Abstract
Idiopathic generalized epilepsies account for 30% of all epilepsies. Despite a predominant genetic aetiology, the genetic factors predisposing to idiopathic generalized epilepsies remain elusive. Studies of structural genomic variations have revealed a significant excess of recurrent microdeletions at 1q21.1, 15q11.2, 15q13.3, 16p11.2, 16p13.11 and 22q11.2 in various neuropsychiatric disorders including autism, intellectual disability and schizophrenia. Microdeletions at 15q13.3 have recently been shown to constitute a strong genetic risk factor for common idiopathic generalized epilepsy syndromes, implicating that other recurrent microdeletions may also be involved in epileptogenesis. This study aimed to investigate the impact of five microdeletions at the genomic hotspot regions 1q21.1, 15q11.2, 16p11.2, 16p13.11 and 22q11.2 on the genetic risk to common idiopathic generalized epilepsy syndromes. The candidate microdeletions were assessed by high-density single nucleotide polymorphism arrays in 1234 patients with idiopathic generalized epilepsy from North-western Europe and 3022 controls from the German population. Microdeletions were validated by quantitative polymerase chain reaction and their breakpoints refined by array comparative genomic hybridization. In total, 22 patients with idiopathic generalized epilepsy (1.8%) carried one of the five novel microdeletions compared with nine controls (0.3%) (odds ratio = 6.1; 95% confidence interval 2.8–13.2; χ2 = 26.7; 1 degree of freedom; P = 2.4 × 10−7). Microdeletions were observed at 1q21.1 [Idiopathic generalized epilepsy (IGE)/control: 1/1], 15q11.2 (IGE/control: 12/6), 16p11.2 IGE/control: 1/0, 16p13.11 (IGE/control: 6/2) and 22q11.2 (IGE/control: 2/0). Significant associations with IGEs were found for the microdeletions at 15q11.2 (odds ratio = 4.9; 95% confidence interval 1.8–13.2; P = 4.2 × 10−4) and 16p13.11 (odds ratio = 7.4; 95% confidence interval 1.3–74.7; P = 0.009). Including nine patients with idiopathic generalized epilepsy in this cohort with known 15q13.3 microdeletions (IGE/control: 9/0), parental transmission could be examined in 14 families. While 10 microdeletions were inherited (seven maternal and three paternal transmissions), four microdeletions occurred de novo at 15q13.3 (n = 1), 16p13.11 (n = 2) and 22q11.2 (n = 1). Eight of the transmitting parents were clinically unaffected, suggesting that the microdeletion itself is not sufficient to cause the epilepsy phenotype. Although the microdeletions investigated are individually rare (<1%) in patients with idiopathic generalized epilepsy, they collectively seem to account for a significant fraction of the genetic variance in common idiopathic generalized epilepsy syndromes. The present results indicate an involvement of microdeletions at 15q11.2 and 16p13.11 in epileptogenesis and strengthen the evidence that recurrent microdeletions at 15q11.2, 15q13.3 and 16p13.11 confer a pleiotropic susceptibility effect to a broad range of neuropsychiatric disorders.
Keywords: idiopathic generalized epilepsy, microdeletions, association, genetics
Introduction
The idiopathic generalized epilepsies (IGEs) affect up to 0.3% of the general population and account for 30% of all epilepsies (Jallon and Latour, 2005). The clinical features are characterized by the age-related occurrence of recurrent unprovoked generalized seizures in the absence of detectable brain lesions or metabolic abnormalities (ILAE, 1989). Childhood and juvenile absence epilepsy, juvenile myoclonic epilepsy and epilepsies with generalized tonic–clonic seizures alone represent the most common IGE syndromes (ILAE, 1989). The electroencephalographic signature of IGE seizures is marked by generalized spike–wave discharges, which reflect a synchronized hyperexcitable state of thalamocortical circuits (Blumenfeld, 2005).
Genetic factors play a predominant role in the aetiology of common IGE syndromes. Heritability estimates are >80% and recurrence risk for first-degree relatives varies between 4% and 9% (Helbig et al., 2008). Molecular genetic approaches have identified causative gene mutations in mainly rare monogenic forms of idiopathic epilepsies. Most of the currently known genes for human idiopathic epilepsies encode voltage- or ligand-gated ion channels (Reid et al., 2009). Despite extensive research, the genetic variants predisposing to common IGE syndromes remain elusive. The genetic architecture is likely to display a biological continuum, in which a small fraction follows monogenic inheritance, whereas the majority of IGE patients presumably display an oligo-/polygenic predisposition.
The role of copy number variations (CNVs) in human disease, and especially in neuropsychiatric disorders, is becoming increasingly evident (Cook and Scherer, 2008; Slavotinek, 2008; Mefford and Eichler, 2009; Sharp, 2009). While many of the observed structural genomic variations have been detected only in individual patients, other CNVs are found recurrently at low frequencies, either de novo or inherited (Itsara et al., 2009a; Mefford and Eichler, 2009; Sharp, 2009). In particular, pathogenic significance of CNVs has been shown for genomic rearrangements flanked by segmental duplications, which promote non-allelic homologous recombinations resulting in recurrent microdeletions or microduplications (Gu et al., 2008; Itsara et al., 2009a). Structural genomic variations in these rearrangement hotspot regions represent many of the genomic disorders identified to date (Slavotinek, 2008; Mefford and Eichler, 2009). It is therefore possible that a bulk of rare CNVs occurring in excess in common disorders collectively explain a substantial fraction of the disease heritability.
A recurrent microdeletion at 15q13.3 was recently shown to constitute a genetic risk factor for common IGE syndromes and was found in 1% of IGE patients whereas it was not detected in controls (Helbig et al., 2009). This association was confirmed in an independent IGE sample (Dibbens et al., 2009). This microdeletion was originally described in patients exhibiting mental retardation associated with seizures (Sharp et al., 2008), and subsequently in patients with schizophrenia (Schizophrenia Consortium, 2008; Stefansson et al., 2008; Kirov et al., 2009), psychotic disorder (Miller et al., 2009), autism (Miller et al., 2009; Pagnamenta et al., 2009) and developmental delay (van Bon et al., 2009). The broad phenotypic spectrum associated with the 15q13.3 microdeletion suggests that shared mechanisms might be involved in the pathogenesis of seemingly unrelated neuropsychiatric disorders. Accordingly, the question arises whether additional recurrent microdeletions associated with neuropsychiatric disorders also confer risk to common IGE syndromes.
Five additional large microdeletions at 1q21.1, 15q11.2, 16p11.2, 16p13.11 and 22q11.2 are found recurrently in patients either affected by schizophrenia, psychotic disorder, autism or mental retardation (Sebat et al., 2007; Ullmann et al., 2007; Basset et al., 2008; Brunetti-Pierri et al., 2008; Cook and Scherer, 2008; Kumar et al., 2008; Marshall et al., 2008; Mefford et al., 2008; Schizophrenia Consortium, 2008; Sharp et al., 2009; Slavotinek, 2008; Stefansson et al., 2008; Weiss et al., 2008; Hannes et al., 2009; Itsara et al., 2009b; Kirov et al., 2009; Need et al., 2009) and some of the patients reported in the previous studies are also affected by seizures. This association study examined the role of these five recurrent microdeletions in the aetiology of common IGE syndromes.
Subjects and methods
Choice of candidate microdeletions
The selection of microdeletions at 1q21.1, 15q11.2, 16p11.2, 16p13.11 and 22q11.2 was based on previous large-scale copy number variation analyses (1q21.1: Brunetti-Pierri et al., 2008; Mefford et al., 2008; Schizophrenia Consortium, 2008; Stefansson et al., 2008; Need et al., 2009; 15q11.2: Stefansson et al., 2008; Kirov et al., 2009; 16p11.2: Sebat et al., 2007; Kumar et al., 2008; Marshall et al., 2008; Weiss et al., 2008; 16p13.11: Ullmann et al., 2007; Hannes et al., 2009; Need et al., 2009; 22q11.2: Basset et al., 2008; Need et al., 2009; Schizophrenia Consortium, 2008) and a recent meta-analysis (Itsara et al., 2009b) in neuropsychiatric disorders including autism, intellectual disability and schizophrenia (Table 1). The following inclusion criteria for the selection of candidate microdeletions were applied: (i) recurrent non-allelic homologous recombination-generated microdeletion (equal size and defined breakpoints); (ii) previous association of the microdeletion with neuropsychiatric disorders (P < 0.05) and (iii) size of the deletion larger than >400 kb to ensure a reliable detection by the Affymetrix single nucleotide polymorphism (SNP) 6.0 array (coverage: 200–1500 probe sets). Extending our previous studies (Dibbens et al., 2009; Helbig et al., 2009), this study focussed on five additional candidate microdeletions at 1q21.1, 15q11.2, 16p11.2, 16p13.11 and 22q11.2. To evaluate the relative impact of the six candidate microdeletions on epileptogenesis, nine IGE patients with 15q13.3 deletions observed in this IGE sample were also included in the overall comparison and transmission analysis.
Table 1.
Recurrent microdeletions reported in neuropsychiatric disorders
| Chrom. segment | Chrom. position (Mb)a | MicroDel size (Mb) | Candidate gene | Neuropsychiatric disorder |
|---|---|---|---|---|
| 1q21.1 | 145.0–146.35 | 1.35 | GJA5, GJA8, HYDIN2 | ID, SZ |
| 15q11.2 | 20.3–20.75 | 0.45 | CYFIP1 | SZ |
| 16p11.2 | 29.5–30.1 | 0.7 | KCTD13, SEZ6L2 | ASD, ID |
| 16p13.11 | 14.7–16.3 | 1.6 | NDE1 | ID, SZ, ASD |
| 22q11.2 | 17.5–20.5 | 3.0 | COMT, SNAP29 | SZ, ID, ASD |
| 15q13.3 | 28.7–30.3 | 1.5 | CHRNA7 | ID/EPI, SZ, ASD |
a NCBI build 36.
ASD = autism spectrum disorder; EPI = epilepsy; ID = intellectual disability; SZ = schizophrenia.
The candidate approach applied in this study tried to avoid the inclusion of a large number of mainly neutral CNVs/CNPs or artificial CNVs detected by a genome-wide CNV scan, which would drastically reduce the power to detect rare pathogenic CNVs.
Study participants
All study participants gave informed consent according to the regulations at their local institutional review boards. Phenotyping and diagnostic classification of IGE syndromes were carried out according to standardized phenotyping protocols available at the Cologne Center for Genomics website (http://www.ccg.uni-koeln.de/epilepsygenetics1.html) (ILAE, 1989). According to the exclusion criteria, individuals with a history of major psychiatric disorders (autism spectrum disorder, schizophrenia and affective disorder) or severe intellectual disability were excluded. In a multi-centre effort, 1234 unrelated IGE patients (458 males, 776 females) were collected from Austria (n = 166), Belgium (n = 35), Denmark (n = 72), Germany (n = 755) and the Netherlands (n = 206). The epilepsy sample comprised the following IGE syndromes: childhood/juvenile absence epilepsy (n = 576); juvenile myoclonic epilepsy (n = 487), epilepsy with generalized tonic–clonic seizures alone (EGTCS; n = 171). Notably, 884 of the IGE patients and 1202 of the International Database on the Legal and Socio-ethical Aspects of Population Genetic (PopGen) sector of controls were investigated in a previous study, including eight IGE patients carrying a 15q13.3 deletion (Helbig et al., 2009). In addition, 134 IGE patients from the present cohort were part of a replication study (Dibbens et al., 2009), but did not carry 15q13.3 deletions.
Affymetrix SNP 6.0 data from 3022 German population controls (1550 males, 1472 females) were obtained from two datasets, the first from the Cooperative Health Research in the Region of Augsburg (KORA: n = 1786; Wichmann et al., 2005) and the second from PopGen (n = 1236; Krawczak et al., 2006). The population controls were not screened for epilepsy or major neuropsychiatric disorders, and consequently a small proportion (<1%) of controls might be affected. All samples were checked for ancestry matching on genotype by EIGENSTRAT analysis (Price et al., 2006).
Genotyping and copy number variation detection
Samples were typed for 1.8 million probe sets on the Affymetrix Genome-Wide Human SNP Array 6.0. The selected microdeletions were covered by 200–1500 probe sets each on the Affymetrix SNP 6.0 array. CNV analysis was performed by the algorithm implemented in the Affymetrix Genotyping Console version 3.0.2. Changes of the heterozygosity state and log2 ratios along with candidate deletions were visually inspected to exclude technical artefacts.
Microdeletions were considered to match the published deletions if they overlapped at least 85% of the genomic region of the candidate microdeletion (Table 1). All deletions identified by Affymetrix SNP 6.0 arrays were verified by real-time quantitative PCR, using a novel Duplex TaqMan CNV assay (Applied Biosystems, TaqMan CN early access program; TaqMan probe sequences are available on request) and/or array comparative genomic hybridization, as described previously (Itsara et al., 2009). Array comparative genomic hybridization data were used for refining deletion breakpoints.
Statistical analysis
Association analyses between genotype and phenotype were carried out by two-sided χ2-tests or Fisher's exact tests where appropriate.
Results
Detection of microdeletions in patients with IGE and controls
Altogether, we detected deletions at the five candidate loci (1q21.1, 15q11.2, 16p11.2, 16p13.11 and 22q11.2) in 22 (1.8%) out of 1234 patients and in nine (0.3%) out of 3022 controls [odds ratio (OR) = 6.1; 95% confidence interval (CI) 2.8–13.2; χ2 = 26.7; 1 degree of freedom; P = 2.4 × 10−7] (Fig. 1, Table 2). Microdeletions at 15q11.2 were observed in 12 (1%) out of 1234 IGE patients representing the most common microdeletion site, but were also observed in six (0.2%) out of 3022 controls (OR = 4.9; 95% CI 1.8–13.2; χ2 = 12.5; 1df; P = 4.2 × 10−4). In addition, an association with IGE was obtained for 16p13.11 microdeletions, which were found in six (0.5%) out of 1234 IGE patients and two (0.07%) out of 3022 controls (OR = 7.4; 95% CI 1.3–74.7; Fisher's exact test, P = 0.0094). Microdeletions at 22q11.2 were observed in two patients and microdeletions at 16p11.2 and 1q21.1 in a single patient each (Fig. 1). A 1q21.1 microdeletion was also identified in one control subject, whereas the other two deletions were not detected in controls. Microdeletions at 15q13.3 were found in nine (0.7%) out of 1234 patients and in none of the 3022 controls (Fisher's exact test, P = 1.4 × 10−5). Eight of the IGE patients with 15q13.3 deletions have been previously reported (Helbig et al., 2009) and one patient was identified in the extended IGE sample. Including the 15q13.3 deletions, we detected microdeletions in 31 (2.5%) of 1234 patients and in nine (0.3%) of 3022 controls (OR = 8.6; 95% CI 4.1–18.2; χ2 = 46.1; 1df; P = 1.1 × 10−11) (Fig. 1, Table 2).
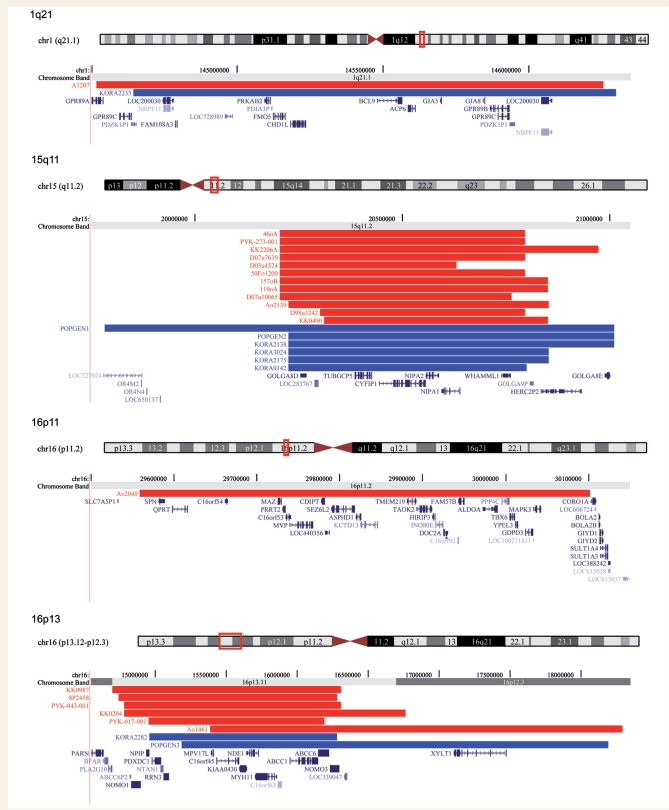
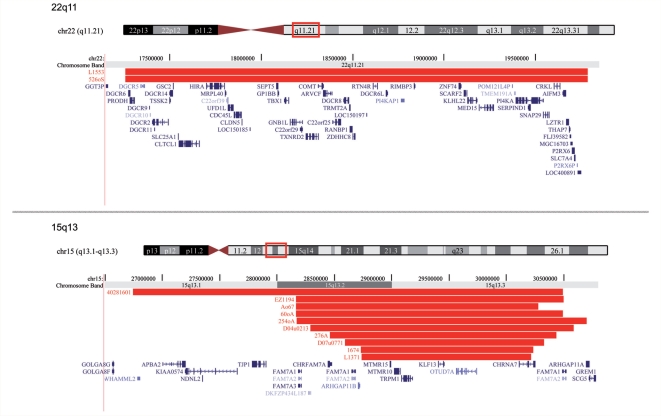
Figure 1.
Genomic position of the microdeletions at the genomic hot spot regions 1q21.1, 15q11.2, 16p11.2, 16p13.11, 22q11.2 and 15q13.3. Red = IGE patients; blue = controls. The positions of genes are also shown. Produced with the University of California, Santa Cruz Genome Browser (http://www.genome.ucsc.edu).
Table 2.
Recurrent microdeletions in IGE patients and controls
| Chromosome | IGE | Controls | P-value | USA sample |
|---|---|---|---|---|
| region | n = 1234 | n = 3022 | two-sided** | n = 2493a |
| 1q21.1 | 1 | 1 | – | 0 |
| 15q11.2 | 12 | 6 | 4.2 × 10−4 | 4 |
| 16p11.2 | 1 | 0 | – | 0 |
| 16p13.11 | 6 | 2 | 0.0094* | 0 |
| 22q11.2 | 2 | 0 | – | 0 |
| Microdels w/o 15q13.3 | 22 | 9 | 2.4 × 10−7 | 4 |
| 15q13.3 | 9 | 0 | 1.4 × 10−5* | 0 |
| Microdels total | 31 | 9 | 1.1 × 10−11 | 4 |
a Samples of 1607 USA subjects of European ancestry and 886 subjects from the Human Genome Diversity Panel (Itsara et al., 2009a).
*Fisher's exact test; **significant P-values.
Bold values indicate statistical significance.
Cosegregation analysis
DNA samples from both parents were available for 14 out of 31 patients with identified microdeletions (Fig. 2). For segregation analysis, all available family members were typed by quantitative PCR and/or array comparative genomic hybridization.
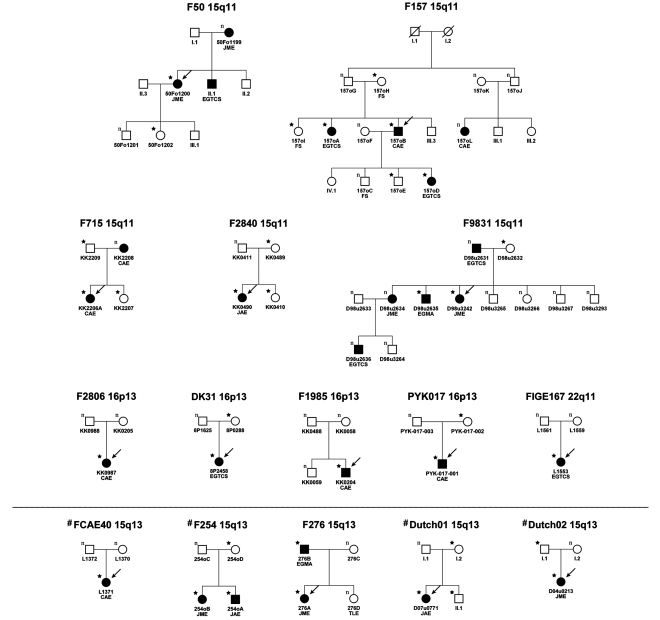
Figure 2.
Familial segregation of the microdeletions at 15q11.2, 16p13.11, 22q11.2 and 15q13.3. Arrows denote the index-IGE patient typed by the Affymetrix SNP 6.0 array. Black symbols = individuals affected by IGE. FS = febrile seizure; CAE = childhood absence epilepsy; JAE = juvenile absence epilepsy; JME = juvenile myoclonic epilepsy; EGTCS = epilepsy with generalized tonic–clonic seizures alone; EGMA = epilepsy with generalized tonic–clonic seizures on awakening; TLE = temporal lobe epilepsy; copy number state: n = normal/two copies; filled star indicates deletion carrier. #Families of IGE patients with 15q13.3 deletions reported previously (Helbig et al., 2009).
While 10 out of 14 microdeletions were inherited (seven maternal and three paternal transmissions), four de novo deletions were identified in 14 IGE patients (Table 3, Fig. 2). DNA from both parents was available for four out of 12 IGE patients carrying a 15q11.2 microdeletion. Maternal inheritance was seen in three and paternal inheritance in one out of four patients. For 16p13.11 microdeletions, DNA samples from both parents were available for four out of six families. De novo deletions were observed in two out of four patients and maternal inheritance in two out of four patients. In five out of nine patients with 15q13.3 microdeletions, parental DNA was available. Paternal and maternal inheritances were each found in two out of five transmissions. One de novo microdeletion occurred in these five families. Parental DNA was also available for one out of two patients with 22q11.2 microdeletions, in whom a de novo deletion was observed.
Table 3.
Characteristics of study participants carrying a candidate deletion
| Sample ID | Sex | Chrom. | Start | End | Size (kb) | Array | Age-at-onset (years) | Seizure types | IGE syndrome | Intellectual status | Inheritance |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1207 | F | 1q21.1 | 144 516 731 | 146 256 137 | 1 739 | Array CGH | 23 | abs., GTCS | JAE | Normal | Unknown |
| 46oA | M | 15q11.2 | 20 203 685 | 20 795 813 | 592 | Array CGH | 15 | abs., myocl., GTCS | JME | Normal | Unknown |
| D07u7639 | F | 15q11.2 | 20 203 685 | 20 795 813 | 592 | Array CGH | 14 | myocl., GTCS | JME | Normal | Unknown |
| KK2206A | F | 15q11.2 | 20 203 685 | 20 971 169 | 767 | Array CGH | 4 | abs. | CAE | Normal | Paternal |
| PYK-273-001 | F | 15q11.2 | 20 203 685 | 20 795 813 | 592 | Array CGH | 5 | abs. | CAE | Normal | Unknown |
| 119oA | F | 15q11.2 | 20 204 077 | 20 850 488 | 646 | Array CGH | 12 | GTCS | EGTCS | Normal | Unknown |
| 157oB | M | 15q11.2 | 20 204 077 | 20 850 488 | 646 | Array CGH | 4 | abs. | CAE | Normal | Parental |
| 50Fo1200 | F | 15q11.2 | 20 204 077 | 20 795 813 | 592 | Array CGH | Puberty | myocl. | JME | Normal | Maternal |
| D03u4524 | M | 15q11.2 | 20 204 077 | 20 629 367 | 425 | Array CGH | 16 | abs., myocl. | JME | Normal | Unknown |
| D07u10065 | F | 15q11.2 | 20 204 077 | 20 763 914 | 560 | Array CGH | 10 | abs., GTCS | JAE | Normal | Unknown |
| Ao2139 | F | 15q11.2 | 20 224 751 | 20 852 202 | 1 485 | Affy 6.0 array | 7 | abs., myocl. | JME | Normal, legasthenia | Unknown |
| D98u3242 | F | 15q11.2 | 20 301 665 | 20 795 813 | 494 | Array CGH | 4 | abs., myocl., GTCS | JME | Normal | Maternal |
| KK0490 | F | 15q11.2 | 20 310 606 | 20 850 488 | 540 | Array CGH | 15 | abs., GTCS | JAE | Normal | Maternal |
| Ao2040 | F | 16p11.2 | 29 559 251 | 30 101 408 | 542 | Array CGH | <20 | GTCS | EGTCS | Normal | Unknown |
| 8P2458 | F | 16p13.11 | 14.742.556 | 16.285.151 | 1.543 | Affy 6.0 array | 9 | GTCS | EGTCS | Normal | Maternal |
| KK0987 | F | 16p13.11 | 14 699 106 | 16 308 654 | 1 610 | Array CGH | Childhood | abs., FS | CAE | Learning disability | De novo |
| PYK-043-001 | F | 16p13.11 | 14 785 031 | 16 308 654 | 1 524 | Array CGH | 5 | abs. | CAE | Normal | Unknown |
| KK0204 | M | 16p13.11 | 14 785 031 | 16 767 009 | 1 982 | Array CGH | 8 | abs., GTCS | CAE | Normal | De novo |
| PYK-017-001 | M | 16p13.11 | 14 956 201 | 16 193 208 | 1 237 | Array CGH | 7 | abs. | CAE | Normal | Maternal |
| Ao1461 | F | 16p13.11 | 15 386 338 | 18 291 982 | 2 906 | Array CGH | 17 | myocl., GTCS | JME | Learning disability | Unknown |
| 526oS | M | 22q11.2 | 17 258 339 | 19 786 713 | 2 528 | Array CGH | 9 | GTCS | EGTCS | Minor developmental delay | Unknown |
| L1553 | F | 22q11.2 | 17 258 339 | 19 786 713 | 2 528 | Array CGH | 20 | GTCS | EGTCS | Normal | De novo |
| D07u0771a | F | 15q13.3 | 28.595.222 | 30.326.817 | 1.732 | Affy 6.0 array | 14 | abs. | JAE | Normal | Maternal |
| 40281601a | F | 15q13.3 | 26 745 821 | 30 494 518 | 3 749 | Array CGH | 3 | abs., GTCS | CAE | Normal | Unknown |
| 60oAa | M | 15q13.3 | 28 168 397 | 30 491 740 | 2 323 | Array CGH | 12 | abs., GTCS | JAE | Normal | Unknown |
| Ao67a | F | 15q13.3 | 28 168 397 | 30 276 525 | 2 108 | Array CGH | 16 | myocl., GTCS | JME | Normal | Unknown |
| EZ1194a | M | 15q13.3 | 28 168 397 | 30 498 257 | 2 330 | Array CGH | 6 | abs., myocl., GTCS | JME | Normal | Unknown |
| 254oAa | M | 15q13.3 | 28 171 483 | 30 701 463 | 2 530 | Array CGH | 9 | abs., GTCS | JAE | Normal | Maternal |
| 276A | F | 15q13.3 | 28 461 375 | 30 436 131 | 1 975 | Array CGH | 14 | myocl. | JME | Normal | Paternal |
| L1371a | F | 15q13.3 | 28 738 025 | 30 215 571 | 1 478 | Array CGH | 4 | abs. | CAE | Minor developmental delay | De novo |
| KORA2235 | M | 1q21.1 | 144 643 813 | 146 297 795 | 1 654 | Affy 6.0 array | – | – | Control | Unknown | Unknown |
| POPGEN1 | M | 15q11.2 | 19 781 829 | 21 010 631 | 1 229 | Affy 6.0 array | – | – | Control | Unknown | Unknown |
| KORA0142 | F | 15q11.2 | 20 224 751 | 20 799 862 | 575 | Affy 6.0 array | – | – | Control | Unknown | Unknown |
| KORA2138 | F | 15q11.2 | 20 224 751 | 21 010 631 | 786 | Affy 6.0 array | – | – | Control | Unknown | Unknown |
| KORA2175 | F | 15q11.2 | 20 224 751 | 20 852 202 | 627 | Affy 6.0 array | – | – | Control | Unknown | Unknown |
| KORA3024 | M | 15q11.2 | 20 224 751 | 20 852 202 | 627 | Affy 6.0 array | – | – | Control | Unknown | Unknown |
| POPGEN2 | M | 15q11.2 | 20 224 751 | 21 010 631 | 786 | Affy 6.0 array | – | – | Control | Unknown | Unknown |
| KORA2282 | F | 16p13.11 | 14 961 214 | 16 285 151 | 1 324 | Affy 6.0 array | – | – | Control | Unknown | Unknown |
| POPGEN3 | M | 16p13.11 | 15 186 307 | 18 192 575 | 3 006 | Affy 6.0 array | – | – | Control | Unknown | Unknown |
a Patients carrying a 15q13.3 deletion reported previously (Helbig et al., 2009).
comparative genomic hybridization (CGH), abs. = absence seizures; myocl. = bilateral myoclonic seizures on awakening; GTCS = generalized tonic–clonic seizures; FS = febrile seizures; CAE = childhood absence epilepsy; JAE = juvenile absence epilepsy; JME = juvenile myoclonic epilepsy; EGTCS = epilepsy with generalized tonic–clonic seizures; F = female; M = male.
Deletions in 15 IGE probands were shared with five affected and 14 unaffected first-degree relatives, while four first-degree relatives affected with IGE did not carry the deletion (Fig. 2). These four affected family members without a deletion were all found in families exhibiting the 15q11.2 deletion. In 10 out of 15 families, the microdeletions were inherited (three paternally and seven maternally). Eight of the transmitting parents (15q11.2: n = 3; 15q13.3: n = 3; 16p13.11: n = 2) were clinically unaffected, one father carrying a 15q13.3 deletion was affected by IGE and one mother with a 15q11.2 deletion had a history of febrile seizures (Fig. 2). Cosegregation between the microdeletions and the phenotype was not consistent with autosomal dominant inheritance, particularly in three large families with 15q11.2 microdeletions (Fig. 2; families: F50, F157 and F9831).
Genotype–phenotype correlations
The deletions investigated in this study are flanked by highly homologous segmental duplications and non-allelic homologous recombination is thought to promote genomic rearrangements between the putative breakpoints (Hastings et al., 2009; Itsara et al., 2009a; Sharp, 2009). The exact positions of the deletion breakpoints inside the segmental duplication clusters are difficult to determine and results may vary between array platforms depending on the genomic coverage of the array probe sets across the flanking segmental duplications. We designed a customized oligonucleotide microarray to refine further the breakpoints of the microdeletions characterized in this study (Itsara et al., 2009a). The individual breakpoint estimates and the resulting sizes of the microdeletions are shown in Table 3, Fig. 1 and Supplementary Fig. S1. The individual breakpoints and sizes of the observed microdeletions consistently corresponded with those described previously (Table 1).
Patients with the different deletions (n = 31) displayed a representative distribution of IGE syndromes (childhood absence epilepsy/juvenile absence epilepsy 48.4%, juvenile myoclonic epilepsy 35.5%, epilepsy with generalized tonicclonic seizures 16.1%; males 29%, females 71%) similar to that observed in the entire IGE sample (childhood absence epilepsy/juvenile absence epilepsy 46.6%, juvenile myoclonic epilepsy 39.5%, epilepsy with generalized tonic–clonic seizures 13.9%; males 37%, females 63%). We found no evidence that patients with microdeletions had refractory seizures and there was no preponderance of a particular seizure type or a shift towards an early age of onset (Table 3). We did not observe other neuropsychiatric phenotypes in family members carrying a deletion.
Discussion
In the present study, we investigated whether five large recurrent microdeletions (at 1q21.1, 15q11.2, 16p11.2, 16p13.11 and 22q11.2), previously associated with neuropsychiatric disorders, also confer risk to common IGE syndromes. Recurrent microdeletions were found at 15q11.2 (n = 12), 16p13.11 (n = 6) and 22q11.2 (n = 2) in 1234 IGE patients. The microdeletions at 1q21.1 and 16p11.2 occurred in one IGE patient each. Altogether, the five microdeletions showed a significant excess in the IGE patients compared with controls (P = 2.4 × 10−7) and the present association results indicate an involvement of microdeletions at 15q11.2 and 16p13.11 in the aetiology of IGE (Table 2). Including the 15q13.3 deletions (IGE/control: 9/0), recurrent microdeletions were present in 2.5% of 1234 IGE patients versus 0.3% of 3022 population controls (P = 1.1 × 10−11). IGE patients carrying a microdeletion display typical clinical features of IGE regarding seizure types and age of onset. Although the microdeletions investigated are individually rare (<1%) in patients with IGE, they collectively account for a significant fraction of the genetic variance of common IGE syndromes.
The investigated microdeletions seem to differ with regard to the magnitude of the epileptogenic effect (e.g. point estimates of odds ratio), occurrence of de novo deletions and familial segregation patterns (Tables 2 and 3, Fig. 2). The 15q13.3 microdeletion emerged as the major genetic risk factor with a point estimate of OR > 50 (95% CI 21.7–139.6), assuming a frequency <0.02% in the general population (Schizophrenia Consortium, 2008; Sharp et al., 2008; Stefansson et al., 2008; Dibbens et al., 2009; Helbig et al., 2009; Kirov et al., 2009). In contrast, 15q11.2 (OR = 4.9) and 16p13.11 (OR = 7.4) microdeletions seem to confer a lower genetic risk to IGE, also reflected by the higher frequency in controls (0.20% and 0.07%, respectively).
Segregation between microdeletions and the IGE trait was investigated if family members were available for testing. Particularly, the 15q11.2 microdeletion did not cosegregate with the IGE trait in three large families (Fig. 2). For the other deletions, large families were not available for segregation analysis. Consistent with two small families in which 15q13.3 deletions segregated with affected family members in the present study, Dibbens and colleagues (2009) found incomplete penetrance of the 15q13.3 microdeletion in four out of seven pedigrees and three pedigrees included family members with IGE lacking the 15q13.3 deletion. Despite the remarkable odds ratio (OR > 50), 15q13.3 deletions are not sufficient to express a disease phenotype, which might also vary considerably depending on the genetic background and possible environmental effects. De novo microdeletions at 15q13.3, 16p13.11 and 22q11.2 were observed in four out of 14 families, for which DNA was available from both parents. The presence of de novo deletion events in conjunction with low population frequencies implicates purifying selection and thus may suggest a strong influence on the disease phenotype.
Taking into account all published studies, remarkable phenotypic variability is observed for carriers of the six recurrent microdeletions assessed in our study, ranging from apparently unaffected carriers to individuals with severe cognitive deficits, dysmorphisms and various neuropsychiatric features. The present epilepsy sample was ascertained by the IGE phenotype excluding those patients affected by major psychiatric and mental disorders. Moreover, carriers of microdeletions were re-evaluated for the presence of intellectual disability or other neuropsychiatric disorders. It is therefore unlikely that the excess of microdeletions found in our study is caused by unobserved comorbidity of neuropsychiatric disorders and IGE.
The mechanisms by which microdeletions mediate their pathogenic effects remain unknown (Itsara et al., 2009; Sharp, 2009). Haploinsufficiency of the deleted segment seems the most likely mechanism (Itsara et al., 2009a; Sharp, 2009) and several plausible candidate genes have been suggested (Table 1). Besides purely stochastic or environmental effects, other genetic mechanisms such as imprinting, unmasking of different recessive allelic mutations on the intact homologous chromosomal segment and background genomic variation may contribute to the highly variable phenotypic expression (Sharp, 2009).
Overall, emerging evidence suggests that recurrent microdeletions may confer a pleiotropic effect underlying various neuropsychiatric disorders. The complex interaction with additional factors might determine the specific phenotype. For example, the 16p13.11 candidate gene NDE1 (encoding NudE nuclear distribution gene E homologue 1) is known to interact with DISC1 (gene disrupted in schizophrenia) and LIS1 (gene causing lissencephaly 1, PAFAH1B1). Deficiency of the LIS1–NDE1 complex impairs cortical neurogenesis and neuronal migration (Pawlisz et al., 2008) frequently leading to epilepsy, whereas DISC1-NDE1 deficiency appears to play a role in neuropsychiatric disorders, including schizophrenia and bipolar affective disorder (Hennah et al., 2009). Together, our findings support the role of neurodevelopmental processes in epileptogenesis. Given the frequency of recurrent microdeletions in various neuropsychiatric and neurodevelopmental disorders, identification of genetic and non-genetic factors determining phenotype specificity will be a major focus of future research.
Despite the high heritability of IGE, the genetic architecture remains elusive. Relatively few epilepsy genes have been identified thus far, mainly in rare monogenic forms of idiopathic epilepsies (Helbig et al., 2008; Reid et al., 2009). By identifying recurrent microdeletions at 15q11.2, 15q13.3 and 16p13.11 as collectively significant genetic risk factors for IGE, our study provides new insights into the complex genetic predisposition of common epilepsies. Although the risk estimate of microdeletions associated with IGE is considerably higher (OR: 5–50) than that observed for common SNPs in complex traits (OR < 2), it is much lower than that of highly penetrant mutations causing Mendelian diseases (OR > 100). Our present family study revealed a high percentage (>70%) of apparently unaffected parents transmitting the microdeletion to the affected child (Fig. 2), suggesting that the microdeletion alone is not sufficient to cause an epilepsy phenotype in some cases. Likewise, unprecedented phenotypic heterogeneity has been found for seemingly identical microdeletions at 1q21.1, 16p11.2, 15q13.3 and 22q11.2, ranging from severe genomic syndromes (e.g. 22q11.2 microdeletion: DiGeorge syndrome, velocardiofacial syndrome) to a wide range of neuropsychiatric disorders (e.g. schizophrenia, intellectual disability and autism spectrum disorder), as well as in apparently unaffected individuals (for review see Mefford and Eichler, 2009). With regard to the highly variable phenotypic expressivity of the microdeletions investigated, it is difficult to assess the clinical relevance and implications for genetic counselling, and further studies are clearly needed to specify the phenotype–genotype relationship. Advances in large-scale sequencing and high-resolution mapping of structural genomic rearrangements will provide a survey on structural genomic variations at the genome-wide level, allowing for a more comprehensive assessment of the impact of structural genomic variations in common seizure disorders in the near future (Itsara et al., 2009a; Mefford and Eichler, 2009).
Funding
European Community (FP6 Integrated Project EPICURE, LSHM-CT-2006-037315, FP6 MEXCT visual sensitivity, grant no. 024224 to D.K.-N.T.); the German Research Foundation (SA434/4-2 to T.S. and P.N.); the German Federal Ministry of Education and Research, National Genome Research Network (NGFN-2: NeuroNet to E.C.E., T.S. and H.L.; NGFNplus: EMINet, 01GS08120 to P.N and T.S, and 01GS08123 to H.L.); the Belgian National Fund for Scientific Research (Flanders, 0399.08 to P.J.); The Netherlands National Epilepsy Fund (grant no. 04-08 to B.P.C.K. and D.L.); The Netherlands Organization for Scientific Research (grant no. 917.66.315 to B.P.C.K. and C.G.F.d.K.); and the National Institutes of Health (HD043569 to E.E.E., HD043376 to H.C.M., partial); the PopGen biobank (to A.F. and S.S.). The PopGen project received infrastructure support through the German Research Foundation excellence cluster ‘Inflammation at Interfaces’. The KORA research platform (KORA, Cooperative Research in the Region of Augsburg) was initiated and financed by the Helmholtz Zentrum München—German Research Centre for Environmental Health, which is funded by the German Federal Ministry of Education and Research and by the State of Bavaria.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Acknowledgements
We thank all individuals and their families for participating in this study.
Glossary
Abbreviations
- CNV
copy number variation
- IGE
idiopathic generalized epilepsy
- SNP
single nucleotide polymorphism
References
- Basset AS, Marshall CR, Lionel AC, Chow EWC, Scherer SW. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum Mol Genet. 2008;17:4045–53. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenfeld H. Cellular and network mechanisms of spike-wave seizures. Epilepsia. 2005;46(Suppl 9):21–33. doi: 10.1111/j.1528-1167.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–71. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–23. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- Dibbens LM, Mullen S, Helbig I, Mefford HC, Bayly MA, Bellows S, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum Mol Genet. 2009;18:3626–31. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. PathoGenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannes FD, Sharp AJ, Mefford HC, de Ravel T, Ruivenkamp CA, Breuning MH, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–32. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig I, Scheffer IE, Mulley JC, Berkovic SF. Navigating the channels and beyond: unravelling the genetics of the epilepsies. Lancet Neurol. 2008;7:231–45. doi: 10.1016/S1474-4422(08)70039-5. [DOI] [PubMed] [Google Scholar]
- Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–2. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS One. 2009;4:e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International League Against Epilepsy. Proposal for revised classification of epilepsies and epileptic syndromes: Commission on Classification and Terminology of the International League Against Epilepsy. Epilepsia. 1989;30:389–99. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009a;84:148–61. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Addendum. Am J Hum Genet. 2009b;84:550–1. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallon P, Latour P. Epidemiology of idiopathic generalized epilepsies. Epilepsia. 2005;46(Suppl 9):10–4. doi: 10.1111/j.1528-1167.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–38. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–99. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J Med Genet. 2009;46:242–8. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnamenta AT, Wing K, Akha ES, Knight SJ, Bolte S, Schmotzer G, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009;17:687–92. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlisz AS, Mutch C, Wynshaw-Boris A, Chenn A, Walsh CA, Feng Y. Lis1-Nde1-dependent neuronal fate control determines cerebral cortical size and lamination. Hum Mol Genet. 2008;17:2441–55. doi: 10.1093/hmg/ddn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Reid CA, Berkovic SF, Petrou S. Mechanisms of human inherited epilepsies. Prog Neurobiol. 2009;87:41–57. doi: 10.1016/j.pneurobio.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ. Emerging themes and new challenges in defining the role of structural variation in human disease. Hum Mutat. 2009;30:135–44. doi: 10.1002/humu.20843. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–8. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek AM. Novel microdeletion syndromes detected by chromosome microarrays. Hum Genet. 2008;124:1–17. doi: 10.1007/s00439-008-0513-9. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–82. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- van Bon BW, Mefford HC, Menten B, Koolen DA, Sharp AJ, Nillesen WM, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;46:511–23. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Wichmann HE, Gieger C, Illig T MONICA/KORA Study Group. KORA-gen—resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl 1):S26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.