Abstract
Opsins are light-sensitive pigments in the vertebrate retina, comprising a G protein-coupled receptor and an 11-cis-retinaldehyde chromophore. Absorption of a photon by an opsin pigment induces isomerization of its chromophore to all-trans-retinaldehyde. After a brief period of activation, opsin releases all-trans-retinaldehyde and becomes insensitive to light. Restoration of light sensitivity to the apo-opsin involves the conversion of all-trans-retinaldehyde back to 11-cis-retinaldehyde via an enzyme pathway called the visual cycle. The critical isomerization step in this pathway is catalyzed by Rpe65. Rpe65 is strongly associated with membranes but contains no membrane-spanning segments. It was previously suggested that the affinity of Rpe65 for membranes is due to palmitoylation of one or more Cys residues. In this study, we re-examined this hypothesis. By two independent strategies involving mass spectrometry, we show that Rpe65 is not palmitoylated nor does it appear to undergo other post-translational modifications at significant stoichiometry. Instead, we show that Rpe65 binds the acidic phospholipids, phosphatidylserine, phosphatidylglycerol, and cardiolipin, but not phosphatidic acid. No binding of Rpe65 to basic phospholipids or neutral lipids was observed. The affinity of Rpe65 to acidic phospholipids was strongly pH-dependent, suggesting an electrostatic interaction of basic residues in Rpe65 with negatively charged phospholipid headgroups. Binding of Rpe65 to liposomes containing phosphatidylserine or phosphatidylglycerol, but not the basic or neutral phospholipids, allowed the enzyme to extract its insoluble substrate, all-trans-retinyl palmitate, from the lipid bilayer for synthesis of 11-cis-retinol. The interaction of Rpe65 with acidic phospholipids is therefore biologically relevant.
Introduction
Visual perception in vertebrates is mediated by two types of photosensitive cells, rods and cones. Both contain a process called the outer segment that consists of a stack of flattened membranous discs loaded with rhodopsin or cone-opsin visual pigment. Opsin pigments are photo-sensitive members of the G protein-coupled receptor superfamily. The light-absorbing ligand of most opsin pigments is 11-cis-retinaldehyde (11-cis-RAL),2 which is covalently coupled to a Lys residue in the opsin protein via a protonated Schiff-base linkage. Absorption of a photon by 11-cis-RAL induces its isomerization to all-trans-retinaldehyde (all-trans-RAL), transiently activating the opsin pigment. After a brief period, the photobleached pigment decays to yield apo-opsin and free all-trans-RAL. Recombination of apo-opsin with another 11-cis-RAL regenerates the visual pigment. To maintain light sensitivity, the all-trans-RAL released following photobleach of an opsin pigment is converted back to 11-cis-RAL by a multistep enzymatic pathway called the visual cycle (Fig. 1). Most steps of the visual cycle take place within cells of the retinal pigment epithelium (RPE), an epithelial monolayer adjacent to photoreceptor outer segment. The critical step in this pathway is all-trans to 11-cis re-isomerization of the retinoid, catalyzed by Rpe65 isomerase (1–3). Consistently, mice with a knock-out mutation in the rpe65 gene contain no 11-cis-retinoids in their ocular tissues and are blind (4). Similarly, mutations in the human RPE65 gene cause a severe, recessively inherited blinding disease called Leber congenital amaurosis (5).
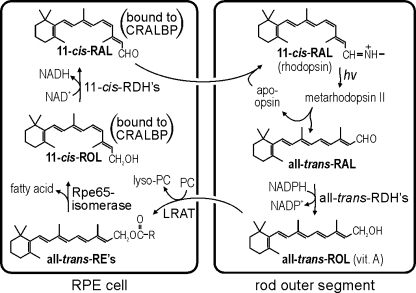
FIGURE 1.
Visual cycle in RPE cells. Absorption of a photon (hv) by a rhodopsin pigment molecule induces isomerization of 11-cis-RAL to all-trans-RAL resulting in activated metarhodopsin II. Decay of metarhodopsin II yields apo-opsin and free all-trans-RAL, which is reduced to all-trans-ROL by one or more all-trans-ROL dehydrogenases (all-trans-RDHs). The all-trans-ROL is released by photoreceptors into the extracellular space and taken up by RPE cells where it is esterified to a fatty acid from phosphatidylcholine by lecithin:retinol acyltransferase (LRAT). Rpe65 isomerase uses all-trans-RE as substrate to form 11-cis-ROL plus a free fatty acid. 11-cis-ROL is oxidized to 11-cis-RAL by one of several 11-cis-ROL dehydrogenases (11-cis-RDHs). Both 11-cis-ROL and 11-cis-RAL are bound to cellular retinaldehyde-binding protein in RPE cells. 11-cis-RAL is released by the RPE and taken up by the photoreceptor where it combines with apo-opsin to re-form rhodopsin.
Rpe65 is an abundant 61-kDa protein in RPE cells. Rpe65 is homologous to β-carotene 15,15′-oxygenase in mammals (6) and apo-carotene oxygenase in cyanobacteria (7). By x-ray diffraction analysis, Rpe65 is a seven-bladed β-propeller with an Fe2+-His4 arrangement at its axis (8). The four His residues that define the Fe2+-binding site are conserved in all members of this protein family (Fig. 2). Rpe65 catalyzes the isomerization and hydrolysis of an all-trans-retinyl ester (all-trans-RE), such as all-trans-retinyl palmitate (all-trans-RP), to 11-cis-retinol (11-cis-ROL), and a free fatty acid (9–11). In this reaction, the energy of ester hydrolysis is used to drive the energetically unfavorable conversion of a planar all-trans-retinoid to the strained 11-cis configuration (12). All-trans-REs are insoluble in water and stored in RPE internal membranes. Rpe65 strongly associates with membranes (13), providing access to its all-trans-RE substrate. The basis for the association of Rpe65 with membranes is unresolved. Rpe65 has no predicted transmembrane segments. It was suggested that Rpe65 is reversibly S-palmitoylated on residues Cys-231, Cys-329, and Cys-330 (14). A “palmitoylation-switch” mechanism for the regulation of isomerase activity was proposed whereby binding of Rpe65 to its all-trans-RP substrate is mediated by reversible S-palmitoylation of these three residues (14). However, it was subsequently shown that Cys-231, Cys-329, and Cys-330 are not modified in Rpe65 (15). More recently, it was suggested that Rpe65 is palmitoylated on Cys-112 (16). In this study, we examined all 12 Cys residues in bovine Rpe65 and found no evidence for significant S-palmitoylation at any site. However, we found that Rpe65 interacts directly with three acidic phospholipids but not with basic phospholipids or neutral lipids, explaining the affinity of Rpe65 for membranes. We show that this interaction is functionally relevant, as it permits Rpe65 to extract all-trans-REs from the lipid bilayer and use them as substrate for synthesis of 11-cis-ROL.
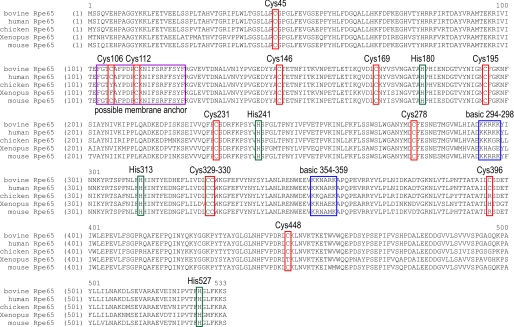
FIGURE 2.
Annotated sequence alignment of Rpe65 from several vertebrate species. Bovine Rpe65 contains 12 Cys residues (red boxes). Eight are conserved in all species, and the remaining four, including Cys-396, are not conserved. The four conserved His residues that form the Fe2+-coordination sphere are indicated by green boxes. The two highly basic sequences that map to the surface of Rpe65 are indicated by blue boxes. The possible hydrophobic membrane-anchor region (residues 103–123) is indicated by the purple box.
EXPERIMENTAL PROCEDURES
Materials
Unless stated otherwise, all chemicals and solvents were purchased from Sigma. 2,3,3-Acrylamide-d3 was purchased from Cambridge Isotope Laboratories. Modified trypsin was purchased from Promega. Endopeptidase Glu-C and Asp-N were purchased from Roche Diagnostics. Phospholipids (PC and PE from egg yolk, PG from egg yolk lecithin, PS from bovine brain, and PI from bovine liver) were purchased from Sigma as chloroform solutions. The membrane lipid strip was purchased from Echelon Bioscience Inc. CompleteTM EDTA-free proteinase inhibitor mixture tablets were purchased from Roche Diagnostics. Protein concentration was detected by a mini BCA kit from Pierce (part of Thermo Fisher Scientific). GelCode Coomassie Blue was also purchased from Pierce. The Millipore Montage In-Gel Digestzp kit was purchased from Fisher.
Preparation of Bovine RPE Homogenates and Microsomes
Bovine eyes were obtained from a local slaughterhouse and used within 4 h. After dark adaptation for 1 h, bovine RPE cells were collected by gentle brushing from dissected cow eyeballs after removal of the retina. RPE cells were suspended in 30 ml of 20 mm HEPES (pH 7.4), 150 mm NaCl with addition of CompleteTM EDTA-free proteinase inhibitor and homogenized by nitrogen cavitation at 1000 p.s.i. The supernatant was collected after centrifugation at 3000 × g as a bovine RPE homogenate. The homogenate was lyophilized to a dry powder and resuspended in 2 ml of distilled H2O and stored at −80 °C. Bovine RPE microsomes were collected by centrifugation of the supernatant mentioned above at 100,000 × g, and the pellet was resuspended in HEPES or Tris-HCl buffer and stored in −80 °C.
Preparation of Acrylamide-modified Bovine Rpe65
Two aliquots of bovine RPE microsomal protein solutions (225 mg of protein in each) were dissolved in 50 μl of 50 mm Tris-HCl buffer (pH 8.0) containing 0.1% SDS. To one aliquot, 20 μl of 50 mm dithiothreitol (DTT) in 50 mm ammonium bicarbonate (pH 8.4) was added, and the solution was incubated at 70 °C for 3 h. To another aliquot, 20 μl of 50 mm tributylphosphine (TBP) in 50 mm ammonium bicarbonate (pH 8.4) was added, and the solution was incubated at 37 °C for 0.5 h. The DTT sample was incubated with 20 μl of 250 mm acrylamide (AA-d0) in 50 mm ammonium bicarbonate (pH 8.4) at 25 °C for 3 h, and its TBP counterpart was incubated with triple deuterium-labeled acrylamide (AA-d3) instead. The 2 aliquots were pooled and separated by SDS-PAGE. The protein bands representing purified bovine Rpe65 were dissected and subjected to in-gel proteolytic digestion (described below). A control sample incubated with a 1:1 acrylamide-d0/d3 solution was prepared in the same manner.
In-gel Proteolytic Digestion
Following the manufacturer's procedure in the Montage kit, gel slices containing purified Rpe65 were diced and placed in a 96-well plate. 100 μl of destain solution (50:50 acetonitrile/water) was added to each well and incubated for 20 min. After removal of the destaining solution by vacuum, 100 μl of 100% acetonitrile was added to dehydrate the gel slices. After incubation for 10 min, the solution was removed by aspiration. The gel slices were re-hydrated in a 25–30 μl of solution containing the selected protease (weight ratio of substrate to enzyme, 100:1 for trypsin and 200:1 for endoproteinase Glu-C and Asp-N) in 50 mm ammonium bicarbonate (pH 8.0), and incubated at 37 °C for 15 h. The digested peptides were extracted from the gel slice by addition of 200 μl of extraction solution (50 mm ammonium bicarbonate) and incubated for 30 min. After washing with 100 μl of washing buffer (0.1% formic acid), the peptides were recovered by addition of elution solution (50:50 0.1% formic acid/methanol) with vacuum. The final peptide solutions were lyophilized in a centrifugal vacuum concentrator and dissolved into 10 μl of 0.1% formic acid, 5% acetonitrile in water.
Synthesis of S-Palmitoyl-N-acetyl-l-cysteine Methyl Ester
In a glass flask equipped with a stir bar, 17.7 mg (0.1 mmol) of N-acetyl-l-cysteine methyl ester (S-palm-N-Ac-Cys-Me) was incubated with palmitoyl chloride (20 eq), N-hydroxybenzotriazole (20 eq), dicyclohexylcarbodiimide (20 eq), and 4-dimethylaminopyridine (22 eq) in 2 ml of anhydrous tetrahydrofuran for 15 h. After filtration, the reaction was stopped with addition of 5 ml of 0.1 m HCl, and the mixture was extracted twice with CH2Cl2. The pooled extracts were washed with brine and dried with anhydrous Na2SO4. Pure S-palm-N-Ac-Cys-Me was separated by reversed phase HPLC and characterized by tandem mass spectrometry (MS/MS).
Confirmation of Cysteine De-palmitoylation by DTT but Not TBP
To 3 aliquots of purified S-palm-N-Ac-Cys-Me in 50 mm ammonium bicarbonate (pH 8.4) were added 20 μl of 50 mm DTT, 20 μl of 50 mm TBP in 50 mm ammonium bicarbonate (pH 8.4), and 20 μl of 50 mm ammonium bicarbonate (pH 8.4), respectively. The aliquot with DTT was incubated at 70 °C for 3 h, and the other 2 aliquots were incubated at 37 °C for 0.5 h. The 3 aliquots were then incubated with 20 μl of 1:1 mixture of 250 mm acrylamide (AA-d0/d3) in 50 mm ammonium bicarbonate (pH 8.4) at 25 °C for 3 h. The final samples were subjected to capillary LC/MS analysis.
Liquid Chromatography with Electrospray Ionization-Mass Spectrometry (LC-MS) of Intact Rpe65 Protein
Proteins were fractionated by reverse-phase liquid chromatography as described below. The eluent was split between a fraction collector, collecting 1-min fractions, and an electrospray ionization (ESI) mass spectrometer, API III+ mass spectrometer (PE Sciex, Applied Biosystem, Foster City, CA), as described previously (17, 18). For HPLC fractionation, a PLRP-S polymeric column was used with a pore size of 300 Å, bead size of 5 μm, and dimensions of 150 × 2 mm (Polymer Laboratories, Amherst, MA). The column was maintained at 40 °C and initially equilibrated in solvent A (0.1% trifluoroacetic acid in water) at 95% and solvent B (0.1% trifluoroacetic acid in acetonitrile) at 5%. Rpe65 protein was eluted at a flow rate of 100 μl/min using a program of 5 min at 5% B, followed by a 25-min gradient from 5 to 30% B, and ending with a 100-min gradient from 30 to 100% B, for a total gradient time of 130 min. The API III+ mass spectrometer was tuned and calibrated as described previously (19) to yield a mass accuracy of 0.01% (1.0 Da at 10 kDa). The 50-μl fractions collected during chromatography were stored at −20 °C. The data were analyzed using BioMultiview 1.3.1 software (Applied Biosystems).
Nano-liquid Chromatography with Data-dependent Tandem Mass Spectrometry
Samples were analyzed by nano-liquid chromatography-MS/MS with data-dependent acquisition (Q STAR XL, Applied Biosystems, Foster City, CA) as described previously (20). After dissolution in 10 μl of 0.1% formic acid and 5% acetonitrile (v/v), samples were injected onto a trapping column (3 cm, 75 μm, C18, Micro-Tech) previously equilibrated in 100% solvent A (0.1% formic acid, 5% acetonitrile in water) at a flow rate of 2 μl/min. Following 10 min of washing, the trapping column was eluted through a pre-equilibrated analytical column (15 cm, 75 μm, C18, Micro-Tech) at a flow rate of 300 nl/min using the compound linear gradient of solvents A and B (0.1% formic acid in acetonitrile) as follows: 3 min at 100% A; 80% A, 20% B at 8 min; 65% A, 35% B at 13 min; 25% A, 75% B at 23 min; and 90% A, 10% B at 23.1 min. The column eluent was directed to an uncoated, pulled silica nanospray tip (Picotip FS360-20-10-N-5-C12, New Objective) at 2.4 kV for ionization without nebulizer gas. The mass spectrometer was operated in “information-dependent acquisition” mode with a survey scan (350–1600 m/z), data-dependent MS/MS on the two most abundant ions with exclusion after two MS/MS experiments. The Cys peptide sequences were detected using BioAnalyst program (Applied Biosystem) with help of the characteristic doublet isotopic tag.
Algorithm for Calculation of d3/d0 from the MS Data
The relative isotopic peak abundances of Cys peptides were obtained via integrated peak areas of the isotopic mass peaks by Analyst QS (version 1.1, Applied Biosystems, Carlsbad, CA). Starting from the first highest isotopic peak, their relative abundances were assigned as I0, I1, I2, …, etc. According to the identities of the peptides analyzed by tandem mass, their theoretical (native) isotopic abundance of peptide isotopic peaks was calculated as I0*, I1*, I2*, …, etc., based on their molecular formulas. The following were generated thereafter: d3/d0 = x; I0 = I0*, I1 = I1*, I2 = I2*; I3 = I3* + xI0*, I4 = I4* + xI1*, I5 = I5* + xI2*; I6 = I6* + xI3*. I6* ≅ 0, in most of the cases, then I6 = xI3* as shown in Equation 1,
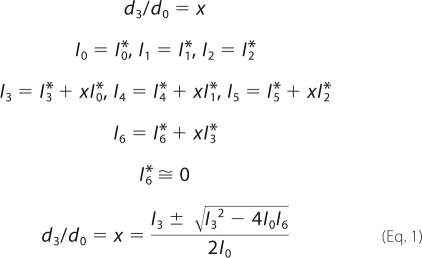 |
The d3/d0 ratios were calculated as the product of the equation involving I0, I3, and I6. The final d3/d0 ratios were normalized to the peptide containing Cys-231, which was shown in a previous study to be nonpalmitoylated (15).
Membrane Lipid Binding Assay
The membrane lipid strips were blocked by incubation in PBS + 3% nonfat dry milk and gently agitated for 1 h at 25 °C. After discarding the block solution, the strips were incubated with samples of bovine RPE homogenates containing the indicated total protein concentrations (20, 40, and 80 mg/ml) in PBS + 3% nonfat-dry milk for 1 h at 25 °C. The protein solutions were removed, and the strips were washed with three changes of PBS/Tween 20 for 10 min per wash with gentle agitation. The strips were then incubated with antisera against Rpe65 (see below) diluted 1:2000 in blocking solution for 1 h at 25 °C. The strips were washed three times for 10 min with PBS/Tween 20 and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) for 1 h at 25 °C. Rpe65 that reacted with the antibodies was visualized with the enhanced ECL-Plus Western blot detection system (Amersham Biosciences).
Effect of pH on the Association of Rpe65 with Membranes
Ten-ml aliquots containing 2–3 mg/ml bovine RPE homogenate (see above) were dissolved in 990 ml of PBS (pH 7.0, 8.0, 10.0, and 11.0). The aliquots were homogenized by nitrogen cavitation at 1000 p.s.i. and subjected to centrifugation at 100,000 × g. The pellets and the lyophilized powders of the supernatants were dissolved in 50 ml of denatured SDS-PAGE sample buffer. The samples were separated by a 10% SDS-PAGE, and Rpe65 was visualized by GelCode Coomassie Blue stain and by immunoblotting.
Immunoblot Analysis
Proteins in Laemmli sample buffer were heated for 10 min at 75 °C, separated by SDS-PAGE in a 10% polyacrylamide gel, and transferred to an Immobilon-P membrane (Millipore). The membrane was incubated in blocking buffer (PBS (pH 7.4), 5% nonfat milk) for 2 h at 37 °C and then overnight at 4 °C with rabbit polyclonal antiserum against the Rpe65 peptide, NFITKINPETLETIK (residues 150–164). After washing three times in PBS/Tween 20 for 10 min per wash, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) for 1 h and washed again. Rpe65 that reacted with the antibodies was visualized with the enhanced ECL-Plus Western blot detection system (Amersham Biosciences).
In Vitro Isomerase Assay in the Presence of Liposomes
For assays that used all-trans-ROL substrate, the isomerase assay mixtures contained 20 mm HEPES (pH 7.5), 150 mm NaCl, 6% bovine serum albumin, 10 μm all-trans-ROL, and 50–100 μg of bovine RPE homogenate. For assays that used all-trans-RP substrate, the isomerase assay mixtures contained 20 mm HEPES (pH 7.5), 150 mm NaCl, 6% bovine serum albumin, 10 μm all-trans-RP, and 50–100 μg of bovine RPE homogenate. The final volume for each reaction was 400 μl. To make liposomes, 20 μl of 10 mg/ml phospholipid in chloroform was dried under a stream of nitrogen gas. HEPES buffer without substrate and proteins in the isomerase assay mixture were added, and the mixture was sonicated on ice under nitrogen for 20 min. For all-trans-RP-containing liposomes, 10 μl of 400 mm all-trans-RP in hexane was added to the phospholipids before drying. Incubations were started by adding the bovine serum albumin and bovine RPE homogenate. In some assays, all-trans-RP substrate was dissolved in sodium cholate at 6 mm final concentration in the assay mixture. After incubation for 1 h in the dark at 37 °C, the reactions were quenched by adding 0.2% SDS and 2 volumes of methanol. Retinoids were extracted with hexane and analyzed by HPLC, as described below. Isomerase-specific activities were calculated after subtracting endogenous 11-cis-ROL, determined by hexane extraction of representative reaction mixtures before addition of substrate. Bovine RPE homogenates were bleached by UV light for 5 min to remove the endogenous retinoids.
Cloning of Chicken Rpe65
Chicken Rpe65 was cloned by primer amplification of the gene from chicken retina cDNA based on the previously reported sequence (NCBI accession NM204884). Briefly, RNA was prepared from freshly dissected chicken retinas (Absolutely RNA, Stratagene) and used as a template for first-strand cDNA synthesis (Superscript III, Invitrogen). The complete Rpe65 coding region was amplified by PCR using primers 5′-GAGACAATGTACAGCCAGGTG (forward) and 5′-GTGCTTGCAAATGGAGACTG (reverse) under the following conditions: 25 cycles of 94 °C for 30 s; 60 °C for 30 s; and 72 °C for 60 s in a PTC-200 gradient thermocycler (MJ Research). The amplified fragment was cloned into the Topo-TA cloning vector (Invitrogen). The chicken Rpe65 cDNA insert was released by restriction digestion and subcloned into the pcDNA3.1 mammalian expression vector (Invitrogen). The full coding region was checked by DNA sequencing (DYEnamic ET Terminator Cycle Sequencing, ABI 3100 DNA Sequencer, GE Healthcare). HEK-293T cells were transfected with the expression plasmid for protein expression (Polyfect, Qiagen).
Preparation of Homogenates from Cells Expressing Chicken Rpe65
HEK-293T cells expressing chicken Rpe65 were suspended in 5 ml of 20 mm HEPES (pH 7.4), 150 mm NaCl with addition of CompleteTM EDTA-free proteinase inhibitor and homogenized by nitrogen cavitation at 1000 p.s.i. The supernatant was collected after centrifugation at 3000 × g and stored at −80 °C.
In Vitro Isomerase Assay of Chicken Rpe65 Using All-trans-RP as Substrate in Phospholipid Liposomes
The isomerase assay mixtures contained 20 mm HEPES (pH 7.5), 150 mm NaCl, 6% bovine serum albumin, 10 μm all-trans-RP, and 50–100 μg of chicken Rpe65 cell homogenate. The final volume for each reaction was 500 μl. To make all-trans-RP-containing liposomes, 20 μl of 10 mg/ml mixture of phospholipids in chloroform were mixed with 10 μl of 400 μm all-trans-RP in hexane and dried under a stream of nitrogen gas. HEPES buffer in isomerase assay mixture (without proteins) was added and sonicated on ice under nitrogen for 20 min in the dark. Incubations were started with addition of the chicken Rpe65 cell homogenate. After incubation for 0.5 h in the dark at 37 °C, the reactions were quenched by adding 0.2% SDS and 2 volumes of methanol. Retinoids were extracted with hexane and analyzed by HPLC, as described below. Isomerase-specific activities were calculated after subtracting endogenous 11-cis-ROL, determined by hexane extraction of representative reaction mixtures before addition of substrate.
HPLC Analysis of Retinoids
Retinoids were analyzed by normal-phase HPLC as described previously. In brief, after drying under a stream of nitrogen, samples were dissolved in 100 μl of hexane, and retinoids were separated by chromatography on a silica column (Supelcosil LS-SI 5 μm, 4.6 × 250-mm inner diameter) by gradient elution (mobile phase 0.2–10% dioxane in hexane, 2.0 ml per min flow rate) in an Agilent 1100 liquid chromatography equipped with a UV photodiode-array detector. Identified peaks were confirmed by spectral analysis and co-elution with authentic retinoid standards.
Surface Electrostatic Potential Map
Electrostatic surface potential of bovine Rpe65 was calculated and viewed using DeepView/Swiss-PDBViewer version 4.01 (21). The Protein Data Bank file of bovine Rpe65 (code 3FSN) was obtained from RCSB. The surface electrostatic potential computation was performed using the Poisson-Boltzmann equation method (22).
RESULTS
Identification of 19 Peptides Containing the 12 Cys Residues in Bovine Rpe65
Based on the amino acid sequence, we defined a set of 19 proteolytic peptides in bovine Rpe65 that collectively contain the 12 Cys residues (Table 1). To identify these Cys-containing peptides, we incubated a sample of bovine RPE microsomes with dithiothreitol (DTT) under conditions that reduce disulfides and deacylate modified Cys residues (23). Next, we incubated the sample with a 1:1 mixture of triple deuterium-labeled acrylamide (AA-d3) and nondeuterated acrylamide (AA-d0). Under the conditions used, acrylamide efficiently S-alkylates free thiol groups (24). The sample was separated by SDS-PAGE, and gel slices containing Rpe65 were excised. The purified Rpe65 was subjected to in-gel digestion using trypsin, endoproteinase Glu-C, and/or Asp-N. Every Cys residue in the resulting peptide mixtures should then be modified with AA-d3 and AA-d0 at a 1:1 ratio. We separated these peptide mixtures by reverse-phase liquid chromatography (LC) and analyzed by mass spectrometry (MS). Fig. 3A shows the chromatogram for quadruple-charged ion of m/z = 621.7059, corresponding to the Cys-112-containing, AA-d0-modified peptide, AN110–129 (Table 1). Fig. 3D shows the chromatogram for a double-charged m/z = 860.2878 ion, corresponding to the Cys-231-containing, AA-d0-modified peptide, AN218–232 (Table 1). Similar chromatograms were acquired for ions of m/z = 622.4657 and 861.7873, corresponding to the AA-d3-modified AN110–129 and AN218–232 peptides, respectively (data not shown). Using the protease combinations indicated in Table 1, we performed LC/MS analysis of the remaining 17 peptides (data not shown). After correcting for charge, the observed masses were in good agreement with the predicted masses for all 19 AA-d0- and AA-d3-modified peptides.
TABLE 1.
Observed peptides in the proteolytic digestion of AA-d0/d3-treated bovine Rpe65
The cysteine residues are underlined. T, trypsin; G, endoproteinase Glu-C; AN, Asp-N.
| Peptide | Mass d0/d3 | Cys no. | Sequence |
|---|---|---|---|
| T223–234 | 1448.68/1451.70 | 231 | SEIVVQFPCSDR |
| G22–51 | 3260.76/3263.78 | 45 | LSSPLTAHVTGRIPLWLTGSLLRCGPGLFE |
| G162–184 | 2578.27/2581.29 | 169 | TIKQVDLCNYVSVNGATAHPHIE |
| G168–184 | 1893.89/1896.91 | 169 | LCNYVSVNGATAHPHIE |
| G185–224 | 4468.21/4471.23 | 195 | NDGTVYNIGNCFGKNFSIAYNIVKIPPLQADKEDPISKSE |
| G218–233 | 1832.87/1835.89 | 231 | DPISKSEIVVQFPCSD |
| G321–348 | 3610.71/3616.75 | 329, 330 | HEFLIVDLCCWKGFEFVYNYSYLANLRE |
| G323–352 | 3902.81/3908.85 | 329, 330 | FLIVDLCCWKGFEFVYNYSYLANLRENWEE |
| G365–399 | 3884.06/3887.08 | 396 | VRRYVLPLNIDKADTGKNLVTLPNTTATAILCSDE |
| G418–462 | 5461.76/5464.78 | 448 | FPQINYQKYGGKPYTYAYGLGLNHFVPDRLCKLNVKTKETWVWQE |
| AN87–109 | 2687.33/2690.35 | 106 | DAYVRAMTEKRIVITEFGTCAFP |
| AN110–129 | 2481.20/2484.22 | 112 | DPCKNIFSRFFSYFRGVEVT |
| AN142–160 | 2291.05/2294.07 | 146 | DYYACTETNFITKVNPETL |
| AN167–185 | 2122.96/2125.98 | 169 | DLCNYVSVNGATAHPHIEN |
| AN186–214 | 3225.64/3228.66 | 195 | DGTVYNIGNCFGKNFSIAYNIVKIPPLQA |
| AN218–232 | 1717.85/1720.87 | 231 | DPISKSEIVVQFPCS |
| AN277–292 | 1920.83/1923.85 | 278 | DCFESNETMGVWLHIA |
| AN378–397 | 2102.08/2105.10 | 396 | DTGKNLVTLPNTTATAILCS |
| AN445–463 | 2442.26/2445.28 | 448 | DRLCKLNVKTKETWVWQEP |
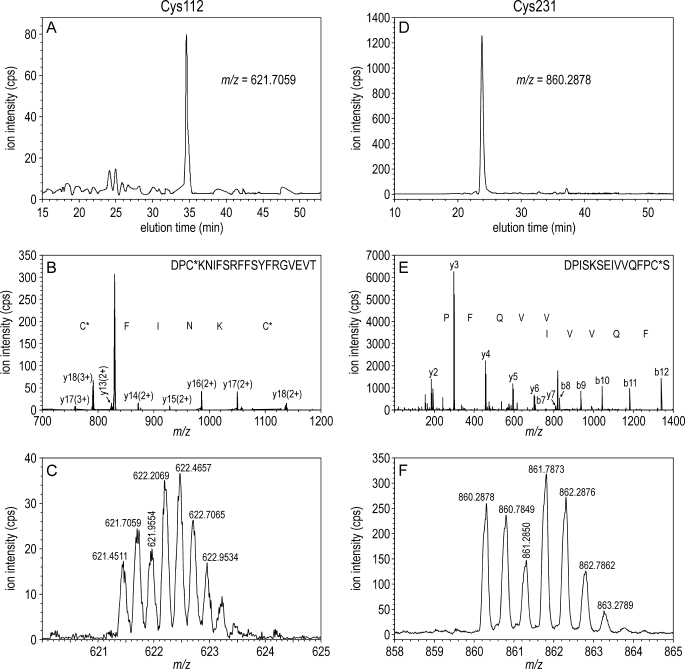
FIGURE 3.
ESI-MS/MS data showing similar abundance of AA-d0- and AA-d3-modified peptides containing residues Cys-112 and Cys-231. A, selected ion monitoring chromatogram of an ion with m/z 621.7059. This ion corresponds to tetra-charged Asp-N peptide AN110–129 from bovine Rpe65 after modification of Cys-112 with AA-d0. B, tandem mass spectrum of the 621.7059 m/z ion with the corresponding observed b- and y-ions indicated. C, zoom spectrum of the indicated m/z values acquired at 35 min. Note the deviation from the ideal d0/d3 distribution shown on Fig. 4 due to the overlap of native isotopic d0 ions on the d3 ion species. The similar abundance of the corresponding AA-d0 and AA-d3-labeled ions indicates that Cys-112 is not palmitoylated. D, selected ion monitoring chromatogram of an ion with m/z 860.2878 corresponding to the double-charged Asp-N peptide AN218–232 from bovine Rpe65 after modification of Cys-231 with AA-d0. E, tandem MS chromatogram of the m/z 860.2878 ion with the corresponding b- and y-ions indicated. F, zoom spectrum of the indicated m/z values acquired at 24 min. The similar abundances of the corresponding AA-d0- and AA-d3-labeled ions indicate that Cys-231 is not palmitoylated. Note that the b- and y-ions containing Cys residues show similar d0/d3 doublet peaks, whereas ions lacking Cys show only singlet peaks.
To confirm identification of these peptides, we performed MS/MS analysis following in-gel digestion of Rpe65 with the indicated proteases. The resulting y- and b-ion series are shown in Fig. 3B for peptide AN110–129 and Fig. 3E for AN218–232. These data yielded the partial amino acid sequences, identifying both peptides. We performed similar MS/MS analysis on the remaining Rpe65 peptides. For each of the 19 peptides, the observed and predicted sequences were in full agreement. The ion peaks from Cys-containing peptide fragments were doublets versus singlets from non-Cys-containing fragments (Fig. 3, B and E) due to AA-d0 plus AA-d3 modifications of the Cys residues. Together, these results validate our identification of the 19 Cys-containing Rpe65 peptides.
Rpe65 Is Not S-Palmitoylated
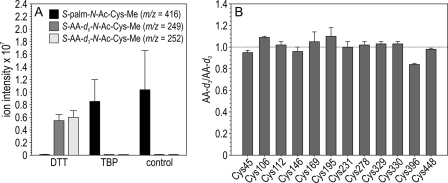
We tested for palmitoylation of the 12 Cys residues in bovine Rpe65 by the strategy shown in Fig. 4. This strategy employs two disulfide-reducing agents. As discussed, DTT reduces disulfides and deacylates Cys residues (23). TBP reduces disulfides but does not react with acylated Cys residues (25). To confirm these activities of DTT and TBP on palmitoylated Cys residues, we synthesized S-palm-N-Ac-Cys-Me as a model reagent. We incubated aliquots of S-Palm-N-Ac-Cys-Me with DTT, TBP, or without any reducing agent under the same conditions as those used with Rpe65. Next, we incubated the samples with a 1:1 mixture of AA-d0/d3, and subjected them to LC/MS analysis. Virtually no m/z = 416 ion corresponding to S-palm-N-Ac-Cys-Me was detectable in the sample treated with DTT (Fig. 5A). Instead, this sample contained m/z = 249 and 252 ions of approximately equal intensities, corresponding to S-AA-d0-N-Ac-Cys-Me and S-AA-d3-N-Ac-Cys-Me. In contrast, samples incubated with TBP or no reducing agent contained only the m/z = 416 ion (S-palm-N-Ac-Cys-Me) with virtually no detectable m/z = 249 or 252 ions (Fig. 5A). These data suggest that incubation with DTT should fully de-palmitoylate Rpe65 were it so modified, whereas incubation with TBP should have no effect on the palmitoylation state of Rpe65.
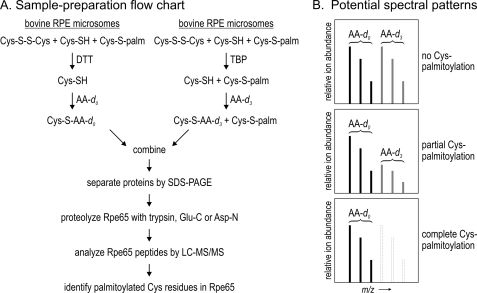
FIGURE 4.
Strategy to determine palmitoylation status of Cys residues in Rpe65. A, flow chart for the preparation of Rpe65 peptide samples for LC-MS analysis. One aliquot of bovine RPE membranes is incubated with DTT, which reduces disulfides and de-palmitoylates Cys residues in all proteins, including Rpe65. The resulting free sulfhydryl groups are alkylated by incubating with nondeuterated acrylamide (AA-d0). A second aliquot is incubated with TBP to reduce disulfide without de-palmitoylating Cys residues. Next, the nonpalmitoylated sulfhydryls are alkylated by incubating with triple-deuterated acrylamide (AA-d3). These two aliquots are pooled and the constituent proteins separated by SDS-PAGE. The Rpe65 band is excised and digested with trypsin, endopeptidase Glu-C, and/or Asp-N, and the resulting peptides are analyzed by LC-MS/MS. B, diagram showing predicted spectral patterns for the different potential palmitoylation states of a Cys residue. If a Cys-containing peptide is not palmitoylated, it would incorporate equal amounts of AA-d0 and AA-d3. Therefore, the two sets of native isotopic peaks that differ by three atomic mass units would be equally abundant in the spectrum. If the Cys residue is partially palmitoylated, the triple-deuterated ions would be correspondingly less abundant than the nondeuterated ions. If the Cys residue is fully palmitoylated, no triple-deuterated ions would be present in the spectrum.
FIGURE 5.
Analysis of Rpe65 S-palmitoylation. A, effects of DTT and TBP on Cys palmitoylation. S-Palm-N-Ac-Cys-Me was treated with DTT, TBP, or without a reducing agent (control) followed by reaction with a 1:1 mixture of AA-d0/d3. This histogram shows the ion intensities of S-palm-N-Ac-Cys-Me (m/z = 416), S-AA-d0-N-Ac-Cys-Me (m/z = 249), and S-AA-d3-N-Ac-Cys-Me (m/z = 252) in the different samples. Note the virtual absence of palmitoylated-Cys in the sample incubated with DTT and the presence of only palmitoylated Cys in the samples incubated with TBP or no reducing agent (control). B, AA-d3/d0 ratio for each Cys residue in bovine Rpe65. This ratio is equivalent to the fraction of nonpalmitoylated Cys in each peptide. The values were normalized to the peptide containing Cys-231, which is known to be nonpalmitoylated. Except for Cys-396, the other 11 Cys residues show AA-d3/d0 values close to 1, indicating nonpalmitoylation of these residues in bovine Rpe65. Error bars show standard deviations (n = 3).
We prepared bovine RPE microsomes and split them into 2 equal aliquots. One aliquot was incubated with DTT, to reduce and fully de-palmitoylate Rpe65, and then with AA-d0, to alkylate all the Cys residues in Rpe65. The other aliquot was incubated with TBP, to reduce disulfides without altering any palmitoylated Cys residues, and then with AA-d3, to alkylate the free but not palmitoylated Cys residues in Rpe65. The 2 aliquots were pooled and separated by SDS-PAGE, and gel slices containing Rpe65 were excised. The purified Rpe65 was subjected to in-gel digestion using trypsin, endoproteinase Glu-C, and/or Asp-N (Table 1) for MS analysis. The zoomed in spectrum for peptide AN110–129 is shown in Fig. 3C. Here, the 621.4511, 621.7059, and 621.9554 m/z peaks correspond to the quadruple-charged AA-d0, AA-d0+1, and AA-d0+2 isotopic ions, respectively. The 622.2069 peak contains the AA-d0+3 and AA-d3 ions. The 622.4657 peak contains the AA-d0+4 and AA-d3+1 ions; the 622.7065 peak contains the AA-d0+5 and AA-d3+2 ions, and the 622.9534 peak contains the AA-d0+6 and AA-d3+3 ions. Fig. 3E shows the spectrum for peptide AN218–232. The 860.2878, 860.7849, and 861.2850 m/z peaks correspond to double-charged AA-d0, AA-d0+1, and AA-d0+2 isotopic ions, respectively. The 861.7873 peak contains the AA-d0+3 and AA-d3 ions. The 862.2876 peak contains the AA-d0+4 and AA-d3+1 ions, the 862.7862 peak contains the AA-d0+5 and AA-d3+2 ions, and the 863.2789 peak contains the AA-d0+6 and AA-d3+3 ions. We performed similar analysis of the remaining 17 peptides (data not shown). The ratio of AA-d3 to AA-d0, which is equivalent to the fraction of unmodified Cys, was calculated from these MS data for each peptide using the algorithm described under “Experimental Procedures.” These ratios are plotted in Fig. 5B. The only residue in bovine Rpe65 that showed greater than 5% modification was Cys-396. However, Cys-396 is not conserved within mammals (Fig. 2), suggesting that modification of this residue is not required for the association of Rpe65 with membranes. It is notable that Cys-231, Cys-329, and Cys-330, which were suggested to be palmitoylated in an earlier study (14), and Cys-112, which was suggested to be palmitoylated more recently (16), show no evidence of palmitoylation in this study.
Rpe65 Does Not Undergo Post-translational Modifications at Significant Stoichiometry
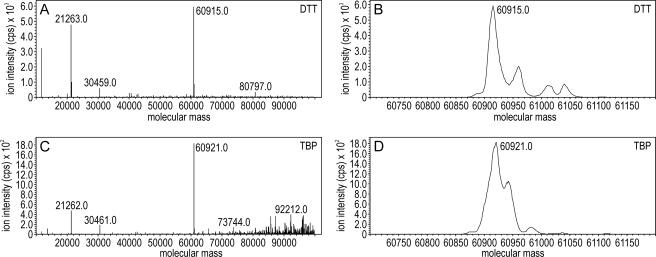
To test further for Rpe65 post-translational modifications, we performed ESI-MS analysis on the intact (nonproteolyzed) protein purified from bovine RPE microsomes after reduction with TBP or DTT. For both samples, greater than 80% of total Rpe65 immunoreactivity eluted from the PLRP-S polymeric column in a single peak (data not shown). The major ion in this peak fraction was 60,921 m/z for the TBP and 60,915 m/z for the DTT sample (Fig. 6, A–D). These m/z values are similar to the calculated molecular mass of unmodified bovine Rpe65 (60,905). The mass accuracy of this method is 100 ppm or ∼6 Da. By comparison, the mass of a single palmitoyl group is 238. These observations establish that the maximum possible stoichiometry for palmitoylation of Rpe65 is 20%. A similar argument can be applied for phosphorylation and most other post-translational modifications. Low mass modifications of Rpe65 cannot be ruled out by these data.
FIGURE 6.
ESI-MS deconvolution analysis to determine the molecular mass of intact Rpe65. After incubation with DTT or TBP, undigested Rpe65 was separated by reverse-phase HPLC and analyzed by ESI-MS. A and B show the deconvoluted molecular mass spectra for Rpe65 treated with DTT. C and D show the deconvoluted spectra for Rpe65 treated with TBP. A and C show the full mass range, and B and D show a zoomed in mass range. Note the absence of an ion peak at 61,143 Da, which would indicate palmitoylation of Rpe65.
Rpe65 Binds to Phospholipids with Negatively Charged Headgroups
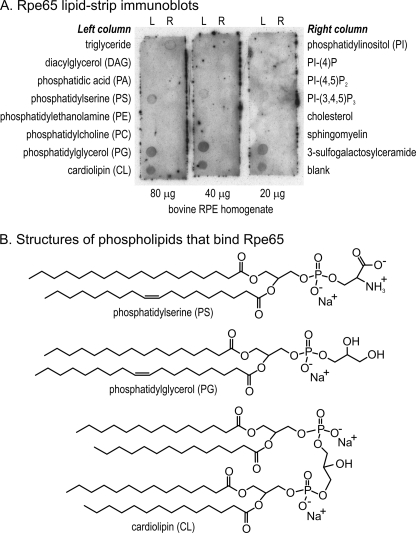
An alternative explanation for the association of Rpe65 with membranes is a direct interaction between the protein and phospholipid headgroups. To test this possibility, we incubated RPE homogenates with membrane strips containing immobilized lipids and the major phospholipids. After washing, the strips were incubated with antisera against bovine Rpe65 and developed as immunoblots. We observed Rpe65 immunoreactivity associated with spots containing three acidic phospholipids, PS, PG, and cardiolipin (CL) (Fig. 7A). We observed no binding of Rpe65 to phosphatidic acid, nor to the positively charged phospholipids, phosphatidylethanolamine (PE) and PC, nor to any neutral lipids (Fig. 7A).
FIGURE 7.
Binding of Rpe65 to negatively charged phospholipids. A, membranes containing the indicated immobilized lipids were incubated with 80, 40, or 20 μg of bovine RPE homogenates. After washing, bound Rpe65 was detected by immunoblotting. Note the Rpe65 immunoreactivity on the spots containing PS, PG, and CL. B, molecular structures of PS, PG, and CL. Note that CL contains two net negative charges, versus one negative charge for PS and PG.
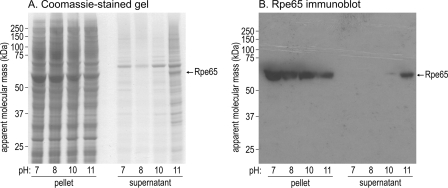
Association of Rpe65 with Membranes Is pH-dependent
We incubated bovine RPE microsomes in buffers of pH 7–11. Next, we pelleted the microsomes and performed immunoblot analysis of Rpe65 in the pellet and supernatant fractions. Rpe65 remained tightly associated with membranes up to pH 10 (Fig. 8, A and B). At pH 11, we observed significant solubilization of Rpe65. These results are consistent with an electrostatic interaction between a basic region on the surface of Rpe65 and acidic headgroups on the membrane surface.
FIGURE 8.
Rpe65 dissociates from microsomal membranes at high pH. Bovine RPE microsomes were incubated in buffers at the indicated pH values and then disrupted by nitrogen cavitation. After centrifugation, the resulting high speed pellets and supernatants were separated by SDS-PAGE. A, Coomassie-stained gel of the pellet and supernatant fractions. B, Rpe65 immunoblot of the same fractions. Note the release of Rpe65 from membranes into the supernatant between pH 10 and 11.
Nonsubstrate-containing PS, PG, and CL Liposomes Inhibit Rpe65 Isomerase Activity
Addition of all-trans-ROL to an RPE homogenate results in synthesis of all-trans-REs by lecithin: retinol acyltransferase in ER membranes (15). These membrane-bound all-trans-REs serve as substrate for Rpe65 isomerase. To confirm binding of Rpe65 to PS, PG, and CL, we prepared liposomes from these and other phospholipids, added the liposomes to Rpe65 assay mixtures containing bovine RPE homogenates and all-trans-ROL, and assayed for synthesis of 11-cis-ROL. Compared with control assay mixtures with no added liposomes, we observed significant inhibition of 11-cis-ROL synthesis in assays containing PS, PG, and CL but not PC or PE liposomes (Fig. 9A). Here, the PS, PG, and CL-containing liposomes are inhibiting isomerase activity by binding Rpe65 and restricting its access to the substrate-containing ER membranes.
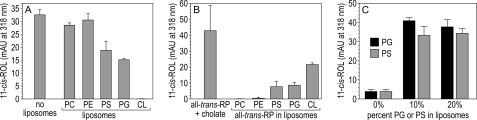
FIGURE 9.
Inhibition of Rpe65 isomerase activity and delivery of all-trans-RP substrate to Rpe65 by acidic liposomes. A, synthesis of 11-cis-ROL (in milli-absorbance units at 318 nm) from all-trans-ROL by bovine RPE homogenates containing either no liposomes (positive control) or liposomes constituted from the indicated phospholipids. Note the partial inhibition of 11-cis-ROL synthesis by liposomes containing PS and PG, complete inhibition by liposomes containing CL, and no inhibition by liposomes containing PC and PE. B, synthesis of 11-cis-ROL from all-trans-RP by bovine RPE homogenates that had been stripped of endogenous retinoids by exposure to UV light. The all-trans-RP substrate was delivered in sodium cholate detergent (positive control) or in liposomes constituted from the indicated phospholipids. Note the synthesis of 11-cis-ROL when all-trans-RP was delivered in PS, PG, or CL liposomes, but not liposomes containing PC or PE. C, isomerase activities of expressed chicken Rpe65 with liposomes of different phospholipid composition that also contain all-trans-RP substrate. Liposomes in the experiments labeled 0% contained PC, PE, and PI at the same molar ratios as human RPE membranes with no added PG or PS. Liposomes in the experiments labeled 10% contained the same phospholipids plus 10% PG or PS. Liposomes in the experiments labeled 20% contained the same phospholipids plus 20% PG or PS. Note the dramatic activation of isomerase activity (11-cis-ROL synthesis) with addition of PG or PS. Error bars shown show standard deviations (n = 3).
PS, PG, and CL Liposomes Containing All-trans-RP Serve as a Source of Substrate for Rpe65
To test the functional significance of Rpe65 binding to phospholipids, we prepared liposomes from PC, PE, PS, PG, and CL that also contain all-trans-RP. These were added to bovine RPE homogenates stripped of endogenous retinoids by exposure to UV light. We assayed for synthesis of 11-cis-ROL. As a positive control for substrate delivered in liposomes, we used all-trans-RP solubilized in sodium cholate. We observed virtually no isomerase activity when all-trans-RP was delivered in liposomes containing PC or PE (Fig. 9B). However, delivery of all-trans-RP in PS, PG, or CL liposomes resulted in significant 11-cis-ROL synthesis (Fig. 9B). These results suggest that the interaction of Rpe65 with acidic phospholipid headgroups is functionally relevant, providing access to its all-trans-RP substrate within the bilayer.
Negatively Charged Phospholipids Are Required for Membrane Association and Isomerase Activity of Rpe65
We investigated the effect on isomerase activity of increasing PG or PS concentrations in liposomes that otherwise contain the same molar ratios of phospholipids as human RPE membranes (PC/PE/PI = 2:6:1) (26). We prepared liposomes containing PC, PE, and PI at these molar ratios plus 0, 10, or 20% PG or PS. The liposomes also contained 10 μm all-trans-RP. We incubated these liposomes with homogenates of HEK-293T cells expressing chicken Rpe65 and measured 11-cis-ROL synthesis. Interestingly, we observed a 10.5-fold increase in isomerase activity when the HEK-293T cell homogenate was incubated with PC/PE/PI liposomes that also contained 10% PG versus liposomes with no added PG (Fig. 9C). No further change in isomerase activity was seen when the PG content was increased from 10 to 20% (Fig. 9C). We observed a similar pattern with PS. Addition of 10% PS to PC/PE/PI liposomes increased isomerase activity by 8.5-fold (Fig. 9C). No further increase in 11-cis-ROL synthesis was seen when the PS content of the substrate liposomes was increased from 10 to 20%. These data show that Rpe65 isomerase activity is strongly stimulated by addition of a relatively small fraction of the acidic phospholipids, PG or PS, to liposomes containing the same major phospholipids as RPE membranes.
DISCUSSION
Lecithin:retinol acyltransferase, Rpe65, and 11-cis-RDH act sequentially in RPE cells to convert all-trans-ROL into 11-cis-RAL (Fig. 1). These enzymes are located in the ER of RPE cells. Lecithin:retinol acyltransferase and 11-cis-RDH are integral membrane proteins (27, 28). The mechanism whereby Rpe65 associates with membranes is the subject of this study. We performed MS analysis on Rpe65 from bovine RPE microsomes and found that it is not significantly palmitoylated on any Cys residue. We observed minor modification of Cys-396 by our MS assay. However, because Cys-396 is not conserved (Fig. 2), palmitoylation of this residue is unlikely to explain the affinity of Rpe65 for membranes. Furthermore, the extent of Cys-396 modification (∼16%) is insufficient to account for the ∼85% association of Rpe65 with membranes (15). The starting material for our MS analysis was RPE microsomes. Accordingly, we analyzed the membrane-associated form of Rpe65. If palmitoylation is required for Rpe65 to associate with membranes, we should have observed this modification at a stoichiometry approaching 100%. As a further test for palmitoylation or other post-translational modifications, we determined the mass of undigested Rpe65 by ESI-MS following reduction with DTT or TBP. The resulting masses were 60,915 and 60,921 Da, respectively, and the calculated mass of unmodified bovine Rpe65 is 60,905 Da. The error in these determinations is ∼6 Da. Given the close agreement between the measured mass of Rpe65 from bovine eyes and the calculated mass of unmodified Rpe65, this protein is unlikely to undergo palmitoylation, which would increase its mass by 238 Da per modification, at a net stoichiometry of greater than 20%. Fatty acylation does not significantly alter ionization efficiency under the conditions used. Other post-translational modifications, such as phosphorylation, which would increase the mass by 80 Da per modified residue, are also unlikely to occur at greater than 20% stoichiometry. Low mass modifications of non-Cys residues in Rpe65, such as N-methylation or N-acetylation, may occur at high stoichiometry. However, none of these possible modifications could explain the association of Rpe65 with membranes.
In a previous study, a palmitoylation-switch mechanism was proposed whereby Rpe65 undergoes reversible palmitoylation by lecithin:retinol acyltransferase on residues Cys-231, Cys-329, and Cys-330 (14). Palmitoylation of Rpe65 was suggested as a mechanism to regulate isomerase catalytic activity. The data supporting this hypothesis were challenged in subsequent studies that showed no change in the catalytic activity of Rpe65, or its affinity for membranes, when Cys-231, Cys-329, and Cys-330 were substituted with Ala or Ser (15, 29). Furthermore, Cys-231, Cys-329, and Cys-330 were found by MS analysis not to be palmitoylated. The affinity of Rpe65 for membranes was shown to be similar in wild-type and lrat−/− knock-out mice that lack the proposed palmitoyltransferase (15). Finally, no change in the catalytic activity of Rpe65 or its affinity for membranes was seen after treatment of expressing cells with the global inhibitor of protein palmitoylation, 2-bromopalmitate (15). The results presented here provide further evidence against the palmitoylation-switch hypothesis.
More recently, it was suggested that Cys-112 is palmitoylated in Rpe65, based on MS/MS data and the observation that C112A-substituted Rpe65 was not present in the membrane fraction of expressing cells, whereas the unsubstituted protein was present (16). These data are in disagreement with the results presented here (Figs. 5 and 6). How can this discrepancy be resolved? The MS/MS spectra of the Cys-112-containing peptide presented by Takahashi et al. (16) had very high backgrounds. In such noisy spectra, a series of ions can be found that appear to support a nonexistent post-translational modification, especially when the peptides are large (>2 kDa). To make their data fit the proposed palmitoylation of Cys-112, Takahashi et al. (16) had to assume that within the same 21-residue peptide Lys-113 was acetylated and that Thr-101, Thr-105, and Ser-117 were phosphorylated and to accept cross-correlation (Xcorr) scores below 2.0 (16). Furthermore, in experiments where either Cys-106 or Cys-112 was substituted with Ala, Takahashi et al. (16) then had to assume that Lys-113 was no longer acetylated and that Thr-105 and Ser-117 or Thr-101 and Ser-117 were phosphorylated, respectively. No independent evidence for acetylation or phosphorylation of Rpe65 was ever presented. Finally, C112A-substituted Rpe65 was not present in the soluble fraction of expressing HEK-293T cells, as would be expected if palmitoylation of Cys-112 was responsible for the association of Rpe65 with membranes. Instead, C112A Rpe65 was present in the insoluble cytoskeletal pellet (16), suggesting that the substituted protein was misfolded. These results call into question the conclusion of Takahashi et al. (16) that Cys-112 is palmitoylated in Rpe65.
The crystal structure of bovine Rpe65 was recently published (8). Kiser et al. (8) found evidence against palmitoylation of Cys-231, Cys-329, or Cys-330 within the crystal structure. Unfortunately, Cys-112 was present within a disordered region; hence, their crystallographic data provided no information about the palmitoylation state of Cys-112. Based on a low resolution MS analysis, Kiser et al. (8) suggested that Cys-112 is palmitoylated. However, the data supporting this conclusion are problematic. First, if the Cys-112-containing peptide were palmitoylated, it should have eluted much later from the C18 column than the observed 14 min (8). Furthermore, ESI-MS analysis of a Cys-palmitoylated peptide should include an ion resulting from neutral loss of palmitoyl group (238 Da). No such neutral loss ions were visible in the published MS spectra of Kiser et al. (8). Finally, the MS/MS spectrum of the purported Cys-112-containing peptide only showed clear b7 ions. These ions, due to loss of Leu/Ile, are present in many peptides of a given m/z following digestion by pepsin; therefore, they provide little useful sequence information. The other b-and y-series ions, which would have identified this peptide, were either weak or noninformative. Accordingly, it appears that the peptide analyzed by Kiser et al. (8) was misidentified. In this study, the MS/MS spectrum for the Cys-112-containing peptide showed a clear y-ion series that permitted its unambiguous identification (Fig. 3B).
In a previous study, intact Rpe65 was purified from membrane and soluble fractions of expressing Sf9 cells and analyzed by matrix-assisted laser desorption ionization-MS (30). The molecular mass of Rpe65 from the membrane fractions was 0.7–1.0 kDa higher than the mass of Rpe65 from the soluble fractions (30). These results are also in disagreement with the results presented here. The mass spectra of intact Rpe65 in the previous study had peak widths of ∼10 kDa (30), indicating very low mass resolution. At such low resolution, a mass difference of 1 kDa in a 61-kDa protein cannot be reliably determined. The MS peak width of intact Rpe65 in this study was ∼50 Da (Fig. 6). Thus, the mass resolution here was ∼200-fold higher than in the previous study. Rpe65 was prepared from microsomal membranes in this study, which increased the likelihood of detecting any palmitoylated forms. However, the measured mass of Rpe65 after TBP reduction (60,921 Da) was within 16 Da of the predicted mass for unmodified Rpe65 (60,905 Da), further suggesting that Rpe65 is not significantly palmitoylated.
The affinity of Rpe65 for membranes appears instead to be mediated by a direct interaction between the protein and the negatively charged phospholipids PS, PG, and CL. We demonstrated this interaction by showing direct binding of Rpe65 to immobilized phospholipids (Fig. 7A). We also showed that PS-, PG-, or CL-containing, but not PC-or PE-containing, liposomes added to RPE homogenates inhibited synthesis of 11-cis-ROL by Rpe65 from all-trans-REs synthesized by lecithin:retinol acyltransferase in the endogenous ER membranes (Fig. 9A). Finally, we showed that Rpe65 can utilize all-trans-RP contained within PS, PG, or CL liposomes, but not PC or PE liposomes, as substrate for the synthesis of 11-cis-ROL (Fig. 9B). The latter observation establishes the physiological relevance of the interaction between Rpe65 and acidic phospholipids.
We investigated the effect of adding PG or PS to liposomes that otherwise contain the same phospholipid composition as human RPE membranes (PC/PE/PI = 2:6:1) (26) plus all-trans-RP substrate. Here, we used chicken Rpe65 expressed in HEK-293T cells as an enzyme source. When the liposomes were constituted with the addition of 10% PG, synthesis of 11-cis-ROL increased 10-fold compared with PC/PE/PI liposomes without PG (Fig. 9C). Addition of 10% PS to the same RPE/membrane phospholipid mixture yielded an 8.5-fold increase in 11-cis-ROL synthesis (Fig. 9C). This stimulation of isomerase activity appeared to saturate at or below 10% PG or PS in the substrate-containing liposomes, because we observed no further stimulation with addition of 20% PG or PS (Fig. 9C). Consistently, native RPE membranes contain less than 10% acidic phospholipids (26). These results suggest that the stimulation effect of acidic phospholipids on isomerase activity and Rpe65 binding to membranes occurs with normal RPE phospholipid composition.
Rpe65 is located in ER membranes (31). Although PS and PG are minor phospholipids of the ER, CL is thought to be present exclusively in mitochondria (32–37). Thus, the observed in vitro affinity of Rpe65 for CL liposomes (Fig. 7A) may be due to the net double-negative charge of CL versus the single-negative charges of PS and PG (Fig. 7B), and therefore may not reflect the in vivo situation. The observed affinity of Rpe65 for PS and PG probably does reflect the in vivo situation, especially considering the results presented in Fig. 9C. These interactions are likely to involve positively charged residues on the surface of Rpe65. We generated an electrostatic surface-potential map for Rpe65 based on its structure (8). Most of the protein surface is predicted to be neutral or negatively charged (Fig. 10A). However, two strongly basic segments, KKRKK (residues 294–298) and KKNARK (residues 354–359) (Fig. 2) are also present on the surface and may represent interacting structural elements. The predominant basic residue in these segments is Lys, which has a pKa of 10.5. Interestingly, Rpe65 dissociated from RPE membranes between pH 10 and 11 (Fig. 8), suggesting titration of these Lys residues. Rpe65 can also be dissociated from membranes by incubating in a buffer of high ionic strength (38). If the affinity of Rpe65 for membranes was due to palmitoylation, its association with membranes would be stronger, not weaker, at high ionic strength. These observations provide further evidence for an electrostatic interaction between membranes and Rpe65. We observed no binding of Rpe65 to phosphatidic acid (Fig. 7A), suggesting that the affinity of Rpe65 for acidic phospholipids is not exclusively electrostatic but may involve hydrogen bonding as well.
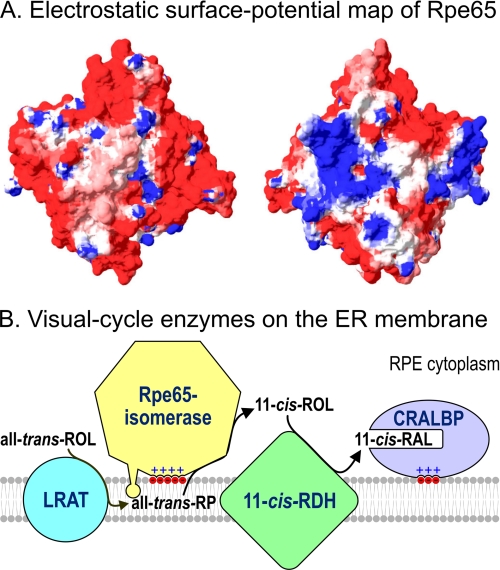
FIGURE 10.
A, electrostatic surface potential of bovine Rpe65. Regions with negative electrostatic potential (acidic) are shown in red, neutral regions are shown in white, and regions with positive electrostatic potential (basic) are shown in blue. The left and right images are from opposite sides of the protein. Note the strongly basic (blue) regions, corresponding to residues 294–298, 354–359, and other basic residues scattered throughout the sequence. B, hypothesized interactions of visual cycle proteins on the ER membrane of an RPE cell. Lecithin:retinol acyltransferase (LRAT) and 11-cis-RDH are integral-membrane proteins, whereas both Rpe65 and CRALBP (40) are soluble proteins with affinity for acidic phospholipid headgroups. Following synthesis by lecithin:retinol acyltransferase, Rpe65 extracts all-trans-RP from the membrane and isomerizes it to 11-cis-ROL. 11-cis-RDH oxidizes it to 11-cis-RAL, which binds to CRALBP and is transferred to the apical plasma membrane of the RPE cell for export to the photoreceptors.
The two basic stretches in Rpe65 are situated near the mouth of a predicted hydrophobic channel (8). The Fe2+-His4 coordination sphere, which is required for isomerase activity (3), is situated deeper in the channel and probably represents part of the catalytic site. Association of Rpe65 with acidic headgroups in the ER membrane may facilitate the extraction of an all-trans-RE from the bilayer into the hydrophobic channel and catalytic site. Lipophilic all-trans-REs may “float” within the bilayer with their polar carboxylate groups protruding into the aqueous phase. Alternatively, all-trans-REs may be located in the central cleavage plane of the bilayer, as was shown for the polyisoprenes (39). The latter possibility would require Rpe65 to extend a loop into the bilayer to facilitate extraction of the all-trans-RE (Fig. 10B). Rpe65 contains at least one disordered hydrophobic loop that could serve this function (FFSYF, residues 119–123). Penetration of this loop into the bilayer would stabilize the interaction of Rpe65 with membranes.
Acknowledgments
We gratefully acknowledge Nawajes A. Mandal and Gene Anderson for analysis of phospholipids in RPE cells and Roxana A. Radu for valuable comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants EY01584 and EY11713 from NEI.
- 11-cis-RAL
- 11-cis-retinaldehyde
- all-trans-RAL
- all-trans-retinaldehyde
- TBP
- tributylphosphine
- PG
- phosphatidylglycerol
- PS
- phosphatidylserine
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- RPE
- retinal pigment epithelium
- CL
- cardiolipin
- DTT
- dithiothreitol
- PBS
- phosphate-buffered saline
- HPLC
- high pressure liquid chromatography
- ESI
- electrospray ionization
- MS/MS
- tandem mass spectrometry
- ER
- endoplasmic reticulum
- all-trans-RP
- all-trans-retinyl palmitate
- all-trans-RE
- all-trans-retinyl ester
- 11-cis-ROL
- 11-cis-retinol
- S-palm-N-Ac-Cys-Me
- S-palmitoyl-N-acetyl-l-cysteine methyl ester
- LC
- liquid chromatography
- AA
- acrylamide.
REFERENCES
- 1.Jin M., Li S., Moghrabi W. N., Sun H., Travis G. H. (2005) Cell 122, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moiseyev G., Chen Y., Takahashi Y., Wu B. X., Ma J. X. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redmond T. M., Poliakov E., Yu S., Tsai J. Y., Lu Z., Gentleman S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redmond T. M., Yu S., Lee E., Bok D., Hamasaki D., Chen N., Goletz P., Ma J. X., Crouch R. K., Pfeifer K. (1998) Nat. Genet. 20, 344–351 [DOI] [PubMed] [Google Scholar]
- 5.Gu S. M., Thompson D. A., Srikumari C. R., Lorenz B., Finckh U., Nicoletti A., Murthy K. R., Rathmann M., Kumaramanickavel G., Denton M. J., Gal A. (1997) Nat. Genet. 17, 194–197 [DOI] [PubMed] [Google Scholar]
- 6.Redmond T. M., Gentleman S., Duncan T., Yu S., Wiggert B., Gantt E., Cunningham F. X., Jr. (2001) J. Biol. Chem. 276, 6560–6565 [DOI] [PubMed] [Google Scholar]
- 7.Kloer D. P., Ruch S., Al-Babili S., Beyer P., Schulz G. E. (2005) Science 308, 267–269 [DOI] [PubMed] [Google Scholar]
- 8.Kiser P. D., Golczak M., Lodowski D. T., Chance M. R., Palczewski K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17325–17330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gollapalli D. R., Rando R. R. (2003) Biochemistry 42, 5809–5818 [DOI] [PubMed] [Google Scholar]
- 10.Moiseyev G., Crouch R. K., Goletz P., Oatis J., Jr., Redmond T. M., Ma J. X. (2003) Biochemistry 42, 2229–2238 [DOI] [PubMed] [Google Scholar]
- 11.Mata N. L., Moghrabi W. N., Lee J. S., Bui T. V., Radu R. A., Horwitz J., Travis G. H. (2004) J. Biol. Chem. 279, 635–643 [DOI] [PubMed] [Google Scholar]
- 12.Rando R. R. (1991) Biochemistry 30, 595–602 [DOI] [PubMed] [Google Scholar]
- 13.Tsilou E., Hamel C. P., Yu S., Redmond T. M. (1997) Arch. Biochem. Biophys. 346, 21–27 [DOI] [PubMed] [Google Scholar]
- 14.Xue L., Gollapalli D. R., Maiti P., Jahng W. J., Rando R. R. (2004) Cell 117, 761–771 [DOI] [PubMed] [Google Scholar]
- 15.Jin M., Yuan Q., Li S., Travis G. H. (2007) J. Biol. Chem. 282, 20915–20924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi Y., Moiseyev G., Ablonczy Z., Chen Y., Crouch R. K., Ma J. X. (2009) J. Biol. Chem. 284, 3211–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitelegge J. P., Zhang H., Aguilera R., Taylor R. M., Cramer W. A. (2002) Mol. Cell. Proteomics 1, 816–827 [DOI] [PubMed] [Google Scholar]
- 18.Whitelegge J. P. (2004) Methods Mol. Biol. 251, 323–340 [DOI] [PubMed] [Google Scholar]
- 19.Whitelegge J. P., Gundersen C. B., Faull K. F. (1998) Protein Sci. 7, 1423–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie J., Marusich M. F., Souda P., Whitelegge J., Capaldi R. A. (2007) FEBS Lett. 581, 3545–3549 [DOI] [PubMed] [Google Scholar]
- 21.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 22.Honig B., Nicholls A. (1995) Science 268, 1144–1149 [DOI] [PubMed] [Google Scholar]
- 23.Jackson C. S., Magee A. I. (2002) in Current Protocols in Protein Science (Coligan J. E. ed) Vol. 2, pp. 14.12.11–14.12.19, John Wiley & Sons, Inc., New York [Google Scholar]
- 24.Brune D. C. (1992) Anal. Biochem. 207, 285–290 [DOI] [PubMed] [Google Scholar]
- 25.Rüegg U. T., Rudinger J. (1977) Methods Enzymol. 47, 111–116 [DOI] [PubMed] [Google Scholar]
- 26.Gülcan H. G., Alvarez R. A., Maude M. B., Anderson R. E. (1993) Invest. Ophthalmol. Vis. Sci. 34, 3187–3193 [PubMed] [Google Scholar]
- 27.Moise A. R., Golczak M., Imanishi Y., Palczewski K. (2007) J. Biol. Chem. 282, 2081–2090 [DOI] [PubMed] [Google Scholar]
- 28.Lidén M., Romert A., Tryggvason K., Persson B., Eriksson U. (2001) J. Biol. Chem. 276, 49251–49257 [DOI] [PubMed] [Google Scholar]
- 29.Takahashi Y., Moiseyev G., Chen Y., Ma J. X. (2006) Invest. Ophthalmol. Vis. Sci. 47, 5191–5196 [DOI] [PubMed] [Google Scholar]
- 30.Ma J., Zhang J., Othersen K. L., Moiseyev G., Ablonczy Z., Redmond T. M., Chen Y., Crouch R. K. (2001) Invest. Ophthalmol. Vis. Sci. 42, 1429–1435 [PubMed] [Google Scholar]
- 31.Huang J., Possin D. E., Saari J. C. (2009) Mol. Vis. 15, 223–234 [PMC free article] [PubMed] [Google Scholar]
- 32.Kuge O., Nishijima M. (2003) J. Biochem. 133, 397–403 [DOI] [PubMed] [Google Scholar]
- 33.Bleasdale J. E., Tyler N. E., Snyder J. M. (1985) Lung 163, 345–359 [DOI] [PubMed] [Google Scholar]
- 34.Poorthuis B. J., Hostetler K. Y. (1976) J. Biol. Chem. 251, 4596–4602 [PubMed] [Google Scholar]
- 35.Matsuzawa Y., Hostetler K. Y. (1980) J. Lipid Res. 21, 202–214 [PubMed] [Google Scholar]
- 36.Houtkooper R. H., Vaz F. M. (2008) Cell. Mol. Life Sci. 65, 2493–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlame M., Rua D., Greenberg M. L. (2000) Prog. Lipid Res. 39, 257–288 [DOI] [PubMed] [Google Scholar]
- 38.Choo D. W., Cheung E., Rando R. R. (1998) FEBS Lett. 440, 195–198 [DOI] [PubMed] [Google Scholar]
- 39.Hauss T., Dante S., Haines T. H., Dencher N. A. (2005) Biochim. Biophys. Acta 1710, 57–62 [DOI] [PubMed] [Google Scholar]
- 40.Saari J. C., Nawrot M., Stenkamp R. E., Teller D. C., Garwin G. G. (2009) Mol. Vis. 15, 844–854 [PMC free article] [PubMed] [Google Scholar]