Abstract
Although the biogenesis of ribosomal subunits occurs predominantly in the nucleus, final remodeling steps take place in the cytosol. One cytosolic step has two components: 1) the removal of the maturation factor Arx1, which transits from the nucleus to the cytosol with the pre-60 S subunit, and 2) its subsequent transport back into the nucleus. Two cytosolic proteins, Rei1 and Jjj1, are required, but their individual contributions to this step are not understood. Here we report that Rei1 and Jjj1 directly interact. This interaction is mediated by a C-terminal segment of Jjj1 encompassing a region rich in charged residues, flanked by C2H2-type zinc fingers. Deletion of the charged region results in defects in 60 S subunit biogenesis in vivo. In addition, we report resolution of an apparent contradiction in the literature regarding the association of Arx1 with the pre-60 S subunit in the absence of Rei1. The association of Arx1 with ribosomes is sensitive to the concentration of magnesium ions when Rei1 is absent. At near physiological concentrations, Arx1 remains associated with the pre-60 S particle, as it does in the absence of Jjj1; at higher concentrations, Arx1 dissociates in the absence of Rei1 but not in the absence of Jjj1. As both Rei1 and Jjj1 are required for dissociation of Arx1 from the pre-60 S subunit, and the region of Jjj1 that mediates interaction with Rei1 is required in vivo for 60 S subunit biogenesis, our data support the idea that the primary role of both Rei1 and Jjj1 is the first step of the Arx1 removal/recycling process.
Keywords: Arx1, Hsp40, J-protein, Jjj1, Rei1, Molecular Chaperone, Protein synthesis, Ribosome Biogenesis
Introduction
The eukaryotic ribosome, the site of protein synthesis, is a remarkably complex machine. The 60 S subunit, the larger of the two that compose a ribosome, consists of the 25 S, 5.8 S, and 5 S rRNAs, as well as ∼49 r-proteins (1, 2). Given this degree of structural complexity, it is not surprising that the process of ribosome biosynthesis is very intricate, involving the correct assembly and orientation of these components to form a properly functioning ribosome (1). In addition to the structural components that make up the mature ribosome, multiple auxiliary or maturation factors are required in the biogenesis process. In the yeast Saccharomyces cerevisiae, over 80 such factors have been identified for the 60 S subunit alone (1–4). Most of these factors associate with and dissociate from the pre-60 S particle in the nucleus. However, several, including Arx1, Alb1, Tif6, Nmd3, and Rlp24, are transported with the particle to the cytosol (5–9). These “shuttling” factors must subsequently be removed and recycled back into the nucleus to allow them to participate in further rounds of biogenesis (2). Most relevant to this report are Arx1 and Alb1, which form a stable heterodimer, with Arx1 serving to tether the complex to the ribosome (10). Although the role of Alb1 is unknown, Arx1 has recently been reported to act as a nuclear export receptor for the ribosome, facilitating transport of the pre-60 S subunit through nuclear pore complexes via interaction with FG repeats of nucleoporin proteins (10, 11).
Several cytosolic proteins have been shown to be important for the release and/or recycling to the nucleus of particular pre-60 S biogenesis factors (8, 9, 12–15). In the case of the Arx1/Alb1 heterodimer, Rei1 and Jjj1 are required. In wild-type (WT)3 cells, the majority of Arx1/Alb1 complexes are associated with pre-60 S particles in the nucleus, consistent with efficient cytosolic dissociation and transport back through the nuclear membrane (8). However, in the absence of either Jjj1 or Rei1, Arx1 and Alb1 accumulate in the cytosol (8, 9, 14). Δrei1 and Δjjj1 cells, as well as Δrei1 Δjjj1 cells lacking both proteins, grow poorly at low temperatures, with Δjjj1 growing slightly better than Δrei1 Δjjj1 and Δrei1, which grow very similarly (14, 16). Other phenotypes of Δjjj1 and Δrei1 are very similar as well, and all are hallmarks of dysfunctional 60 S ribosomal subunit biogenesis. Both strains accumulate half-mer ribosomes (polysomes having one 40 S subunit lacking a 60 S subunit partner (17)) and have a reduced total level of 60 S subunits (8, 9, 14, 16, 18). In addition, deletion of ARX1 suppresses the cold sensitive growth phenotype of both Δrei1 and Δjjj1 strains, suggesting that the failure of Arx1 to dissociate or recycle to the nucleus from the cytosol is more detrimental than the complete absence of the Arx1 protein (8, 9, 14).
Although little information is available regarding Rei1, Jjj1 is known to be a member of a class of proteins called J-proteins, which work in concert with Hsp70-type molecular chaperones. Hsp70/J-protein chaperone machineries participate in multiple cellular processes including protein folding, protein import into organelles, and remodeling of protein-protein complexes (19). Apart from the highly conserved 70-amino acid-long J-domain, which is required for cooperation with the Hsp70, many J-proteins have little in common. Previously, we have shown that the J-domain of Jjj1 is required for its function in ribosome biogenesis and that Jjj1 cooperates with the major cytosolic Hsp70, Ssa (14). The C terminus of Jjj1, however, is unique among J-proteins of S. cerevisiae. This 286-amino acid-long region contains two C2H2-type zinc fingers that flank a 168-amino acid-long region rich in charged residues, the function of which is unknown (14).
There is general acceptance that both Rei1 and Jjj1 are important factors in 60 S subunit biogenesis, specifically affecting the dynamics of the cytosolic interactions of Arx1. However, the roles the two proteins play and the functional relationship between them are not understood. In particular, a contradiction exists in the literature concerning the effect of the absence of Rei1 on Arx1/Alb1 recycling. On one hand, it has been reported that Arx1 and Alb1 accumulate in a small cytoplasmic complex when Rei1 is absent (8, 16). On the other hand, results of a different study indicate that Arx1 remains associated with the pre-60 S subunit in a Δrei1 strain (9). However, there is agreement that the heterodimer remains ribosome-associated with pre-60 S subunits in the cytosol in the absence of Jjj1. Here we report that Arx1 remains associated with pre-60 S ribosomes in cell lysates made from either Δrei1 or Δjjj1 strains when physiological conditions are mimicked by maintaining low concentrations of magnesium ions. In combination with our demonstration of a direct physical interaction between Rei1 and Jjj1, our data support the hypothesis that Rei1 and the Jjj1-Ssa chaperone system work together to facilitate the release of Arx1/Alb1 from the pre-60 S subunit.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Genetic Techniques
REI1 was obtained by PCR amplification of genomic DNA from positions −232 to 1446 using Pfu Turbo polymerase (Stratagene, La Jolla, CA) and cloned into the HindIII and XbaI sites in pRS315 (20). All REI1 variants were generated by QuikChange mutagenesis (Stratagene) in this same vector. To create HA-REI1 WT and variants, three tandem copies encoding the hemagglutinin tag were inserted after the start codon in pRS315 REI1.
To obtain pRS316 jjj1Δ363–534, amino acids 363–534 were removed from pRS316 JJJ1 (14) by QuikChange mutagenesis (Stratagene). To create the HA-tagged version, three tandem copies encoding the hemagglutinin tag were then inserted prior to the stop codon. For purification of Jjj1-His6, JJJ1 followed by six CAC codons was isolated by PCR amplification and cloned into the NdeI and BamHI sites of pET3a (Novagen). Variants used for analysis of binding to Rei1 were generated by subsequent QuikChange mutagenesis (Stratagene) on pET3a JJJ1 His6. For purification of MBP-Rei1, the coding sequence of Rei1 was cloned into the XbaI and HindIII sites of the pMAL-HIS-TEV vector, a gift from A. W. Johnson.
Null alleles of JJJ1 and REI1 were obtained in DS10 (GAL2 his3-11,15 leu2-3112 lys1 lys2 Δtrp1 ura3-52) as described previously (14). To generate Δjjj1 Δrei1, haploid strains of Δrei1 and Δjjj1 were mated and sporulated. Strains having ARX1 tagged with three tandem copies of the HA epitope or GFP inserted in the chromosome at the ARX1 locus were made by homologous recombination in Δjjj1 or Δrei1 (21).
Analysis of Cell Extracts
Lysate preparation and sucrose gradient centrifugation for polysome analysis were as follows. For analysis of Arx1-HA in Δrei1, 1 liter of culture was grown in yeast peptone dextrose (YPD) medium at 23 °C to an optical density of 0.5–1 at 600 nm. Cells were treated with 100 μg/ml cycloheximide and harvested by centrifugation at 4 °C. The cells were then washed with 14 ml of ice-cold Buffer I (20 mm Tris-HCl (pH 7.5), 50 mm KCl/5 mm MgCl2)4 or II (10 mm Tris-HCl (pH 7.4)/100 mm NaCl/30 mm MgCl2) (8, 16) and centrifuged at 4,000 × g for 5 min and resuspended in 3.5 ml of Buffer I or II plus 140 units of rNAsin (Promega, Madison, WI). In some experiments, as indicated in text, MgCl2 concentrations were changed. Lysis of cells and clearing of lysate were performed as described (22). Ten A254 units of cleared lysate were layered on 10-ml 5–50% sucrose gradients prepared in Buffer I or II containing the appropriate concentration of MgCl2. Gradients were centrifuged in an SW40 Ti rotor at 4 °C at 40,000 rpm for 4.5 h. Fractions of 0.6 ml were collected, and proteins were precipitated with 86% acetone overnight at −20 °C before immunoblot analysis. For analysis of WT Jjj1-HA and Jjj1Δ363–534-HA, as well as Jjj1 and HA-Rei1 in Δrei1 and Δjjj1 strains, cells were grown at 23 °C in medium lacking uracil or leucine, unless noted otherwise, and lysis was done in Buffer I containing 5 mm MgCl2. Gradients were performed essentially as described (22). For analysis of HA-Rei1 WT and mutant levels, as well as WT Jjj1-HA and Jjj1Δ363–534-HA levels, cells were grown at 23 °C in medium lacking leucine or uracil, respectively, and lysis was performed in Buffer I as above. Lysate was examined for the presence of Jjj1 or Rei1 by anti-HA Western blotting.
Rei1 and Jjj1 Purification
Preparation of WT Jjj1 and variants was essentially as described (14). For Rei1, pMAL-HIS-TEV REI1 was transformed into BL21 pLysS cells lacking DnaK and DnaJ for protein expression. Proteins were induced at 30 °C with the addition of 1 mm isopropyl-β-d-thiogalactopyranoside at an A600 of 0.5 and incubated for an additional 4 h. The cell lysates were prepared using a French press, and purification was performed following the His tag protein purification protocols from Novagen.
Jjj1-Rei1 Interaction
Pulldown assays using purified recombinant Jjj1 and Rei1 proteins were performed by incubating purified MBP-Rei1 in 100 μl of binding buffer (300 mm sorbitol, 20 mm HEPES (pH 7.5), 5 mm MgCl2, 300 mm KCl, 10% glycerol), 0.5% bovine serum albumin, and 20 μl of a 50:50 slurry of equilibrated amylose resin for 4 h, rotating, at 4 °C. The resin was washed twice with 1 ml of binding buffer, and the Rei1-bound resin was then incubated with purified Jjj1-His6, again in 100 μl of buffer with 0.5% bovine serum albumin for 1 h, rotating, at 4 °C. The resin was then washed three times with 1 ml of binding buffer, and proteins bound to the beads were loaded on an SDS-polyacrylamide gel and analyzed by Western blotting. For binding curves, immunoblot signals were quantified by densitometry and plotted against Rei1 concentrations. All curves were plotted in GraphPad Prism (GraphPad Software Inc., San Diego CA) using a single binding hyperbola to fit data, except that of Jjj14C-4A was fit by linear regression.
Other Methods
For microscopy, overnight cultures were diluted into fresh medium to an A600 of 0.1 and cultured at 23 °C to mid-log phase. Fluorescence was visualized on a Nikon Eclipse E800 microscope fitted with a ×60 objective and MetaMorph software (GE Healthcare). Images were captured using a Photometrics CoolSnap HQ camera (Roper Scientific, Tuscon AZ). Immunoblot detection was carried out by using the ECL system (Amersham Biosciences) according to the manufacturer's suggestion. Purified maltose-binding protein (MBP), anti-MBP antisera, and amylose resin were purchased from New England Biolabs (Ipswich, MA). The anti-Jjj1 antibody was produced as described (14). The anti-HA antibody was purchased from Covance (Denver, PA). All chemicals were obtained from Sigma unless otherwise stated.
RESULTS
Rei1 Directly Interacts with Jjj1
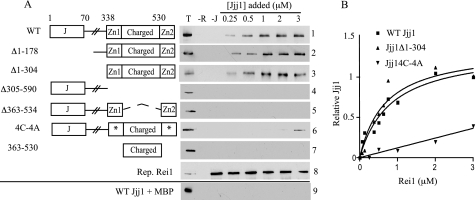
As a step toward a better understanding of how Rei1 and Jjj1 function in ribosome biogenesis, we asked whether they physically interact, following up on a report of a two-hybrid interaction (16). To this end, we first purified full-length Rei1 and Jjj1 fused to MBP and a 6-residue histidine tag, respectively. We then proceeded to develop a binding assay. MBP-Rei1 or, as a control, MBP alone, was prebound to amylose resin prior to addition of various concentrations of Jjj1. Jjj1 was retained by the MBP-Rei1-bound resin at all concentrations tested (Fig. 1A, panel 1). Binding was saturable as the amount of Jjj1 retained was similar at the three highest concentrations tested (Fig. 1A). Jjj1 binding was not detected when only MBP was bound to the resin, indicating that the observed interaction between Rei1 and Jjj1 was specific (Fig. 1A, panel 9). We conclude that Rei1 and Jjj1 can directly interact in the absence of other cellular components.
FIGURE 1.
Jjj1 interacts directly with Rei1. A, to assay Jjj1-Rei1 binding, maltose-binding protein-tagged Rei1 (MBP-Rei1) was prebound to amylose resin and then incubated with the indicated concentrations of WT Jjj1 (panel 1) or Jjj1 variants (panels 2–7). The J domain (J), zinc fingers (Zn1 and Zn2), and charged region, which are designated in the adjacent diagrams, are described more fully under “Results.” Asterisks indicate zinc finger 1 and 2 cysteine to alanine alterations. After pelleting of the resin, Jjj1 binding was assessed by immunoblotting using anti-Jjj1 antibody. Three control lanes were included in each panel: 0.25 μm of Jjj1 protein (T); resin with no Rei1 added (−R); and resin with no Jjj1 added (−J). In addition, a representative blot of MBP-Rei1 was incubated with MBP-specific antibody (panel 8), and MBP alone was prebound to resin and incubated with WT Jjj1 (panel 9). B, concentrations of MBP-Rei1 ranging from 0.06 to 3 μm were incubated with amylose resin prior to incubation with 1 μm WT Jjj1, Jjj1Δ1–304, or Jjj14C-4A. Bound Jjj1 was quantified by densitometry. The amount of WT Jjj1 pulled down by 3 μm Rei1 was set as 1. Values were plotted in GraphPad Prism using a single binding hyperbola to fit data obtained for WT Jjj1 and Jjj1Δ1–304 and linear regression to fit Jjj14C-4A.
Amino Acids 305–590 of Jjj1, Encompassing the Zinc Fingers and Charged Region, Are Sufficient for Interaction with Rei1
Having established that Jjj1 and Rei1 directly interact, we sought to determine what regions of the two proteins are important for this interaction. To this end, we attempted to purify variants of both Jjj1 and Rei1. We were unsuccessful in purification of Rei1 variants. However, we were able to isolate and test several Jjj1 variants. First, we tested two N-terminal truncations, one lacking the first 178 amino acids, Jjj1Δ1–178, and a second lacking the first 304 amino acids, Jjj1Δ1–304. Both bound Rei1 very similarly to full-length Jjj1 (Fig. 1A, panels 2 and 3). However, we observed no binding with a C-terminal truncation lacking residues 305–590, Jjj1Δ305–590 (Fig. 1A, panel 4).
These results suggest that the C-terminal 286 residues are necessary and sufficient for binding of Jjj1 to Rei1. To better understand what sequences within this region play a role in binding, we proceeded to isolate and test additional Jjj1 constructs, focusing on the two C2H2-type zinc finger motifs and the region of highly charged amino acids that they flank. We were able to purify two Jjj1 C-terminal variants. One, Jjj1Δ363–534, lacked the charged region. The second bore alanine residues in place of the conserved cysteines in each zinc finger, at positions 340, 343, 551, and 554. This variant is referred to as Jjj14C-4A throughout. No binding of Jjj1Δ363–534 to Rei1 was detected, suggesting that the charged region of Jjj1 is important for interaction with Rei1 (Fig. 1A, panel 5). Binding of Jjj14C-4A was observed, but only at the two highest concentrations tested, thus implicating the zinc fingers in binding as well (Fig. 1A, panel 6). We also tested a fragment consisting of the charged region alone, Jjj1363–530. Consistent with a role for the zinc fingers in facilitating the Jjj1-Rei1 interaction, no binding of this fragment was detected (Fig. 1A, panel 7).
To extend and confirm these results, we assessed binding again, this time keeping the concentration of Jjj1 constant while varying Rei1 levels. Results similar to those reported above were obtained with full-length Jjj1 and Jjj1Δ1–304. Association of both approached saturation, with 50% of observed maximal binding obtained at ∼0.5 μm (Fig. 1B). Binding of Jjj14C-4A was again detectable but did not reach saturation, with only ∼40% of the maximum binding obtained with full-length WT protein or the Jjj1Δ1–304 fragment observed at the highest concentration tested (Fig. 1B). Together, our results suggest that the 286-amino acid-long fragment containing the zinc fingers and the charged region of Jjj1 is necessary and sufficient for mediating the Jjj1-Rei1 interaction, with both features being important for an efficient interaction.
The Region of Jjj1 Sufficient for Interaction with Rei1 Is Important for 60 S Subunit Biogenesis
Amino acids 305–590 of Jjj1 are sufficient for its interaction with Rei1, but is this interaction important in vivo for biogenesis of the 60 S ribosomal subunit? To address this question, we assessed strains expressing alterations in the C-terminal region of Jjj1 for slow growth at low temperatures, mislocalization of Arx1, and the presence of half-mer ribosomes, three hallmarks of defects in ribosome biogenesis.
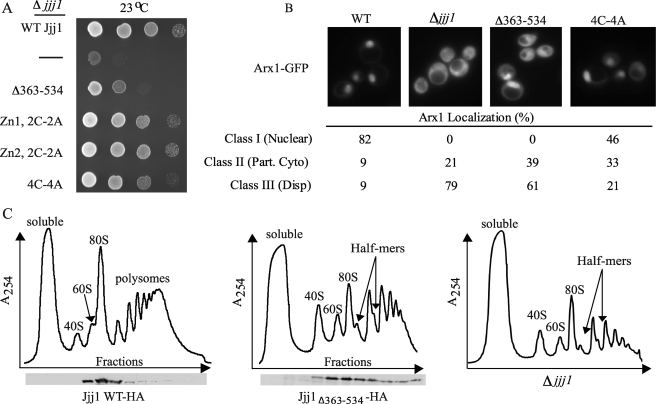
We first tested the ability of C-terminal Jjj1 variants to rescue the growth defect of Δjjj1 cells at 23 °C. Jjj1 having cysteine to alanine substitutions in one zinc finger rescued Δjjj1 as well as the WT protein, whereas cells expressing Jjj1 having alterations in both, Jjj14C-4A, exhibited a slight growth defect (Fig. 2A). However, growth of cells expressing Jjj1Δ363–534 was significantly compromised (Fig. 2A). To assure that the phenotype observed was not due to a low level of expression, we wanted to monitor protein expression levels. As our Jjj1-specific antibody would not be expected to recognize Jjj1Δ363–534 and full-length protein equally, both Jjj1Δ363–534 and full-length Jjj1 were tagged with HA. We found that WT and variant proteins were expressed at similar levels and that cells expressing the HA-tagged Jjj1 variants grew at a rate similar to those expressing untagged proteins (supplemental Fig. S1, data not shown). Thus, we conclude that the poor growth of jjj1Δ363–534 cells is not due to low levels of expression of the deletion variant.
FIGURE 2.
The region of Jjj1 required for interaction with Rei1 is necessary for efficient ribosome biogenesis. A, 10-fold serial dilutions of Δjjj1 cells carrying WT JJJ1, the indicated JJJ1 mutant gene, or no JJJ1 gene (−) on a plasmid were spotted on a minimal medium plate and incubated at 23 °C for 2 days. B, Δjjj1 cells expressing Arx1-GFP and Jjj1 (WT), no Jjj1 (−), or the indicated variants were grown to an A600 of 0.3–0.6 at 23 °C prior to imaging. Upper, representative images showing the Arx1-GFP localization. Lower, the percentages of cells with Class I, II, or III Arx1-GFP localization are shown. A minimum of 100 cells was counted for each. Part. Cyto, partial cytosolic localization; Disp, dispersed localization. C, Δjjj1 cells expressing WT Jjj1-HA, Jjj1Δ363–534-HA, or no Jjj1 (Δjjj1) were grown at 23 °C and lysed in Buffer I. Ten A254 units of lysate were centrifuged through a 5–50% sucrose gradient. Upper, the A254 was monitored and plotted versus the time course of fraction collection. The positions of 40 S, 60 S, 80 S, polysomes, and half-mer migration are labeled. Lower, fractions were analyzed for the presence of Jjj1Δ363–534-HA and WT Jjj1-HA by immunoblot analysis using HA-specific antibody.
Next, we monitored the localization of Arx1, concentrating on the two mutants having growth defects, jjj1Δ363–534 and jjj14C-4A. Making use of an Arx1-GFP fusion protein, we microscopically observed the distribution of the fluorescent signal between the nucleus and cytosol. Each cell observed was placed into one of three classes: Class I, predominantly nuclear signal; Class II, obvious cytosolic signal but with a more intense nuclear signal; and Class III, dispersed fluorescence, with approximately equal intensity in the nucleus and cytoplasm. As expected (8, 9), the vast majority of WT cells, 82%, showed a predominantly nuclear Arx1-GFP signal and were placed in Class I, whereas the majority of Δjjj1 cells, 79%, showed disperse cytosolic fluorescence and were placed in Class III (Fig. 2B). Although a significant portion (46%) of jjj14C-4A cells were classified as belonging to Class I, 33% showed partial cytosolic localization (Class II) and 21% showed equal cytosolic and nuclear localization (Class III) (Fig. 2B). Thus, Jjj14C-4A cells have a significant defect in Arx1 localization. jjj1Δ363–534 cells displayed even more cytosolic mislocalization of Arx1-GFP than did jjj14C-4Acells, with 39 and 61% of cells placed in Class II and Class III, respectively (Fig. 2B).
To assess the presence of half-mers, extracts from WT cells and cells expressing Jjj1Δ363–534-HA were analyzed by centrifugation through sucrose gradients to separate species of ribosomes and their subunits. In addition to an accumulation of half-mer ribosomes, a decrease in the level of free 60 S subunits when compared with 40 S subunits at 23 °C was apparent in jjj1Δ363–534-HA cells when compared with WT cells (Fig. 2C). Both the presence of half-mers and the decrease in 60 S subunits are typically observed in cells having defects in 60 S subunit biogenesis. Thus, by all three criteria, we found Jjj1Δ363–534 to be deficient in promoting 60 S subunit biogenesis; Jjj14C-4A was less so, but defects in growth at low temperatures and Arx1 localization were observed. We reasoned that a combination of the charged deletion and point mutations of the cysteines in one or both of the zinc fingers might lead to an enhanced defect as the zinc fingers contribute to Rei1 binding in vitro. Unfortunately, these mutant proteins were not stably expressed.
We note two additional observations regarding Jjj1. First, similar to WT Jjj1, Jjj1Δ363–534-HA co-migrated with the 60 S subunit in the sucrose gradient, suggesting that the charged region of Jjj1 is not simply required for anchoring Jjj1 to the pre-60 S particle (Fig. 2C). Also, we found that a small percentage of Jjj1Δ363–534-HA migrated farther into the gradient, implying that ribosomes having Jjj1 associated are able to participate in protein synthesis, at least at some level. In sum, the data described above are consistent with the hypothesis that the interaction between Jjj1 and Rei1 is important for proper 60 S subunit maturation in vivo.
The Jjj1-Rei1 Interaction Is Not Required for Ribosome Association of Either Protein
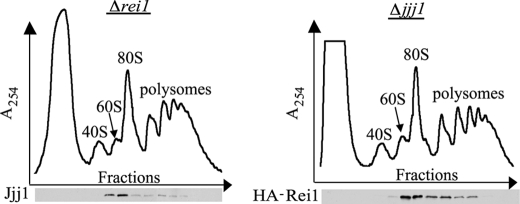
Having established that Rei1 and Jjj1 physically interact, we wanted to begin to address how Rei1 and Jjj1 function and the importance of the physical interaction between them. First, we asked whether Jjj1 is required for binding of Rei1 to the ribosome, or vice versa. Sucrose gradient analysis revealed that Jjj1 and Rei1 co-migrated with ribosomes in the absence of Rei1 or Jjj1, respectively (Fig. 3). This lack of interdependence for tethering to ribosomal particles indicates that direct interaction between Jjj1 and Rei1 is not important for recruitment Rei1 or Jjj1 to the 60 S subunit. Rather it points to the idea that their interaction may be important for their role in ribosome biogenesis per se.
FIGURE 3.
Neither Jjj1 nor Rei1 is required for tethering of its binding partner to the ribosome. Δrei1 and Δjjj1 cells, grown at 30 °C, were lysed in Buffer I. Ten A254 units were centrifuged through 5–50% sucrose gradients. Upper, the A254 was monitored and plotted versus the time course of fraction collection. The positions of 40 S, 60 S, 80 S, and polysomes are labeled. Lower, fractions were tested by immunoblot analysis for the presence of Jjj1 in fractions from Δrei1 lysates (left) or of HA-tagged Rei1 in Δjjj1 lysates (right). Antibodies specific for Jjj1 and HA, respectively, were used.
Arx1 Remains Associated with the Pre-60 S in the Absence of Rei1
The results described above indicate not only that the Rei1-Jjj1 interaction is biologically relevant but also suggest that the two proteins are functionally coupled. However, the recently reported observation that Arx1 and Alb1 accumulate in a small cytoplasmic complex in the absence of REI1, whereas the heterodimer remains ribosome-bound in the absence of JJJ1 (8, 16), suggests a critical distinction between the function of Rei1 and that of Jjj1. As it has also been reported that Arx1 remains associated with the pre-60 S subunit in a Δrei1 strain (9), we sought to uncover the reason behind this apparent discrepancy.
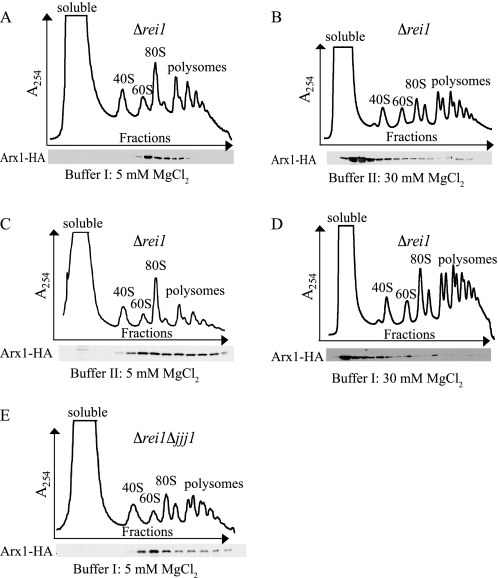
We used sucrose gradient centrifugation to separate particles in cell lysates, employing a Δrei1 strain containing chromosomally integrated, C-terminally HA-tagged Arx1 to allow for detection of Arx1. Cell lysis and centrifugation were carried out in the buffers used in the previous studies. In the case of the experiments reported by Hung and Johnson (9), cells were lysed in 20 mm Tris, pH 7.5, 50 mm KCl, and 5 mm MgCl2; the lysis buffer employed by Lebreton et al. (8) and Demoinet et al. (16) consisted of 10 mm Tris-HCl, pH 7.4, 100 mm NaCl, and 30 mm MgCl2. For simplicity, we refer to these as Buffer I and II, respectively. We were able to reproduce both results. When cells were lysed in Buffer I, Arx1-HA predominantly co-migrated with 60 S subunits, consistent with continued association of Arx1 with the ribosome (Fig. 4A). When cells were lysed in Buffer II, Arx1-HA accumulated near the top of the gradient, consistent with Arx1 being part of the previously described small complex (8, 16) (Fig. 4B). To determine which component(s) contributed to the differences in ribosome association of Arx1, we varied the concentrations of each component of Buffer II individually. Lowering the concentration of NaCl to 50 mm, changing the salt from KCl to NaCl, or increasing the concentration of Tris-HCl did not result in sedimentation of Arx1-HA at the 60 S position (data not shown). However, when the MgCl2 concentration was lowered from 30 to 5 mm, the majority of Arx1-HA co-migrated with free 60 S subunits, suggesting that this protein remained associated with the 60 S subunit under these conditions (Fig. 4C). Consistent with this conclusion, increasing the concentration of MgCl2 in Buffer I resulted in accumulation of Arx1-HA near the top of the gradient when Δrei1 extracts were analyzed (Fig. 4D).
FIGURE 4.
Arx1 remains 60 S-associated in the absence of REI1. Lysates from 23 °C-grown Δrei1 (A–D) or Δrei1 Δjjj1 (E) cells containing chromosomally integrated ARX1-HA were centrifuged through 5–50% sucrose gradients, and the A254 was monitored versus the time course of fraction collection (upper). Fractions were analyzed for the presence of Arx1-HA by immunoblotting (lower). Cells were lysed and centrifuged in the following conditions: A and E, Buffer I containing 5 mm MgCl2; B, Buffer II in the presence of 30 mm MgCl2; C, Buffer II in the presence of 5 mm MgCl2; and D, Buffer I in the presence of 30 mm MgCl2. Note: The small peak on the top of the gradient side of the 40 S peak in B and D is not consistently observed.
On the basis of the results of the experiments described above, we conclude that differences in MgCl2 concentration in the lysis and gradient buffers were responsible for the apparent discrepancy in Arx1 ribosome association in the absence of Rei1. At a concentration of 5 mm, Arx1 is associated; at 30 mm, it is not. The concentration of Mg2+ in the cytosol has been reported to be between 1 and 3 mm (23). For this reason and because magnesium ions are known to significantly affect ribosome structure, we favor the idea that Arx1 remains associated with 60 S subunits in vivo in the absence of either Jjj1 or Rei1 but that high concentrations of Mg2+ during lysis destabilize Arx1 if Rei1 is absent.
Cold Sensitivity of Δrei1 Correlates with Dysfunctional Ribosome Biogenesis
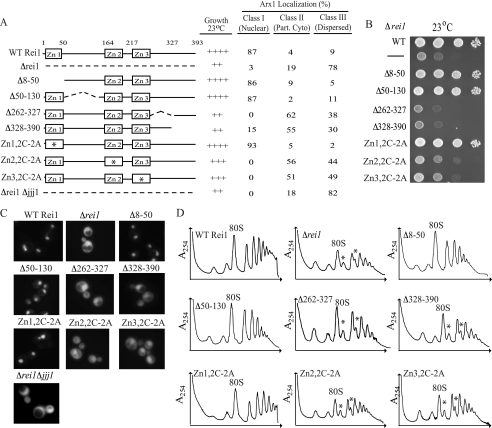
We decided to carry out a mutagenic analysis of REI1, with a goal of determining what regions of Rei1 were critical for its in vivo function and whether the growth defects correlated with effects on ribosome biogenesis. We took two approaches: 1) constructing deletion mutants to remove ∼60-amino acid-long segments of Rei1 and 2) making amino acid substitutions, changing the cysteine residues in the zinc finger motifs to alanines. The deletion mutants constructed that resulted in stable Rei1 variants lacked amino acids 8–50, 50–130, 262–327, or 328–390 (Fig. 5A). rei1Δ8–50 and rei1Δ50–130 cells grew as well as WT cells, whereas rei1Δ262–327 and rei1Δ328–390 grew slowly at 23 °C, similar to cells carrying a complete deletion of REI1, despite being expressed to greater levels than the WT protein (Fig. 5B and supplemental Fig. S2A). To determine whether the function of Rei1 in ribosome biogenesis was affected in these mutants, we monitored the localization of Arx1, again making use of an Arx1-GFP fusion. As in the previous experiment, the vast majority, 87%, of WT cells displayed nuclear fluorescence and were thus placed in Class I (Fig. 5, A and C). Again, as expected, Δrei1 cells displayed an increase in cytosolic fluorescence; 19 and 78% of the cells were placed into Classes II and III, respectively (Fig. 5, A and C). As expected, if slow growth is indicative of defects in ribosome biogenesis, Arx1-GFP was localized predominantly to the nucleus in rei1Δ8–50 and rei1Δ50–130, with 86 and 87%, respectively, falling into Class I (Fig. 5, A and C). On the other hand, fluorescence was detected in the cytoplasm in cells expressing Rei1Δ262–327 and Rei1Δ328–390, the two strains that were compromised for growth at low temperatures, with between 85 and 100% of cells categorized as either Class II or Class III (Fig. 5, A and C).
FIGURE 5.
Zinc fingers 2 and 3, as well as residues 262–390 are important for the role of Rei1 in ribosome biogenesis. A, summary of Rei1 variants tested (left). Asterisks indicate zinc finger (Zn) in which the cysteine residues were altered to alanines. Center, growth observed for each at 23 °C; right, the percentages of cells with Class I, II, or III Arx1-GFP localization. A minimum of 100 cells was counted for each variant. Part. Cyto, partial cytosolic localization. B, 10-fold serial dilutions of cells were spotted on a minimal medium plate and incubated at 23 °C for 3 days. Data are summarized in A. C, representative images showing the Arx1-GFP localization resulting from expression of each Rei1 variant tested, as well as in controls: cells expressing WT Rei1, no Rei1 variant (Δrei1), and cells lacking both Rei1 and Jjj1 (Δrei1 Δjjj1). Cells were grown to an A600 of 0.3–0.6 at 23 °C prior to imaging. Quantification is shown in A. D, 10 A254 units of lysate from cells grown at 30 °C were centrifuged through 5–50% sucrose gradients in Buffer I containing 5 mm MgCl2, and the A254 was monitored. Strains analyzed are as follows: Δrei1 expressing WT Rei1 or the indicated Rei1 variants; no Rei1 (Δrei1). Asterisks denote half-mers.
As we were unable to stably express variants deleted for amino acids 131–262, we decided to individually alter the two putative C2H2-type zinc fingers that are found within this region: Zn2, residues 162–187, and Zn3, residues 211–233. In addition, we also altered zinc finger 1, Zn1, which is found within amino acids 9–31. As expected, based on the WT growth levels observed for rei1Δ8–50, alteration of the two cysteine residues of Zn1 did not result in slow growth at 23 °C or in cytosolic accumulation of Arx1-GFP (Fig. 5, A–C). However, despite being expressed at levels similar to that of WT Rei1, cells expressing variants having substitutions in either Zn2 or Zn3 grew more slowly at 23 °C than did WT cells but slightly faster than cells completely lacking Rei1 (Fig. 5B and supplemental Fig. S2A). Alteration of Zn2 or Zn3 also resulted in mislocalization of Arx1-GFP to the cytosol. Like Δrei1 cells, few if any cells had predominantly nuclear fluorescence, indicating a role of zinc fingers 2 and 3 of Rei1 in ribosome biogenesis (Fig. 5, A and C). Similar results were obtained when cells expressing alterations of both Zn2 and Zn3 were analyzed, suggesting that the deleterious effects of Zn2 and Zn3 alteration are not additive (data not shown).
In addition to the defects in Arx1-GFP recycling discussed above, expression of Rei1Δ262–327, Rei1Δ328–390, and both zinc fingers mutants resulted in accumulation of half-mer ribosomes, further underlying the importance of these amino acids in ribosome biogenesis (Fig. 5D). Thus, overall, our results are consistent with slow growth of REI1 mutant strains being predominantly caused by disruption of ribosome biogenesis as a positive correlation was found between the growth phenotype, the mislocalization of Arx1, and the accumulation of half-mer ribosomes.
DISCUSSION
Several recent reports (8, 9, 14, 16) established that Rei1 and Jjj1 are necessary for one of the final cytosolic steps in the biogenesis of the 60 S ribosomal subunit: the dissociation of the Arx1/Alb1 heterodimer and its subsequent recycling to the nucleus. However, whether both Rei1 and Jjj1 function together by facilitating the dissociation of the heterodimer from the pre-60 S subunit or instead act sequentially, Jjj1 in dissociation of the heterodimer and Rei1 in its subsequent transport into the nucleus, has been a matter of debate. The data reported here favor a model in which the primary role of Jjj1 and Rei1 is to effect the physical dissociation of Arx1/Alb1 from the pre-60 S particle and in which their physical interaction is important for their cooperation in that process.
Both Rei1 and Jjj1 Function in the Dissociation of Arx1 from the Pre-60 S Subunit
The main piece of data that led to the model purporting that Jjj1 and Rei1 function in separate steps of the pathway (e.g. the dissociation of Arx1 from the pre-60 S particle or its transport back into the nucleus) was the observation that Arx1 remained associated with 60 S particles in the absence of Jjj1 but not in the absence of Rei1 (16). However, we found that the migration of Arx1 in Δrei1 cells was dependent on the concentration of MgCl2 used in the lysis or gradient buffer. At 5 mm MgCl2, Arx1 co-migrated with 60 S; at 30 mm MgCl2, Arx1 was near the top of the gradient. As effects of chloride ion levels were ruled out, we conclude that the observed result is due to the high concentration of magnesium ions. Because the total cytosolic concentration of Mg2+ is 1–3 mm (23), the lower, 5 mm concentration more closely mimics physiological conditions.
For decades, it has been known that magnesium ions bind ribosomal subunits (24–29) through interactions with the negatively charged rRNA backbone and, in this manner, facilitate structural alterations in the ribosome (24). Thus, it is easy to envision that high Mg2+ concentrations in cell lysis/gradient buffers might alter the pre-60 S subunit in such a way that the Arx1-pre-60 S subunit interaction is destabilized. This explanation requires that a difference in structure exists between pre-60 S particles from Δjjj1 and Δrei1 cells such that the Arx1 interaction is sensitive to high Mg2+ concentrations in the absence of Rei1 but not in the absence of Jjj1. Testing of such an idea awaits a better molecular understanding of pre-60 S subunit structure. However, that Δrei1 cells show defects in recycling of the pre-60 S factor Tif6, whereas Δjjj1 cells do not (16), is consistent with the idea that structural differences exist. Also relevant to this discussion is the recent report that 60 S subunits from cells lacking Rei1 are more salt-labile than WT ribosomes (30), indicative of a substantial structural difference between 60 S ribosomal particles in WT and Δrei1 strains.
The Rei1-Jjj1 Interaction Is Biologically Important
The data reported here establish that Rei1 and Jjj1 physically interact. Our ability to identify alterations in Jjj1 that affect both its physical interaction with Rei1 and the ability of Jjj1 to function in 60 S subunit biogenesis in vivo supports the idea that this interaction is biologically important. The region rich in charged amino acids (residues 363–534), situated between the two C2H2-type zinc fingers, very likely plays a prominent role in the interaction of Jjj1 with Rei1 as its deletion profoundly affects the interaction. However, this charged region is not sufficient; binding equivalent to that of the full-length protein requires the adjacent zinc fingers. It is possible that the zinc fingers perform a structural role, stabilizing the fold of the charged region. Alternatively, the interaction between Rei1 and Jjj1 may be partially mediated via direct interactions between the zinc fingers of Jjj1 and those of Rei1. Indeed, several C2H2 zinc finger-containing proteins have been shown to dimerize in such a manner (31). Consistent with this hypothesis, we have shown that zinc fingers 2 and 3 of Rei1 are important for its role in ribosome biogenesis.
How Do Rei1 and Jjj1 Facilitate Dissociation of Arx1 from Pre-60 S Subunits?
Previously, we reported that Jjj1 is a functional J-protein that partners with the major cytosolic Hsp70, Ssa (14). Indeed, alteration of the J-domain results in accumulation of half-mer ribosomes as well as failure to release Arx1 (14). As many J-proteins are thought to interact with client proteins and “target” them to their partner Hsp70 (19), the ability of Jjj1 to interact with Rei1 raises the possibility that Rei1 is a client protein of the Jjj1-Ssa chaperone machinery. Although the results presented here do not establish that this is the case, many of the available data are consistent with this idea. As molecular chaperones typically are not absolutely critical for the occurrence of a cellular process but instead act to increase its efficiency, it is perhaps not surprising that overexpression of Rei1 is capable of partially rescuing the growth defects of cells lacking Jjj1, yet the reverse is not the case (16). It is also interesting to note that several other pre-60 S factors have been shown to require GTPases or ATPases for their efficient and proper release (12, 13, 15). Thus, with the involvement of a J-protein and Hsp70 machinery reported here, a common theme involving utilization of the energy of nucleotide hydrolysis for removal of pre-60 S biogenesis factors is emerging.
Regardless of whether Rei1 is a client protein of the Jjj1-Ssa chaperone system, the exact nature of the role of Rei1 in Arx1 removal remains unknown. If Rei1 is a client protein, it is possible that the Hsp70 plays an active role, helping Rei1 to initiate a conformational change in the ribosome, resulting in dissociation of Arx1. Alternatively, Jjj1 and Ssa may aid in removal of Rei1 itself, weakening the association of Arx1 with the ribosome. Consistent with this idea, Rei1 appears to have a prolonged association with 60 S subunits when Jjj1 is not present, as indicated by trailing of Rei1 into the polysome fractions in jjj1Δ cells. As the ribosome association of Arx1 is sensitive to high levels of magnesium in the absence of Rei1, removal of Rei1 may result in alteration of the Arx1-ribosome interaction. At the other end of the spectrum of possibilities, Rei1 may be a part of the chaperone system itself. For example, it might play a role in regulating the ATP hydrolysis cycle of Ssa. Such a role would not be unprecedented as the yeast protein Ssz1, which binds to the J-protein Zuo1, profoundly affects the stimulation via Zuo1 of the ATPase activity of its partner Hsp70 (32).
Future studies will focus on these and other possible mechanisms of action of Rei1 and the Jjj1-Ssa chaperone system in promoting the release of the pre-60 S biogenesis factors Arx1 and Alb1. Much work also needs to be done to understand how the interaction of Rei1 and Jjj1 themselves with the pre-60 S subunit is modulated as their interaction is also transient.
Supplementary Material
Acknowledgment
We thank A. W. Johnson for helpful discussions and for providing us with the pMAL-HIS-TEV vector.
This work was supported, in whole or in part, by National Institutes of Health Grant GM31107 (to E. A. C.) and University of Wisconsin Molecular Biosciences Training Grant T32GM0721532 from the National Institutes of Health (to L. A. H.). This work was also supported by a grant from the Genetech Foundation (to A. E. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
A. W. Johnson, personal communication.
- WT
- wild-type
- HA
- hemagglutinin
- TEV
- tobacco etch virus
- MBP
- maltose-binding protein
- GFP
- green fluorescent protein.
REFERENCES
- 1.Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. (2003) Gene 313, 17–42 [DOI] [PubMed] [Google Scholar]
- 2.Zemp I., Kutay U. (2007) FEBS Lett. 581, 2783–2793 [DOI] [PubMed] [Google Scholar]
- 3.Tschochner H., Hurt E. (2003) Trends Cell Biol. 13, 255–263 [DOI] [PubMed] [Google Scholar]
- 4.Fatica A., Tollervey D. (2002) Curr. Opin. Cell Biol. 14, 313–318 [DOI] [PubMed] [Google Scholar]
- 5.Saveanu C., Bienvenu D., Namane A., Gleizes P. E., Gas N., Jacquier A., Fromont-Racine M. (2001) EMBO J. 20, 6475–6484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho J. H., Kallstrom G., Johnson A. W. (2000) J. Cell Biol. 151, 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nissan T. A., Bassler J., Petfalski E., Tollervey D., Hurt E. (2002) EMBO J. 21, 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebreton A., Saveanu C., Decourty L., Rain J. C., Jacquier A., Fromont-Racine M. (2006) J. Cell Biol. 173, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung N. J., Johnson A. W. (2006) Mol. Cell. Biol. 26, 3718–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradatsch B., Katahira J., Kowalinski E., Bange G., Yao W., Sekimoto T., Baumgärtel V., Boese G., Bassler J., Wild K., Peters R., Yoneda Y., Sinning I., Hurt E. (2007) Mol. Cell 27, 767–779 [DOI] [PubMed] [Google Scholar]
- 11.Hung N. J., Lo K. Y., Patel S. S., Helmke K., Johnson A. W. (2008) Mol. Biol. Cell 19, 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senger B., Lafontaine D. L., Graindorge J. S., Gadal O., Camasses A., Sanni A., Garnier J. M., Breitenbach M., Hurt E., Fasiolo F. (2001) Mol. Cell 8, 1363–1373 [DOI] [PubMed] [Google Scholar]
- 13.Hedges J., West M., Johnson A. W. (2005) EMBO J. 24, 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer A. E., Hung N. J., Yang P., Johnson A. W., Craig E. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1558–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., Liebminger E., Nobis B., Kappel L., van der Klei I., Högenauer G., Fromont-Racine M., Bergler H. (2007) Mol. Cell. Biol. 27, 6581–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demoinet E., Jacquier A., Lutfalla G., Fromont-Racine M. (2007) RNA 13, 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helser T. L., Baan R. A., Dahlberg A. E. (1981) Mol. Cell. Biol. 1, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwase M., Toh-e A. (2004) Cell Struct. Funct. 29, 1–15 [DOI] [PubMed] [Google Scholar]
- 19.Craig E. A., Huang P., Aron R., Andrew A. (2006) Rev. Physiol. Biochem. Pharmacol. 156, 1–21 [DOI] [PubMed] [Google Scholar]
- 20.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 22.Yan W., Schilke B., Pfund C., Walter W., Kim S., Craig E. A. (1998) EMBO J. 17, 4809–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beeler T., Bruce K., Dunn T. (1997) Biochim. Biophys. Acta 1323, 310–318 [DOI] [PubMed] [Google Scholar]
- 24.Shenvi C. L., Dong K. C., Friedman E. M., Hanson J. A., Cate J. H. (2005) RNA 11, 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noah J. W., Wollenzien P. (1998) Biochemistry 37, 15442–15448 [DOI] [PubMed] [Google Scholar]
- 26.Ghysen A., Bollen A., Herzog A. (1970) Eur. J. Biochem. 13, 132–136 [DOI] [PubMed] [Google Scholar]
- 27.Sperrazza J. M., Spremulli L. L. (1983) Nucleic Acids Res. 11, 2665–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamir A., Miskin R., Vogel Z., Elson D. (1974) Methods Enzymol. 30, 406–426 [DOI] [PubMed] [Google Scholar]
- 29.Elson D., Spitnik-Elson P., Avital S., Abramowitz R. (1979) Nucleic Acids Research 7, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parnell K. M., Bass B. L. (2009) Mol. Cell. Biol. 29, 4014–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackay J. P., Crossley M. (1998) Trends Biochem. Sci. 23, 1–4 [DOI] [PubMed] [Google Scholar]
- 32.Huang P., Gautschi M., Walter W., Rospert S., Craig E. A. (2005) Nat. Struct. Mol. Biol. 12, 497–504 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.