific binding of ERα and ERβ to two putative EREs in the neprilysin gene. The EREs also enhance ERα- and ERβ-dependent reporter gene expression in a yeast model system. Therefore, the study described here provides a putative mechanism by which estrogen positively regulates expression of neprilysin to promote degradation of Aβ, reducing risk for AD. These results may lead to novel approaches to prevent or treat AD.
Keywords: Alzheimer Disease, β-Amyloid Peptide, Estrogen, Estrogen Receptor, Estrogen Response Element, Neprilysin
Introduction
Alzheimer disease (AD)3 is a progressive neurodegenerative disease characterized by declarative memory impairment and progressive dementia. The level of β-amyloid peptide (Aβ) is elevated in the brains of AD patients, and Aβ is believed to play a critical role in the pathology of AD (1, 2). Recent studies show that aggregated oligomers of Aβ (protofibrils) play a direct role in neuronal and behavioral deficits in AD patients (3).
The rate of Aβ degradation could influence the risk of developing AD, and it has been proposed that stimulation of proteolytic degradation of Aβ could be used as a therapeutic approach for AD (4, 5). Neprilysin is thought to be the primary Aβ-degrading enzyme in the brain (6) because degradation of radiolabeled synthetic Aβ42 in rat brain is largely inhibited by the neprilysin inhibitor, phosphoramidon (PA) (7, 8) and because neprilysin degrades both monomeric and oligomeric forms of Aβ40 and Aβ42 in intracellular and extracellular compartments of the brain (9). Moreover, the level of neprilysin mRNA and protein is lower in the hippocampus and temporal gyrus of AD patients (10, 11), which correlates with higher levels of Aβ as Aβ tends to accumulate in these regions (12).
Neprilysin activity is also lower in the hippocampus, cerebellum, and caudate of ovariectomized rats than in non-ovariectomized rats, and this effect can be reversed by exogenous 17β-estradiol (13). These data indicate that 17β-estradiol positively regulates neprilysin activity in the brain. 17β-Estradiol was also reported to reduce the generation of Aβ peptides in neuroblastoma cells and neurons (14). The positive regulation of neprilysin by 17β-estradiol might be a crucial factor in protecting normal adult brain from Aβ damage and in improving cognitive performance in menopausal women.
This study reports that 17β-estradiol promotes Aβ clearance by up-regulating neprilysin expression in human neuroblastoma SH-SY5Y cells. In these cells, 17β-estradiol stimulates neprilysin expression in an estrogen receptor (ER)-dependent manner. Furthermore, two functional estrogen response elements (EREs) were identified in the neprilysin gene, which bind ERα and ERβ in vitro and in vivo and which stimulate ER-dependent reporter gene expression in a yeast system. These results provide insight into the neuroprotective effects of estrogen and suggest that neprilysin could have potential as a therapeutic drug target for AD.
MATERIALS AND METHODS
Cell Culture and Ligand Treatments
SH-SY5Y cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 10 units/ml penicillin, and 5 μg/ml amphotericin B (Amresco). At least 3 days before use cells were switched to phenol red-free Dulbecco's modified Eagle's medium (Invitrogen) containing 10% charcoal-stripped fetal bovine serum (Biological Industries) and 5 μg/ml amphotericin B. 17β-Estradiol (Sigma), propyl pyrazole triol (PPT, Tocris Cookson), and diarylpropionitrile (DPN, Tocris Cookson) were diluted in 100% DMSO (Vehicle) (Sigma). Cells were then treated with 17β-estradiol, PPT, DPN, or vehicle at different concentrations for the indicated length of time.
Recombinant Plasmids
YEP-ERα and YEP-ERβ, which express ERα and ERβ in yeast, respectively, YRPC2, containing a CYC1 promoter and a lacZ reporter gene, and YRPC2-2cERE, which has two consensus EREs (cEREs) in front of the CYC1 promoter, were gift from Dr. Dan Noonan (University of Kentucky). Three putative EREs were identified in the neprilysin genomic sequence. Oligonucleotides containing two copies of each putative ERE and a terminal XhoI recognition site were synthesized (Table 1), annealed, phosphorylated, and ligated into YRPC2 plasmid. Recombinant plasmids pET-28b-ERα and pET-28b-ERβ that expressed ERs in Escherichia coli Rosetta (DE3) cells were constructed by inserting ER-coding sequences from YEP-ERα and YEP-ERβ at the NdeI and EcoRI sites. pEGFP-ERα and pEGFP-ERβ that expressed ERs in mammalian cells were obtained by inserting ERs-coding sequences at the XhoI and EcoRI sites.
TABLE 1.
Sequences of oligonucleotides for construction of YRPC2 plasmids containing putative EREs
Initial, single-underlined (TCGA) sequences indicate XhoI sites. Double-underlined sequences are sequences of putative EREs. Bold residues are core sequences of putative EREs.
| Name | Direction | Seqence (5′–3′) |
|---|---|---|
| ERE-I | Sense | TCGAGTGGGTCAGGTCACTGCAACCTCTCTCGAGTCAGGTCACTGCAACCTCTGTGCC |
| Antisense | TCGAGGCACAGAGGTTGCAGTGACCTGACTCGAGAGAGGTTGCAGTGACCTGACCCAC | |
| ERE-II | Sense | TCGAGTGGGCCTGGTCAGTTTCACCTGTCTCGAGCCTGGTCAGTTTCACCTGTGTGCC |
| Antisense | TCGAGGCACACAGGTGAAACTGACCAGGCTCGAGACAGGTGAAACTGACCAGGCCCAC | |
| ERE-III | Sense | TCGAGTGGGTTGGGTCACTGCAACCTCCCTCGAGTTGGGTCACTGCAACCTCCGTGCC |
| Antisense | TCGAGGCACGGAGGTTGCAGTGACCCAACTCGAGGGAGGTTGCAGTGACCCAACCCAC |
Aβ42 Degradation Assay and Neprilysin Activity Assay
SH-SY5Y cells were treated with 100 nm 17β-estradiol or vehicle. 24 h later the medium was removed, and the cells were incubated with medium spiked with Aβ42 (100 pg/ml; Sigma) and 1 μm ZnCl2 without the serum supplemented. The medium was collected 8 h later, and the Aβ42 levels were detected by enzyme-linked immunosorbent assay kits (CUSABIO BIOTECH). After 17β-estradiol treatment for 24 h, SH-SY5Y cells were harvested, and the membrane proteins were extracted by protein extract kit (DBI Research Products). 150-μg membrane fractions were evaluated for neprilysin enzymatic activity as described previously by Huang et al. (13). PA (Sigma) was used as a specific neprilysin inhibitor to block the neprilysin activities.
RNA Isolation and RT-PCR
Total RNA was extracted using TRIZOL reagent from Invitrogen. cDNA was synthesized using a ReverTra Ace-α-cDNA synthesis kit (TOYOBO) and random primers (TaKaRa) following the manufacturer's instructions. Semiquantitative PCR was performed to determine the mRNA levels of neprilysin. Assessment of the PCR conditions ensures that the cycle is in the linear range.
Primer sequences were: glyceraldehyde-3-phosphate dehydrogenase, 5′-ACCACAGTCCATGCCATCAC-3′ and 5′- TCCACCACCCTGTTGCTGTA-3′; neprilysin, 5′-CTTTAACAAAGATGGAGACCTCGT-3′ and 5′-GAGTTCTGCAAAGTCCCAATAATC-3′. The mRNA level of glyceraldehyde-3-phosphate dehydrogenase served as an internal control.
Western Blot and Antibodies
Cells were washed twice in phosphate-buffered saline, harvested in radioimmune precipitation assay buffer buffer, and diluted in SDS-reducing sample buffer. Approximately 300 μg of cell lysate was resolved by 10% SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore), probed with the indicated antibodies, and detected by SuperSignal West Pico substrate (Pierce). The following antibodies were used: monoclonal mouse anti-human CD10 (CALLA) (14-0108, eBioscience); polyclonal rabbit anti-β-actin (sc-47778, Santa Cruz); polyclonal rabbit anti-ERα (sc-7207, Santa Cruz; polyclonal rabbit anti-ERβ (sc-8974, Santa Cruz); peroxidase-conjugated immunopure goat anti-rabbit and anti-mouse IgG (H+L) (Pierce).
Small Interfering RNA (siRNA) Duplexes Specific for Human ERα and ERβ
Two siRNA duplexes specific for human ERα and ERβ were designed according to Musatov et al. (15). The targeted 19-mer oligonucleotides sequence for ERα and ERβ were GGCATGGAGCATCTCTACA (1923–1941 nucleotides, NM_000125.2) and GGCATGGAACATCTGCTCA (1882–1900 nucleotides, NM_001437.2). All siRNA sequences were designed and synthesized according to the manufacturer's recommendations (GenePharma). The siRNA duplex (UUCUCCGAACGUGUCACGUTT and ACGUGACACGUUCGGAGAATT), which had no significant homology to any known gene sequences from mouse, rat, and human, served as a negative control.
RNA Interference and Overexpression of ERs
The transfection of siRNA duplexes into SH-SY5Y cells cultured in six-well plates was performed using Lipofectamine 2000 (Invitrogen) as described by Deng (16). 100 nm siRNA duplexes were transfected to knock down ERα or ERβ, and 48 h later 10 nm 17β-estradiol was added to induce the expression of neprilysin.
Delivery of 1 μg of recombinant pEGFP plasmids to SH-SY5Y cells grown in 6-well plates was carried out using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. 48 h later 10 nm 17β-estradiol was supplemented to induce the expression of neprilysin.
Transcription Factor Binding Site Analysis
The neprilysin gene sequence is available from the NCBI nucleotide data base (www.ncbi.nlm.nih.gov) (GenBankTM accession number AC106724, Homo sapiens 3 BAC RP11-270G15 complete sequence) (Roswell Park Cancer Institute Human BAC Library). The putative EREs were determined using TRANSFAC® 6.0.
Expression and Purification of Recombinant ERs
pET-28b-ERα/ERβ was transformed into E. coli Rosetta (DE3) cells. Transformed cells were cultured in Luria-Bertani medium supplemented with kanamycin (40 μg/ml) and chloramphenicol (34 μg/ml) at 37 °C until A600 reached 0.8. Then 0.5 mm isopropyl 1-thio-β-d-galactopyranoside was added to induce the expression of recombinant His6-ERs for 24 h at 20 °C. Recombinant His6-ERs were purified on nickel-Sepharose (GE Healthcare) according to the GE Healthcare protocol. The column was washed with column balance buffer containing 40 mm imidazole, and the bound protein was eluted with 260 mm imidazole. Dialysis was applied to remove imidazole from the eluants.
Nonradioactive Gel Mobility Shift Assay
Digoxygenin-labeled probes containing putative EREs or cERE was prepared by amplification of recombinant YRPC2 plasmids with the primers 5′-digoxigenin-TGATCATGTGTCGTCGCA-3′ and 5′- TCTGAGTTCCGACAACAATG-3′. Labeled probes were PAGE-purified. Nonradioactive gel mobility shift assay was performed according to Hammer's Protocol for Nonradioactive electrophoretic mobility shift assays. 50 ng of end-labeled oligonucleotide and 50 μg of purified His6-ER were mixed in the binding reaction and incubated for 60 min. After analyzed on 6% non-denaturing polyacrylamide gels and transferred to nylon membrane, the probes were blotted with the anti-digoxigenin-alkaline phosphatase Fab fragment (Roche Applied Science). Ready-to-use substrate nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (Amresco) was used to develop the nylon membrane.
Chromatin Immunoprecipitation (ChIP)
SH-SY5Y cells were grown in phenol red-deprived Dulbecco's modified Eagle's medium supplemented with 10% charcoal-stripped fetal bovine serum for at least 3 days and treated with 10 nm 17β-estradiol for 45 min. The protein-DNA complexes were cross-linked with 1% formaldehyde for 10 min and sonicated into fragments 200–400 bp in length. Immunoprecipitation was performed using Protein A/G-agarose (Santa Cruz), anti-human IgG (Zhongshan Golden Bridge Biotechnology), anti-ERα (sc-7207, Santa Cruz), and anti-ERβ (sc-8974, Santa Cruz) antibodies followed by reverse cross-linking, proteinase K treatment, phenol-chloroform extraction, and ethanol precipitation. Primers used for semiquantitative PCR were: ERE-I, 5′-GCAGTTGTGCAATTTCAGGTCACT-3′ and 5′-TAGAAAAACAAAAGCTGAGTGTGG-3′; ERE-II, 5′-CTTCCACAGCCCTGGTCAGTTTCA-3′ and 5′-CCACCGTCTTCCAGATCCCAGAAT- 3′; ERE-III, 5′-ACCTCAGATGGAAATGCGGAAAT-3′ and 5′-TGGGTGACAGGACAGAGCAAGAC-3′; promoter, 5′-CACCCTCAACCTCCGATG-3′ and 5′-TCCTGCTTTCTCCACCCC-3′.
Yeast Transformation and β- Galactosidase Assay
Recombinant YRPC2 plasmids containing the putative EREs and YEP-ERα or YEP-ERβ were transformed into BJ5409 yeast cells using quick and easy TRAFO Protocol (17). β-Galactosidase assays were performed essentially as we described before (18).
Statistical Analysis
Results are expressed as the mean ± S.D. Statistical analysis was performed using one-way analysis of variance followed by the Bonferroni ad hoc test. The intensities of the bands were quantitated using Quantity One software (Bio-Rad).
RESULTS
17β-Estradiol Enhances Aβ42 Degradation and Up-regulates Neprilysin
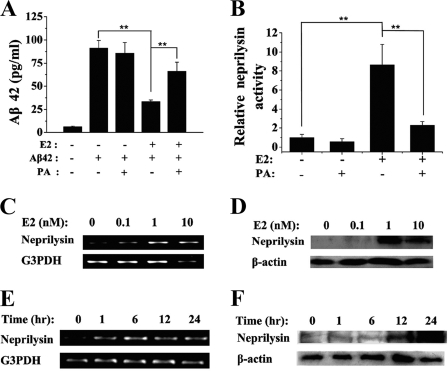
Previously we demonstrated that 17β-estradiol stimulates expression of neprilysin in the hippocampus, cerebellum, and caudate of rat brain (13). Here, we measured degradation of exogenous Aβ in SH-SY5Y cells, a human neuroblastoma cell line widely used for Aβ metabolism (19). As expected, exogenous Aβ42 was degraded more efficiently (reduced to 36% of input substrate) in cells treated with 17β-estradiol (100 nm for 24 h). Increased degradation of Aβ42 was blocked by PA, a specific neprilysin inhibitor (reversed to 73%) (Fig. 1A). In parallel with enhanced degradation of Aβ, neprilysin activity increased ∼8.5-fold, and this increase was 80% inhibited by PA (Fig. 1B). These results suggest that 17β-estradiol might up-regulate expression of neprilysin, which in turn stimulates degradation of Aβ.
FIGURE 1.
Induction of neprilysin and degradation of Aβ42 by 17β-estradiol (E2). SH-SY5Y cells were grown in 17β-estradiol-free medium at least 3 days before use. A, after incubation with 100 nm 17β-estradiol or vehicle for 24 h, cells were incubated with Aβ42 (100 pg/ml). Aβ42 degradation and the reverse effect by PA (75 μm) were detected by enzyme-linked immunosorbent assay kits. Medium without Aβ42 was used as a negative control. B, cells were treated with vehicle or 100 nm 17β-estradiol for 24 h, harvested, and lysed. 150-μg membrane fractions were evaluated for neprilysin enzymatic activity. PA (75 μm) was used to specifically inhibit neprilysin activity. C, cells were treated with vehicle, 100 pm, 1 nm, or 10 nm 17β-estradiol for 6 h. Total RNA was prepared, and RT-PCR was carried out as described under “Materials and Methods.” Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) was an internal control. D, cells were incubated with vehicle, 100 pm, 1 nm, or 10 nm 17β-estradiol for 24 h, harvested, and lysed. Total cellular protein was analyzed by Western blot using anti-CD10 and anti-β-actin (internal control). E, cells were treated with 10 nm 17β-estradiol for 0, 1, 6, 12, or 24 h. Total RNA was prepared, and RT-PCR was carried out. F, cells were treated with 10 nm 17β-estradiol for 0, 1, 6, or 24 h. Western blots were probed as indicated. Data values are represented as the mean ± S.D., and three independent experiments were performed. **, p < 0.01.
RT-PCR and Western blot analysis showed that 17β-estradiol increases neprilysin mRNA and protein in a dose- and time-dependent manner. Neprilysin mRNA increased >2-fold in cells treated for 6 h with 1 nm 17β-estradiol and >5-fold in cells treated with 10 nm 17β-estradiol (Fig. 1C). The level of neprilysin protein increased significantly in cells treated for 24 h with 1 or 10 nm 17β-estradiol (p < 0.01) (Fig. 1D). Neprilysin mRNA reached and was maintained at a maximal level ∼6 h after treatment with 10 nm 17β-estradiol (Fig. 1E), whereas neprilysin protein reached a maximum ∼24 h after treatment (Fig. 1F). These results are consistent with the estrogen activation model proposed by Shang (20), namely, that neprilysin mRNA is relatively unstable, such that ongoing transcription is required to maintain the steady state level of neprilysin mRNA and protein.
ERα/ERβ-selective Agonists Induce Neprilysin Expression
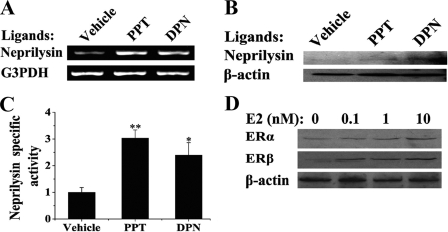
SH-SY5Y cells express both ERα and ERβ (21). Therefore, the roles of ERα and ERβ in regulating of estrogen-stimulated neprilysin expression were examined. This was done by incubating cells in the presence or absence of ERα-specific agonist, PPT (22), or ERβ-specific agonist, DPN (23). As shown in Fig. 2, both PPT and DPN induced neprilysin mRNA (Fig. 2A) and protein (Fig. 2B) in SH-SY5Y cells, and neprilysin activity was elevated by PPT and DPN (Fig. 2C). These subtype-specific agonists were less potent than 17β-estradiol in inducing neprilysin, which is consistent with the fact that 17β-estradiol activates both receptor subtypes. Interesting, 17β-estradiol also appears to stimulate expression of ERα and ERβ in SH-SY5Y cells (Fig. 2D). These results suggest that both ERα and ERβ mediate the effects of 17β-estradiol on expression of neprilysin.
FIGURE 2.
Activation of neprilysin by ERα- or ERβ-selective agonists PPT and DPN. SH-SY5Y cells were grown in 17β-estradiol-depleted medium for at least 3 days before use. A, cells were treated with vehicle, 10 nm PPT, or 10 nm DPN for 6 h followed by total RNA extraction and RT-PCR. The levels of neprilysin and glyceraldehyde-3-phosphate dehydrogenase (G3PDH) mRNA were determined. B, cells were treated with vehicle, 10 nm PPT, or 10 nm DPN for 24 h followed by Western blotting. The relative levels of neprilysin and β-actin were detected by anti-CD10 and anti-β-actin. C, the specific neprilysin activities after 10 nm PPT or 10 nm DPN treatments for 24 h were measured and normalized to vehicle-treated groups. D, cells were harvested and analyzed by Western blot using anti-ERs and anti-β-actin after incubation with the indicated concentration of 17β-estradiol (E2) for 24 h. *, p < 0.05; **, p < 0.01.
Induction of Neprilysin Is Dependent on ERα and ERβ
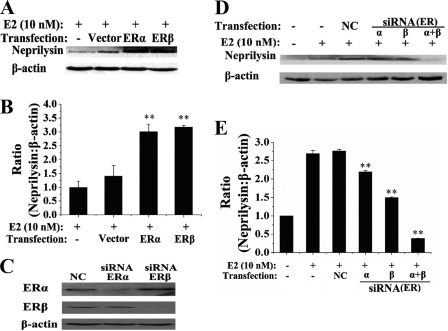
To further investigate the effects of ER subtypes in this process, the expression of ERα or ERβ was up- or down-regulated by transient transfection with ER expression vectors or siRNA-mediated knockdown in SH-SY5Y cells. To overexpress ERs, GFP-tagged ERα or ERβ was expressed from appropriate plasmid vectors, and ER expression was monitored by confocal microscopy (data not shown). As shown in Fig. 3, A and B, transfection of the GFP vector had little effect on neprilysin induction, whereas transfection of GFP-ERα or GFP-ERβ induced neprilysin at least 3-fold.
FIGURE 3.
Overexpression and silence of ERs altered the 17β-estradiol inducibility of neprilysin. A, SH-SY5Y cells were transiently transfected with 1 μg of pEGFP, pEGFP-ERα, or pEGFP-ERβ and incubated for 48 h. The inducibility of neprilysin was detected by anti-CD10 after 10 nm 17β-estradiol (E2) administration for 24 h. The blots were quantified by densitometry (B). C, silence of ERα or ERβ was conducted by transfection with 100 nm corresponding siRNA duplexes. The relative ERs levels were detected by anti-ERs. NC, negative control as described under “Materials and Methods.” D, after 10 nm 17β-estradiol administration for 24 h, the inducibility of neprilysin was detected by anti-CD10 and quantified by densitometry (E). Three independent experiments were performed. The ratios (neprilysin:β-actin) were calculated and normalized to 17β-estradiol-treated (B)/untreated (E) and non-transfection groups. **, p < 0.01.
Previously characterized ERα- or ERβ-targeted siRNAs (15) were used to knock down endogenous ERα or ERβ by ≥70% in SH-SY5Y cells (Fig. 3C). As shown in Fig. 3, D and E, single siRNA knockdown of ERα or ERβ decreased the level of neprilysin slightly (19% or 44%), whereas a much larger effect (86% decrease) was observed when both ERα and ERβ were knocked down. These results confirm that both ERα and ERβ mediate the effects of 17β-estradiol on expression of neprilysin.
Identification of Putative EREs in the Neprilysin Gene and Their Interaction with Recombinant ERα and ERβ in Vitro
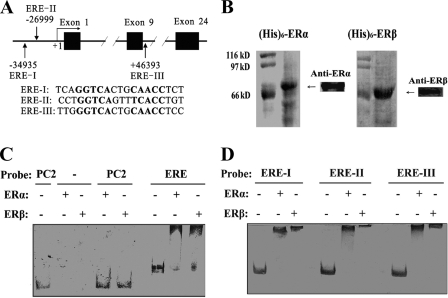
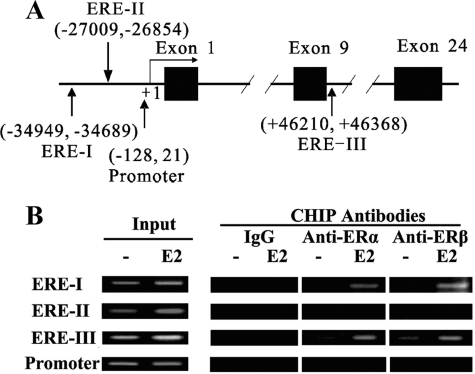
One mechanism to explain the above data is that ligand-activated ER binds to target EREs in the neprilysin gene, which stimulates transcription of neprilysin mRNA. Typically, ligand-activated ER dimerizes, binds to target EREs, recruits RNA polymerase II, and stimulates transcription. This possibility was tested by computationally searching for homology to the cERE, which has the sequence GGTCAnnnTGACC, in the neprilysin gene. This transcription factor binding site analysis identified three putative EREs (ERE-I, ERE-II, and ERE-III) with >80% similarity to the cERE (Fig. 4A). ERE-I and ERE-II are located upstream of the promoter, and ERE-III is in the ninth intron.
FIGURE 4.
Locations of putative EREs in the neprilysin genomic region and their interaction with purified ERs in vitro. A, solid boxes are exons, and solid lines represent introns and adjacent genomic regions. Vertical arrows indicate the locations of putative EREs. The number under or above each putative ERE is its distance from the transcription start site. The detailed sequences of EREs were also listed below. B, recombinant His6-ERs were expressed in Rossetta (DE3) E. coli cells, purified by Ni-Sepharose affinity chromatography, and characterized by immunoblotting with anti-ERα and anti-ERβ. C, the probe from the YRPC2 vector was used as negative control (PC2), and the probe from YRPC2-cERE was used as positive control (ERE). Recombinant ERα or ERβ was included as indicated. D, putative EREs containing probes were incubated with ERα or ERβ as indicated.
To determine whether ERα and ERβ bound to putative EREs I-III from the neprilysin gene in vitro, gel mobility shift assays were performed using recombinant His6-ERα or His6-ERβ, which was expressed in and purified from Rossetta (DE3) E. coli (Fig. 4B). DNA substrates containing cERE, ERE-I, ERE-II, or ERE-III were generated by PCR amplification of target regions from the appropriate plasmid template. As expected, ERα and ERβ did not bind to the negative control DNA probe but did bind to and alter the mobility of a DNA substrate containing cERE (Fig. 4C). Furthermore, ERα and ERβ bind specifically to putative ERE-I, ERE-II, and ERE-III, altering the mobility of a large fraction of the input DNA substrate (Fig. 4D).
In Vivo Occupancy of EREs in the Neprilysin Gene in SH-SY5Y Cells
The significance of the putative EREs in regulating neprilysin expression can only be assessed by testing their functional effects in vivo. Therefore, ChIP assays were performed to evaluate occupancy of the three putative EREs in SH-SY5Y cells. In this experiment SH-SY5Y cells were treated for 45 min with vehicle or 17β-estradiol, and cell lysates were immunoprecipitated using anti-ER antibody or control IgG. Neprilysin gene regions containing ERE-I, ERE-II, or ERE-III were amplified by semiquantitative PCR, as shown in Fig. 5A. ChIP results suggest that ERα and ERβ occupy ERE-I and ERE-III in a 17β-estradiol-dependent manner in SH-SY5Y cells, but ERα and ERβ do not bind ERE-II (Fig. 5B). Thus, there is a discrepancy between in vitro (i.e. gel mobility shift) and in vivo (ChIP assay) binding data for ERE-II, indicating that ERE-II may interact more weakly with ERα and ERβ than ERE-I and ERE-III or that ERE-II may not play a direct role in promoting estrogen-dependent neprilysin expression in vivo. ChIP analysis also showed that the −121 to +28 region of the neprilysin promoter is not bound by ERα or ERβ in 17β-estradiol-treated SH-SY5Y cells (Fig. 5B). These data support the idea that ERE-I and ERE-III, but not ERE-II, are functional EREs that promote 17β-estradiol-stimulated ER-dependent induction of neprilysin in vivo.
FIGURE 5.
Occupancy of target sites in neprilysin gene in SH-SY5Y cells by ERα and ERβ. The cells were grown in estrogen-depleted medium for at least 3 days before use. Cells were treated with 10 nm 17β-estradiol (E2) or vehicle for 45 min, fixed, and harvested. ChIP assay was performed using rabbit anti human IgG, anti-ERα antibody, and anti-ERβ antibody as indicated. Input was used as positive control. A, a map of neprilysin gene and locations of regions amplified for ChIP analysis is shown. +1 is the transcription start site, and the numbers indicate the coordinates of amplified regions in the neprilysin genomic sequence. B, ChIP analysis of putative EREs and promoter region are shown. At least three independent experiments were performed; a representative result is shown.
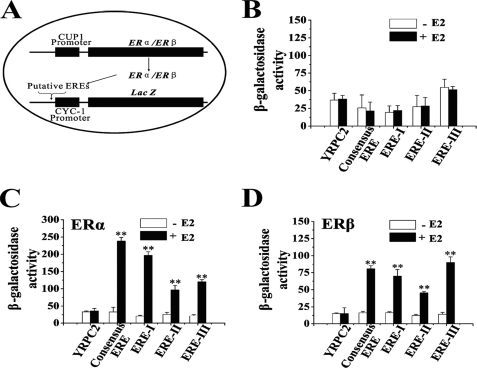
Putative EREs Mediate ERα- and ERβ-dependent Reporter Gene Activity in Yeast
The function of EREs I, II, and III from the neprilysin gene was tested using a yeast reporter gene system. The putative EREs were cloned into the reporter plasmid YRPC2 to generate three recombinant plasmids: YRPC2-ERE-I, YRPC2-ERE-II, and YRPC2-ERE-III. YRPC2-cERE, which carries two copies of cERE, and YRPC2 vector were used as positive and negative controls, respectively. Recombinant YRPC2 plasmids and ERα or ERβ expression plasmids (YEP-ERα and YEP-ERβ) were transformed into yeast BJ5409 cells. Fig. 6A shows the mechanism of the transcriptional activity assay. The β-galactosidase activities indicate the reporter gene activity. Because ER expression plasmids are exogenously expressed in yeast cells, this reporter gene system is free of interference from endogenous ER coregulators. Control experiments showed no reporter gene activity for any of the above constructs in 17β-estradiol-treated cells that do not express ERα or ERβ (Fig. 6B). This result demonstrates the absolute dependence of reporter gene induction on exogenous ER.
FIGURE 6.
Putative EREs could induce yeast reporter gene expression with ERα and ERβ. BJ 5409 yeast cells were transformed with reporter gene constructs as indicated, grown to A600 = 0.60, and treated with 100 nm 17β-estradiol (E2) for 24 h. β-Galactosidase activity was an indicator of reporter gene activity. A, shown is a map of recombinant reporter gene plasmids and the basic mechanism of this transcriptional activity assay. B, shown is β-galactosidase activity in yeast cells that do not express ERs. C, shown is β-galactosidase activity in yeast cells expressing ERα. D, shown is β-galactosidase activity in yeast cells expressing ERβ. Three independent experiments were performed. **, p < 0.01.
However, in 17β-estradiol-treated cells that express ERα, all of the putative EREs showed reporter gene activity. ERE-I, ERE-II, or ERE-III constructs induced 9.6-, 3.7-, or 5.7-fold higher reporter gene activity than the same cells without 17β-estradiol treatment, respectively (Fig. 6C). In 17β-estradiol-treated cells that express ERβ (Fig. 6D), ERE-I, ERE-II, or ERE-III constructs supported a 4.2-, 3.7-, or 6.3-fold increase in reporter gene activity. Thus, these data suggest that ligand-activated ERα and ERβ independently activate reporter gene transcription with promoters containing ERE-I, ERE-II, or ERE-III. ERE-II appears to be a weaker enhancer than ERE-I and ERE-III. There was no evidence for synergism between ERα and ERβ during activation of the three putative EREs (data not shown).
DISCUSSION
This study demonstrates for the first time that 17β-estradiol and two selective ERα/ERβ agonists (PPT and DPN) positively regulate expression of neprilysin, which in turn stimulates Aβ42 degradation in human neuroblastoma SH-SY5Y cells. The data presented here also show that 17β-estradiol stimulates expression of neprilysin in an ERα- and ERβ-dependent manner. Furthermore, we identify two novel putative EREs (ERE-I and ERE-III) located in the neprilysin genomic region that bind ERα and ERβ in vitro and in vivo and convey 17β-estradiol inducibility of a reporter gene in yeast. Together these results indicate a possible novel mechanism for the neuroprotective activity of estrogen.
Previous studies show that estrogen blocks the neurotoxic effects of Aβ42 in SH-SY5Y cells (21). These observations are consistent with our report in this study that 17β-estradiol decreases the neurotoxic Aβ42 in SH-SY5Y cells. Furthermore, we observe that the decrease can be reversed by the specific neprilysin inhibitor (Fig. 1A). Because neprilysin is the principal Aβ degrading enzyme, even modest up-regulation of neprilysin can reduce accumulation of Aβ in the mouse brain (6, 24), or even partial down-regulation of neprilysin activity can contribute to AD development by promoting Aβ accumulation (7). This is consistent with our observation that after 17β-estradiol treatment, the inhibition of neprilysin by the specific neprilysin inhibitor correlates with decreased Aβ degradation. Thus, 17β-estradiol may prevent Aβ-associated neurotoxicity through its ability to stimulate neprilysin-mediated degradation of Aβ peptides. These results provide a possible explanation for the observation that women who receive estrogen replacement therapy soon after menopause have a decreased risk for AD as well as higher cognitive performance.
ERα and ERβ have significant neuroprotective effects in neuronal cells (25, 26). PvuII and XbaI polymorphisms located in ERα (27–29) and five intronic single-nucleotide polymorphisms (30) in the ERβ gene were identified as susceptibility factors for AD in women. Most interestingly, in the hippocampus of female rats, expression of ERs decreases with age (31, 32). If a similar decrease occurs in human brain, it might explain the decreased efficacy of estrogen replacement therapy in older postmenopausal women (33). In this study we show that selective ER agonists (PPT and DPN) positively and independently regulate neprilysin expression (Fig. 2) and that overexpression of either ERα or ERβ significantly enhances the 17β-estradiol inducibility of neprilysin. Also, siRNA knockdown of ERα and ERβ significantly inhibits induction of neprilysin (Fig. 3). Therefore, 17β-estradiol stimulates neprilysin gene expression in an ER-dependent manner, and each ER subtype independently mediates such stimulation.
Previous studies demonstrated that 17β-estradiol exerted its transcriptional effects through genomic pathways and/or non-genomic pathways as reviewed by Björnström and Sjöberg (34). In this study we have identified two putative EREs in the neprilysin gene, ERE-I and ERE-III, that can bind ERs in vitro (Fig. 4) and can be occupied by ERα and ERβ in vivo (Fig. 5). These two EREs confer 17β-estradiol inducibility in a yeast reporter gene system (Fig. 6), suggesting that 17β-estradiol stimulates ERs, which bind EREs in the neprilysin gene and recruit coactivators to initiate neprilysin expression.
Deschênes et al. (35) found that chromatin loops form between multiple EREs spanning 20 kb and a transcriptional start site in the presence of 17β-estradiol, suggesting that a multipartite transcription-regulatory complex could play a role in regulating ER target genes. In our study, although ERE-I is far upstream of the neprilysin promoter and ERE-III is in the ninth intron, it is possible that chromatin loops form between the two EREs, and the neprilysin transcriptional start site in the presence of 17β-estradiol and a similar multipartite transcription-regulatory complex play a role in initiating neprilysin transcription (20, 36).
Last year, Yao et al. (37) found that androgen up-regulates neprilysin gene expression, and their study suggests that androgen treatment could reduce accumulation of Aβ, as a therapeutic approach for AD. This study demonstrates positive regulation of neprilysin expression by 17β-estradiol, leading to Aβ degradation. Undoubtedly, AD may be correlated with the declining level of sex hormones; in fact, depletion of 17β-estradiol has long been regarded as a crucial risk factor for AD in postmenopausal woman. Hence, our data about the regulation mechanism of Aβ and neprilysin by 17β-estradiol may provide a clue to understanding more about the onset of AD and its relation with sex hormones.
However, therapeutic use of estrogen in AD patients could also have adverse effects (such as endometrial carcinoma and breast cancer). The multitarget effect of estrogen may be influenced by distinct expression and differential activities of ERα and ERβ (38). Interestingly, adverse effects of exogenous estrogen tend to involve ERα (39). On the other hand, ERβ activation correlates with stronger anti-tumor activity (40). Some ERβ-selective chemicals (such as genistein) have fewer adverse effects than 17β-estradiol, but confer comparable therapeutic benefits. Most importantly, in cerebral cortex and hippocampus, which are directly related to cognitive behavior and play a greater role in AD pathology (41), ERβ is the predominant expressed ER (42, 43). Results presented here suggest that ERβ independently mediates 17β-estradiol stimulated neprilysin expression (Figs. 2 and 3). These results suggest that ERβ-selective compounds may be extremely valuable agents for Aβ degradation with less adverse effects. Because depletion of 17β-estradiol is an established risk factor for AD in post-menopausal women, the results of this study may have important clinical implications in prevention and/or treatment of AD.
Acknowledgments
We are grateful to Dr. Dan Noonan for generously providing yeast strains and plasmids. We are grateful to Dr. Guo-Min Li (University of Kentucky) for critical reading of the manuscript.
This work was supported by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry of China Grant 2004527, National Basic Science Foundation for Talent Education Grant J0630648, and National Natural Science Foundation of China Grants 30670647 and 30970914.
- AD
- Alzheimer disease
- Aβ
- β-amyloid peptide
- ER
- estrogen receptor
- ChIP
- chromatin immunoprecipitation
- ERE
- estrogen response element
- cERE
- consensus ERE
- PA
- phosphoramidon
- PPT
- propyl pyrazole triol
- DPN
- diarylpropionitrile
- siRNA
- small interfering RNA
- RT
- reverse transcription
- GFP
- green fluorescent protein.
REFERENCES
- 1.Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 2.Tanzi R. E., Bertram L. (2005) Cell 120, 545–555 [DOI] [PubMed] [Google Scholar]
- 3.Martins I. C., Kuperstein I., Wilkinson H., Maes E., Vanbrabant M., Jonckheere W., Van Gelder P., Hartmann D., D'Hooge R., De Strooper B., Schymkowitz J., Rousseau F. (2008) EMBO J. 27, 224–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckman E. A., Eckman C. B. (2005) Biochem. Soc. Trans. 33, 1101–1105 [DOI] [PubMed] [Google Scholar]
- 5.Nalivaeva N. N., Fisk L. R., Belyaev N. D., Turner A. J. (2008) Curr. Alzheimer Res. 5, 212–224 [DOI] [PubMed] [Google Scholar]
- 6.Marr R. A., Guan H., Rockenstein E., Kindy M., Gage F. H., Verma I., Masliah E., Hersh L. B. (2004) J. Mol. Neurosci. 22, 5–11 [DOI] [PubMed] [Google Scholar]
- 7.Iwata N., Tsubuki S., Takaki Y., Shirotani K., Lu B., Gerard N. P., Gerard C., Hama E., Lee H. J., Saido T. C. (2001) Science 292, 1550–1552 [DOI] [PubMed] [Google Scholar]
- 8.Iwata N., Tsubuki S., Takaki Y., Watanabe K., Sekiguchi M., Hosoki E., Kawashima-Morishima M., Lee H. J., Hama E., Sekine-Aizawa Y., Saido T. C. (2000) Nat. Med. 6, 143–150 [DOI] [PubMed] [Google Scholar]
- 9.Wang D. S., Dickson D. W., Malter J. S. (2006) J. Biomed. Biotechnol. 2006, 58406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasojima K., Akiyama H., McGeer E. G., McGeer P. L. (2001) Neurosci. Lett. 297, 97–100 [DOI] [PubMed] [Google Scholar]
- 11.Hellström-Lindahl E., Ravid R., Nordberg A. (2008) Neurobiol. Aging 29, 210–221 [DOI] [PubMed] [Google Scholar]
- 12.Farris W., Schütz S. G., Cirrito J. R., Shankar G. M., Sun X., George A., Leissring M. A., Walsh D. M., Qiu W. Q., Holtzman D. M., Selkoe D. J. (2007) Am. J. Pathol. 171, 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J., Guan H., Booze R. M., Eckman C. B., Hersh L. B. (2004) Neurosci. Lett. 367, 85–87 [DOI] [PubMed] [Google Scholar]
- 14.Xu H., Gouras G. K., Greenfield J. P., Vincent B., Naslund J., Mazzarelli L., Fried G., Jovanovic J. N., Seeger M., Relkin N. R., Liao F., Checler F., Buxbaum J. D., Chait B. T., Thinakaran G., Sisodia S. S., Wang R., Greengard P., Gandy S. (1998) Nat. Med. 4, 447–451 [DOI] [PubMed] [Google Scholar]
- 15.Musatov S., Chen W., Pfaff D. W., Kaplitt M. G., Ogawa S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10456–10460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng H., Jankovic J., Guo Y., Xie W., Le W. (2005) Biochem. Biophys. Res. Commun. 337, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 17.Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 18.Liang K., Yang L., Xiao Z., Huang J. (2009) Mol. Biotechnol. 41, 53–62 [DOI] [PubMed] [Google Scholar]
- 19.Fisk L., Nalivaeva N. N., Boyle J. P., Peers C. S., Turner A. J. (2007) Neurochem. Res. 32, 1741–1748 [DOI] [PubMed] [Google Scholar]
- 20.Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. (2000) Cell 103, 843–852 [DOI] [PubMed] [Google Scholar]
- 21.Bang O. Y., Hong H. S., Kim D. H., Kim H., Boo J. H., Huh K., Mook-Jung I. (2004) Neurobiol. Dis. 16, 21–28 [DOI] [PubMed] [Google Scholar]
- 22.Stauffer S. R., Coletta C. J., Tedesco R., Nishiguchi G., Carlson K., Sun J., Katzenellenbogen B. S., Katzenellenbogen J. A. (2000) J. Med. Chem. 43, 4934–4947 [DOI] [PubMed] [Google Scholar]
- 23.Harrington W. R., Sheng S., Barnett D. H., Petz L. N., Katzenellenbogen J. A., Katzenellenbogen B. S. (2003) Mol. Cell. Endocrinol. 206, 13–22 [DOI] [PubMed] [Google Scholar]
- 24.Marr R. A., Rockenstein E., Mukherjee A., Kindy M. S., Hersh L. B., Gage F. H., Verma I. M., Masliah E. (2003) J. Neurosci. 23, 1992–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H., Bang O. Y., Jung M. W., Ha S. D., Hong H. S., Huh K., Kim S. U., Mook-Jung I. (2001) Neurosci. Lett. 302, 58–62 [DOI] [PubMed] [Google Scholar]
- 26.Zhao L., Wu T. W., Brinton R. D. (2004) Brain Res. 1010, 22–34 [DOI] [PubMed] [Google Scholar]
- 27.Yaffe K., Lui L. Y., Grady D., Stone K., Morin P. (2002) Biol. Psychiatry 51, 677–682 [DOI] [PubMed] [Google Scholar]
- 28.Schupf N., Lee J. H., Wei M., Pang D., Chace C., Cheng R., Zigman W. B., Tycko B., Silverman W. (2008) Dement. Geriatr. Cogn. Disord. 25, 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandi M. L., Becherini L., Gennari L., Racchi M., Bianchetti A., Nacmias B., Sorbi S., Mecocci P., Senin U., Govoni S. (1999) Biochem. Biophys. Res. Commun. 265, 335–338 [DOI] [PubMed] [Google Scholar]
- 30.Pirskanen M., Hiltunen M., Mannermaa A., Helisalmi S., Lehtovirta M., Hänninen T., Soininen H. (2005) Eur. J. Hum. Genet 13, 1000–1006 [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi-Shima N., Yuri K. (2007) Brain Res. 1155, 34–41 [DOI] [PubMed] [Google Scholar]
- 32.Mehra R. D., Sharma K., Nyakas C., Vij U. (2005) Brain Res. 1056, 22–35 [DOI] [PubMed] [Google Scholar]
- 33.Zandi P. P., Carlson M. C., Plassman B. L., Welsh-Bohmer K. A., Mayer L. S., Steffens D. C., Breitner J. C. (2002) JAMA 288, 2123–2129 [DOI] [PubMed] [Google Scholar]
- 34.Björnström L., Sjöberg M. (2005) Mol. Endocrinol. 19, 833–842 [DOI] [PubMed] [Google Scholar]
- 35.Deschênes J., Bourdeau V., White J. H., Mader S. (2007) J. Biol. Chem. 282, 17335–17339 [DOI] [PubMed] [Google Scholar]
- 36.Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 37.Yao M., Nguyen T. V., Rosario E. R., Ramsden M., Pike C. J. (2008) J. Neurochem. 105, 2477–2988 [DOI] [PubMed] [Google Scholar]
- 38.Pettersson K., Gustafsson J. A. (2001) Annu Rev. Physiol. 63, 165–192 [DOI] [PubMed] [Google Scholar]
- 39.Sun M., Paciga J. E., Feldman R. I., Yuan Z., Coppola D., Lu Y. Y., Shelley S. A., Nicosia S. V., Cheng J. Q. (2001) Cancer Res. 61, 5985–5991 [PubMed] [Google Scholar]
- 40.Treeck O., Pfeiler G., Mitter D., Lattrich C., Piendl G., Ortmann O. (2007) J. Endocrinol. 193, 421–433 [DOI] [PubMed] [Google Scholar]
- 41.Rissman E. F., Heck A. L., Leonard J. E., Shupnik M. A., Gustafsson J. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3996–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shughrue P. J., Lane M. V., Merchenthaler I. (1997) J. Comp. Neurol. 388, 507–525 [DOI] [PubMed] [Google Scholar]
- 43.Shughrue P. J., Scrimo P. J., Merchenthaler I. (1998) Endocrinology 139, 5267–5270 [DOI] [PubMed] [Google Scholar]