Abstract
Escherichia coli was isolated from the feces of 122 piglets with diarrhea on 55 farms in Korea. The virulence genes of each isolate were characterized by polymerase chain reaction (PCR). Of the 562 isolates, 191 carried 1 or more of the virulence genes tested for in this study. Of the 191 isolates, 114 (60%) carried 1 or more of the genes for enterotoxigenic E. coli (ETEC) fimbriae F4, F5, F6, F18, and F41 and ETEC toxins LT, STa, and STb, 57 (30%) carried 1 or more of the genes for the Shiga-toxin-producing E. coli (STEC) toxins Stx1, Stx2, and Stx2e, and 21% and 37% carried the gene for enteropathogenic E. coli intimin and for enteroaggregative E. coli toxin, respectively. Collectively, our results indicate that other pathotypes of E. coli as well as ETEC can be strongly associated with diarrhea in piglets. In addition, detection of the genes for Stx1 and Stx2 indicates that pigs are reservoirs of human pathogenic STEC.
Résumé
Escherichia coli a été isolé des fèces de 122 porcelets avec de la diarrhée provenant de 55 fermes situées en Corée. Les gènes de virulence de chaque isolat ont été caractérisés par réaction d’amplification en chaîne par la polymérase (PCR). Parmi les 562 isolats, 191 étaient porteurs d’au moins un des gènes de virulence testés dans la présente étude. Parmi les 191 isolats, 114 (60 %) étaient porteurs d’un gène ou plus des fimbriae F4, F5, F6, F18 et F41 et des toxines LT, STa et STb associés aux souches entérotoxinogéniques de E. coli (ETEC), 57 (30 %) étaient porteurs d’au moins un des gènes Stx1, Stx2 et Stx3e des E. coli producteurs de Shiga-toxine (STEC), et 21 % et 37 % étaient respectivement porteurs du gène de l’intimine des E. coli entéropathogènes et de la toxine des E. coli entéro-aggrégatifs. Dans son ensemble, les résultats indiquent que les ETEC ainsi que des pathotypes différents peuvent être fortement associés à la diarrhée chez les porcelets. De plus, la détection des gènes codant pour Stx1 et Stx2 indique que les porcs sont des réservoirs pour les STEC pathogènes humains.
(Traduit par Docteur Serge Messier)
Pathogenic Escherichia coli is a common agent responsible for a variety of intestinal disorders, such as diarrhea and edema disease syndrome in pigs (1,2). To cause disease, enterotoxigenic E. coli (ETEC) must colonize the mucosal surface of the intestine using surface proteins known as fimbriae and produce enterotoxins, whether heat-stable (STa, STb), heat-labile (LT), or both (3,4). The known porcine fimbriae are F4 (K88), F5 (K99), F6 (987P), F18, and F41 (2,3). Different types of fimbriae can be associated with ETEC diarrhea in animals of different ages (3,4). In piglets under 5 d of age, F4-positive strains of E. coli cause more severe diarrhea, whereas F5, F6, and F41 strains cause milder diarrhea with a later onset (between 4 and 14 d of age) (3,4). Postweaning diarrhea (PWD) is the most constant disease problem at large-scale farms and particularly among piglets weaned at 3 to 4 wk of age (3). The main adhesive virulence factors of ETEC associated with PWD are F4 (mainly K88ac) and F18 fimbriae (3–5).
The serologically diverse group of Shiga-toxin-producing E. coli (STEC), also known as verotoxin-producing E. coli, causes disease in humans and animals (6). The common feature of STEC strains is the production of Shiga toxin 1 (Stx1), Shiga toxin 2 (Stx2), a variant of Stx1 (Stx1c, Stx1d), a variant of Stx2 (Stx2c, Stx2d, Stx2e, Stx2f, Stx2g), or a combination of these toxins (4). The Stx2e variant is produced by the STEC strains that cause edema disease in swine (4). In addition to ETEC and STEC, strains that cause attaching and effacing lesions, similar to those produced by enteropathogenic E. coli (EPEC) in humans, have also been associated with diarrhea in pigs (2). These attaching and effacing E. coli possess the eae gene encoding the outer membrane protein known as intimin, which is involved in attachment of the bacteria to the surface of epithelial cells in the gastrointestinal tract (2). The eae gene is a key marker of EPEC infections (7).
The distinctive aggregative pattern of adhesion to cultured human epithelial cells in vitro defines enteroaggregative E. coli. These strains produce an enterotoxin known as enteroaggregative heat-stable enterotoxin 1 (EAST1) (2,5). The astA gene encoding EAST1 has also been detected in E. coli of different pathogenic strains, such as ETEC, EPEC, and STEC from humans and animals (2,8).
The present study investigated the distribution of pathotypes of enterovirulent E. coli isolated from piglets with diarrhea in Korea and the characteristics of their virulence genes. This kind of work had not been carried out previously in Korea, and the results offer significant epidemiologic information on porcine EPEC.
The strains were isolated from the feces of 122 piglets with diarrhea aged 2 to 72 d on 55 farms in Korea. The fecal samples, obtained per rectum, were inoculated directly onto eosin methylene blue and MacConkey agar (Difco, Sparks, Maryland, USA) and identified by means of the API 32E system (bioMérieux, Marcy l’Etoile, France). Three to eight E. coli colonies per sample were collected randomly for study. The reference E. coli strains used were O157:H7 (American Type Culture Collection no. 43894; intimin+, Stx1+, and Stx2+), G.C.V. (K88+), O9:K35 E-92 (K99+ and F41+), and O141:K85ab E-127 (987P+ and STa+), which were kindly supplied by the National Veterinary Research and Quarantine Service, Anyang, Korea. In addition, the E. coli strains JOL500 (F18+, LT+, STa+, STb+, and Stx2e+) and JOL538 (EAST1+) isolated in this study were used for reference.
The isolates were cultured on blood agar containing 5% sheep blood for 18 h at 37°C, and the presence of hemolysis was determined visually. A polymerase chain reaction (PCR) was used to detect the genes for the adhesins F4, F5, F6, F18, F41 and intimin and the toxins LT, STa, STb, Stx1, Stx2, Stx2e, and EAST1 (Table I), as previously described (2,9,10,11). Fisher’s exact test was used to analyze the correlation between the presence of adhesin genes and the distribution of adhesin genes within weaning and hemolysis status (12). The results were considered significant when P-values were less than 0.05. All statistical analyses were carried out with the SPSS 16.0 program (SPSS, Chicago, Illinois, USA).
Table I.
Polymerase chain reaction primers used in this study to detect genes for Escherichia coli virulence factors and the product sizes
| Virulence factor | Nucleotide sequence | Sizea | Primer coordinates | Accession number | Reference number |
|---|---|---|---|---|---|
| F4 | GCTGCATCTGCTGCATCTGGTAGTG CCACTGAGTGCTGGTAGTTACAGCC |
792 | 31–54 798–822 |
M29374 | 2 |
| F5 | TATTATCTTAGGTGGTATGG GGTATCCTTTAGCAGCAGTATTTC |
314 | 21–40 311–334 |
M35282 | 9 |
| F6 | TCTGCTCTTAAAGCTACTGG AACTCCACCGTTTGTATCAG |
333 | 193–212 506–525 |
M35257 | 2 |
| F18 | GTGAAAAGACTAGTGTTTATTTC CTTGTAAGTAACCGCGTAAGC |
510 | 160–182 649–669 |
M61713 | 10 |
| F41 | GCATCAGCGGCAGTATCT GTCCCTAGCTCAGTATTATCACCT |
380 | 34–51 390–413 |
X14354 | 9 |
| Intimin | GACCCGGCACAAGCATAAGC CCACCTGCAGCAACAAGAGG |
384 | 27–46 391–410 |
A3334567 | 11 |
| LT | ATTTACGGCGTTACTATCCTC TTTTGGTCTCGGTCAGATATG |
281 | 27–47 287–307 |
S60731 | 2 |
| STa | GCTAATGTTGGCAATTTTTATTTCTGTA AGGATTACAACAAAGTTCACAGCAGTAA |
190 | 9–36 171–198 |
M25607 | 9 |
| STb | GCCTATGCATCTACACAATC TGAGAAATCGACAATGTCCG |
279 | 515–534 773–793 |
AY028790 | 2 |
| EAST1 | CCATCAACACAGTATATCCGA GGTCGCGAGTGACGGCTTTGT |
111 | 2–24 94–114 |
S81691 | 2 |
| Stx1 | ATAAATCGCCATTCGTTGACTAC AGAACGCCCACTGAGATCATC |
180 | 424–446 593–603 |
EU754740 | 11 |
| Stx2 | GGCACTGTCTGAAACTGCTCC TCGCCAGTTATCTGACATTCTG |
255 | 606–626 839–860 |
EU816442 | 11 |
| Stx2e | CCACCAGGAAGTTATATTTCC TTCACCAGTTGTATATAAAGA |
759 | 223–243 961–981 |
U72191 | 10 |
Number of base pairs.
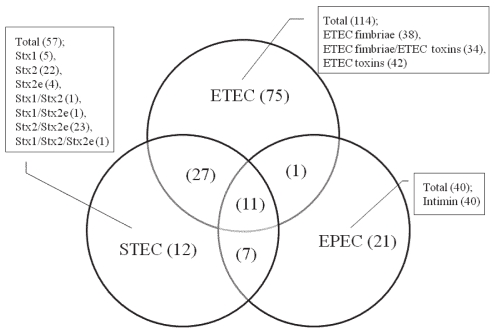
Of the 562 isolates tested, 191 carried 1 or more of the virulence-associated genes, and 114 (60%) of the 191 carried 1 or more of the genes for ETEC fimbriae and toxins (Table II). Of the 114 isolates, 38, 34, and 42 possessed 1 or more of the genes for ETEC fimbriae only, ETEC fimbriae and ETEC toxins, and ETEC toxins only, respectively (Figure 1). Among the 72 isolates carrying the genes for fimbriae, the most prevalent fimbrial gene was that for F18, which was identified in 30 isolates (16% of the 191). Among the 76 isolates carrying the genes for ETEC toxins, the most prevalent toxin gene was that for LT, which was identified in 44 isolates (23% of the 191). The most common combination of the genes for ETEC fimbriae and ETEC toxins was F18/LT/STa/STb, which was detected in 6 (3%) of the 191 isolates. Among the 57 isolates (30%) carrying 1 or more genes for Shiga toxins, the most prevalent Shiga toxin gene was Stx2, which was detected in 47 isolates (25% of the 191). Of the 191 isolates, 40 (21%) carried the intimin gene, which represents EPEC (Figure 1), and 78 (40.8%) possessed the gene for EAST1 (Table II).
Table II.
Distribution of genes for adhesins and toxins among 191 E. coli isolates from piglets with diarrhea in Korea
| Adhesins; no. of isolates carrying the genes |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Toxins | F4 | F5 | F6 | F18 | F41 | Intimin | F4/41 | F5/41 | F6/18 | F18/intimin | F41/intimin | F5/6/18 | F5/18/41 | F5/41/intimin | Noneb | Total |
| LT | 1 | 1 | ||||||||||||||
| STa | 3 | 3 | 1 | 3 | 10 | |||||||||||
| STb | 1 | 8 | 9 | |||||||||||||
| Stx1 | 1 | 1 | 2 | 4 | ||||||||||||
| Stx2 | 1 | 3 | 2 | 4 | 7 | 17 | ||||||||||
| Stx2e | 1 | 2 | 3 | |||||||||||||
| EAST1 | 8 | 37 | 45 | |||||||||||||
| LT/Stx1 | 1 | 1 | ||||||||||||||
| LT/Stx2 | 1 | 1 | ||||||||||||||
| LT/EAST1 | 3 | 1 | 10 | 14 | ||||||||||||
| STa/STb | 4 | 4 | ||||||||||||||
| STa/EAST1 | 1 | 3 | 4 | |||||||||||||
| Stx1/Stx2 | 1 | 1 | ||||||||||||||
| Stx2/Stx2e | 2 | 2 | 1 | 5 | ||||||||||||
| Stx2/EAST1 | 1 | 1 | 2 | |||||||||||||
| Stx2e/EAST1 | 1 | 1 | ||||||||||||||
| LT/STa/STb | 1 | 1 | ||||||||||||||
| LT/STa/EAST1 | 1 | 1 | 2 | |||||||||||||
| LT/STb/Stx2 | 1 | 1 | ||||||||||||||
| LT/Stx2/Stx2e | 1 | 2 | 2 | 1 | 1 | 7 | ||||||||||
| LT/Stx2/EAST1 | 1 | 1 | ||||||||||||||
| STa/STb/EAST1 | 3 | 2 | 5 | |||||||||||||
| LT/STa/STb/EAST1 | 1 | 1 | 2 | |||||||||||||
| LT/STa/Stx2/Stx2e | 2 | 3 | 1 | 6 | ||||||||||||
| LT/STa/STb/Stx2/ | ||||||||||||||||
| Stx2e | 4 | 4 | ||||||||||||||
| LT/STa/Stx1/Stx2/ | 1 | 1 | ||||||||||||||
| Stx2e | ||||||||||||||||
| LT/STa/Stx1/ | ||||||||||||||||
| Stx2e/EAST1 | 1 | 1 | ||||||||||||||
| LT/STa/STb/Stx2/ | 1 | 1 | ||||||||||||||
| Stx2e/EAST1 | ||||||||||||||||
| Nonea | 7 | 4 | 2 | 3 | 8 | 13 | ||||||||||
| Total | 12 | 6 | 9 | 18 | 11 | 32 | 1 | 1 | 3 | 6 | 1 | 2 | 1 | 1 | 87 | 191 |
No genes carried for LT, STa, STb, Stx1, Stx2, or Stx2e.
No genes carried for F4, F5, F6, F18, F41, or intimin.
Figure 1.
Distribution and relationship of the genes for virulence factors of enterotoxigenic Escherichia coli (ETEC), Shiga-toxin-producing E. coli (STEC), and enteropathogenic E. coli (EPEC) among 191 isolates from piglets with diarrhea in Korea. The numbers in parenthesis are the numbers of isolates carrying the genes.
Of the 114 ETEC isolates, the 57 STEC isolates, and the 40 EPEC isolates, 75 (66%), 12 (21%), and 21 (53%) carried the genes for only ETEC, STEC, and EPEC, respectively. Of the 191 isolates, 27 (14%) carried the genes for ETEC/STEC, 1 (0.5%) the genes for ETEC/EPEC, 7 (4%) the genes for STEC/EPEC, and 11 (6%) the genes for ETEC/STEC/EPEC (Figure 1). Of the 71 EAST1-positive isolates, 37 (52%) carried the gene for EAST1 only; 18 (25%), 8 (11%), 5 (7%), 2 (3%), and 1 (1%) carried the genes for ETEC/EAST1, EPEC/EAST1, ETEC/STEC/EAST1, STEC/EPEC/EAST1, and ETEC/STEC/EPEC/EAST1, respectively (Table II). Among these isolates, 5 contained 7 virulence genes, the most complex combinations observed in this study: F18/LT/STa/STb/Stx2/Stx2e/EAST1 (in 1 isolate) and F18/intimin/LT/STa/STb/Stx2/Stx2e (in 4 isolates).
Of the 191 isolates with virulence-associated genes, 86 (45%) and 105 (55%) were isolated from pre- and postweaned piglets, respectively, and 140 (73%) were hemolytic (Table III). The genes for F4, F5, F6, F41, and intimin were isolated from pre- and postweaned piglets (P > 0.1), irrespective of hemolytic activity (P > 0.1). However, the gene for F18 was detected significantly more often among the isolates from pigs with PWD and the hemolytic isolates (P < 0.001).
Table III.
Distribution of adhesin genes according to piglet weaning status and hemolytic activity in the 191 isolates
| Weaning status (no. of isolates) | Hemolysis (no. of isolates) | Adhesins; no. of isolates carrying the genes |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F4 | F5 | F6 | F18 | F41 | Intimin | F4/F41 | F5/F41 | F6/F18 | F18/intimin | F41/intimin | F5/F6/F18 | F5/F18/F41 | F5/F41/intimin | ||
| Preweaned | 47 | 4 | 2 | 2 | 0 | 1 | 9 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | |
| −86 | − (39) | 1 | 0 | 6 | 0 | 4 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Postweaned | 93 | 6 | 4 | 1 | 18 | 4 | 15 | 0 | 0 | 2 | 6 | 1 | 2 | 1 | 0 |
| −105 | − (12) | 1 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total (191) | 12 | 6 | 9 | 18 | 11 | 32 | 1 | 1 | 3 | 6 | 1 | 2 | 1 | 1 | |
This study examined the presence of genes for adhesins, enterotoxins, and Shiga toxins representing the virulence factors and their associations in the E. coli isolated from piglets with diarrhea. The genes for ETEC, STEC, EPEC, and EAST1 were detected in 114, 57, 40, and 71 of the 562 isolates tested, respectively. Isolates containing the specific genes for ETEC were more common than the other types of E. coli. Among ETEC fimbriae, F4 (K88) is the most prevalent contributor to diarrhea in piglets in many countries (2,13). However, in our study the gene for F4 was detected in only 13 (7%) of the 191 isolates with virulence-associated genes. It is believed that the immune pressure elicited by vaccination programs involving mainly F4 antigen may have caused a lower prevalence of F4 fimbriae in ETEC (14,15). Infections with ETEC having the F5, F6, and F41 fimbriae are age-related, occurring primarily in pigs less than 2 wk old (3,4,15). However, this study detected the genes for F4, F5, F6, and F41 fimbriae at similar rates in both pre- and postweaned piglets with diarrhea (P > 0.1), which suggests that these fimbriae are not limited to ETEC causing disease in young piglets. A newly identified adhesin, F18, was the most prevalent type of fimbriae in this study. Moreover, the gene for F18 was isolated significantly more often from piglets with PWD (P < 0.001). Collectively, our data suggest a need for new vaccination programs to protect pre- and postweaned piglets from PWD. In addition, our results imply that F18 has become the main type of E. coli associated with diarrhea and must be considered in vaccination programs.
Among the ETEC toxins, STb is the most frequently isolated from piglets with diarrhea (2,16,17). However, in this study the genes for LT and STa were more widespread than the gene for STb. In addition, this study found the gene combinations for LT/STa, LT/STa/STb, and STa/STb in approximately 5% of isolates each, whereas the gene combination for LT/STb was detected in only 0.5% of the isolates, unlike the rate previously reported (17). Overall, these results suggest that E. coli disease in piglets may be associated not with a single toxin and a major gene combination but with more varied and complex toxins as well as their combinations.
The toxins such as Stx1 and Stx2 produced by human, bovine, and ovine STEC are associated with hemorrhagic colitis and hemolytic–uremic syndrome in humans (2,18). Interestingly, in our study the genes for Stx1 and Stx2 were detected in 4% and 25% of isolates, whereas previous studies could not detect these genes in any pig isolates tested (2,19). In addition, positivity for F6, F18, and intimin was significantly more frequent among the isolates producing STEC toxins (P < 0.05). Thus, our results indicate that piglets are also reservoirs of STEC pathogenic for humans and that intimin as well as F18 must be considered in vaccination programs for piglets.
The eae gene encoding intimin has been recovered from E. coli associated with porcine neonatal diarrhea (1) as well as PWD (2,20). Martins et al (1) reported that most eae-positive isolates were able to produce STa or Stx2e. Similarly, this study detected eae in 20.9% of 191 isolates. On the other hand, the role of EAST1 in swine colio-bacillosis has not been clearly determined. However, several studies have reported that EAST1 is also distributed among other types of E. coli, such as ETEC, and can cause diarrhea in piglets in the presence of other virulence factors, such as enterotoxins (LT, STa, STb), and specific colonization factors, such as F4 (2). We also found a high prevalence (37%) of the EAST1 gene (astA), and approximately 50% of the astA-positive isolates possessed at least 1 of these virulence factors. These results suggest that EAST1-positive strains are also largely distributed and strongly associated with disease in piglets in this area.
Approximately 37% of the 191 isolates harbored genes for more than 1 pathotype-representative factor. These isolates can be classified into 9 pathotypes according to possession of the virulence genes: ETEC/STEC, ETEC/EPEC, ETEC/EAST1, STEC/EPEC, EPEC/EAST1, ETEC/STEC/EPEC, ETEC/STEC/EAST1, STEC/EPEC/EAST1, and ETEC/STEC/EPEC/EAST1. These types are more varied and complex than recent studies have indicated (2,16). The observation of these complex pathotypes of E. coli may indicate the evolution of pathogenic E. coli into forms surviving against the various strategies used for prevention and treatment by acquisition of the necessary virulence genes.
Adhesin genes other than that for F18 fimbriae were not significantly associated with hemolytic activity (P > 0.1), in agreement with the report that hemolysis of E. coli is not an absolute indication for pathogenicity in pigs (1,4). However, all isolates carrying the F18 fimbrial gene were hemolytic. This indicates that hemolytic isolates contained significantly more F18 (P < 0.001), and a test for hemolysis can be a useful indication of the pathogenicity of E. coli causing disease in piglets or at least for the presence of the F18 fimbrial gene.
Acknowledgments
This study was supported by grant RTI05-03-02 from the Regional Technology Innovation Program of the Ministry of Commerce, Industry and Energy (MOCIE) and by the Brain Korea 21 Project in the Republic of Korea.
References
- 1.Martins MDF, Martinez-Rossi NM, Ferreira A, et al. Pathogenic characteristics of Escherichia coli strains isolated from newborn piglets with diarrhea in Brazil. Vet Microbiol. 2000;76:51–59. doi: 10.1016/s0378-1135(00)00223-6. [DOI] [PubMed] [Google Scholar]
- 2.Vu-Khac H, Holoda E, Pilipčcinec E, et al. Serotypes, virulence genes, intimin types and PFGE profiles of Escherichia coli isolated from piglets with diarrhea in Slovakia. Vet J. 2007;174:176–187. doi: 10.1016/j.tvjl.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 3.Nagy B, Fekete PZ. Enterotoxigenic E. coli (ETEC) in farm animals. Vet Res. 1999;30:259–284. [PubMed] [Google Scholar]
- 4.Vu-Khac H, Holoda E, Pilipčcinec E. Distribution of virulence genes in Escherichia coli strains isolated from diarrhoeic piglets in the Slovak Republic. J Vet Med B Infect Dis Vet Public Health. 2004;51:343–347. doi: 10.1111/j.1439-0450.2004.00769.x. [DOI] [PubMed] [Google Scholar]
- 5.Smeds A, Pertovaara M, Timonen T, Pohjanvirta T, Pelkonen S, Palva A. Mapping the binding domain of the F18 fimbrial adhesin. Infect Immun. 2003;71:2163–2172. doi: 10.1128/IAI.71.4.2163-2172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettelheim KA. Non-O157 verotoxin-producing Escherichia coli: A problem, paradox, and paradigm. Exp Biol Med. 2003;228:333–344. doi: 10.1177/153537020322800402. [DOI] [PubMed] [Google Scholar]
- 7.Hornitzky MA, Mercieca K, Bettelheim KA, Djordjevic SP. Bovine feces from animals with gastrointestinal infections are a source of serologically diverse atypical enteropathogenic Escherichia coli and Shiga toxin-producing E. coli strains that commonly possess intimin. Appl Environ Microbiol. 2005;71:3405–3412. doi: 10.1128/AEM.71.7.3405-3412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osek J. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) gene and its relationship with fimbrial and enterotoxin markers in E. coli isolates from pigs with diarrhea. Vet Microbiol. 2003;91:65–72. doi: 10.1016/s0378-1135(02)00262-6. [DOI] [PubMed] [Google Scholar]
- 9.Franck SM, Bosworth BT, Moon HW. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J Clin Microbiol. 1998;36:1795–1797. doi: 10.1128/jcm.36.6.1795-1797.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imberechts H, De Greve H, Schlicker C, et al. Characterization of F107 fimbriae of Escherichia coli 107/86, which causes edema disease in pigs, and nucleotide sequence of the F107 major fimbrial subunit gene, fedA. Infect Immun. 1992;60:1963–1971. doi: 10.1128/iai.60.5.1963-1971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson MP, Newland JW, Holmes RK, O’Brien AD. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb Pathog. 1987;2:147–153. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 12.Niewerth U, Frey A, Voss T, et al. The AIDA autotransporter system is associated with F18 and Stx2e in Escherichia coli isolates from pigs diagnosed with edema disease and postweaning diarrhea. Clin Diagn Lab Immunol. 2001;8:143–149. doi: 10.1128/CDLI.8.1.143-149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexa P, Stouračcova K, Hamrik J, Rychlik I. Gene typing of the colonisation factor K88 (F4) in enterotoxigenic Escherichia coli strains isolated from diarrhoeic piglets. Vet Med-Czech. 2001;46:46–49. [Google Scholar]
- 14.Chen X, Gao S, Jiao X, Liu XF. Prevalence of serogroups and virulence factors of Escherichia coli strains isolated from pigs with postweaning diarrhea in eastern China. Vet Microbiol. 2004;103:13–20. doi: 10.1016/j.vetmic.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Söderlind O, Thafvelin B, Möllby R. Virulence factors in Escherichia coli strains isolated from Swedish piglets with diarrhea. J Clin Microbiol. 1988;26:879–884. doi: 10.1128/jcm.26.5.879-884.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frydendahl K. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhea and edema disease in pigs and a comparison of diagnostic approaches. Vet Microbiol. 2002;85:169–182. doi: 10.1016/s0378-1135(01)00504-1. [DOI] [PubMed] [Google Scholar]
- 17.Osek J, Truszczyñski M. Occurrence of fimbriae and enterotoxins in Escherichia coli strains isolated from piglets in Poland. Comp Immunol Microbiol Infect Dis. 1992;15:285–292. doi: 10.1016/0147-9571(92)90008-f. [DOI] [PubMed] [Google Scholar]
- 18.Coombes BK, Wickham ME, Mascarenhas M, Gruenheid S, Finlay BB, Karmali MA. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing strains. Appl Environ Microbiol. 2008;74:2153–2160. doi: 10.1128/AEM.02566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osek J, Gallien P, Truszczyñski M, Protz D. The use of polymerase chain reaction for determination of virulence factors of Escherichia coli strains isolated from pigs in Poland. Comp Immunol Microbiol Infect Dis. 1999;22:163–174. doi: 10.1016/s0147-9571(98)00083-6. [DOI] [PubMed] [Google Scholar]
- 20.Malik A, Tóth I, Beutin L, et al. Serotypes and intimin types of intestinal and faecal strains of eae+ Escherichia coli from weaned pigs. Vet Microbiol. 2006;114:82–93. doi: 10.1016/j.vetmic.2005.11.044. [DOI] [PubMed] [Google Scholar]