Abstract
Spinocerebellar ataxia type 3 is a neurodegenerative disorder caused by the expansion of the polyglutamine repeat region within the ataxin-3 protein. The mutant protein forms intracellular aggregates in the brain. However, the cellular mechanisms causing toxicity are still poorly understood and there are currently no effective treatments. In this study we show that administration of a rapamycin ester (cell cycle inhibitor-779, temsirolimus) improves motor performance in a transgenic mouse model of spinocerebellar ataxia type 3. Temsirolimus inhibits mammalian target of rapamycin and hence upregulates protein degradation by autophagy. Temsirolimus reduces the number of aggregates seen in the brains of transgenic mice and decreases levels of cytosolic soluble mutant ataxin-3, while endogenous wild-type protein levels remain unaffected. Temsirolimus is designed for long-term use in patients and therefore represents a possible therapeutic strategy for the treatment of spinocerebellar ataxia type 3. Using this disease model and treatment paradigm, we employed a microarray approach to investigate transcriptional changes that might be important in the pathogenesis of spinocerebellar ataxia type 3. This identified ubiquitin specific peptidase-15, which showed expression changes at both the messenger ribonucleic acid and protein level. Ubiquitin specific peptidase-15 levels were also changed in mice expressing another mutant polyglutamine protein, huntingtin. In total we identified 16 transcripts that were decreased in transgenic ataxin-3 mice that were normalized following temsirolimus treatment. In this mouse model with relatively mild disease progression, the number of transcripts changed was low and the magnitude of these changes was small. However, the importance of these transcriptional alterations in the pathogenesis of spinocerebellar ataxia type 3 remains unclear.
Keywords: Usp15, microarray, rapamycin, huntingtin, polyglutamine
Introduction
Spinocerebellar ataxia type 3 (Machado–Joseph disease), is among the commonest of the autosomal dominant spinocerebellar ataxias worldwide (Schols et al., 2004). It is a devastating neurodegenerative disorder resulting from the expansion of the polyglutamine repeat region of the ataxin-3 gene. It shares many features with the eight other diseases caused by polyglutamine expansion mutations, including Huntington’s disease, such as the formation of inclusions within neurons by the mutant protein. The exact mechanism by which mutant ataxin-3 causes cellular toxicity is unknown. One potential pathogenic mechanism involved in polyglutamine repeat disorders is transcriptional dysregulation (reviewed in Sugars and Rubinsztein, 2003; Riley and Orr, 2006). Polyglutamine aggregates, including ataxin-3 aggregates, have been demonstrated to sequester transcription factors (Perez et al., 1998). Ataxin-3 may also have a more direct role in transcription regulation through its interactions with histone acetyltransferases (Li et al., 2002) and histone deacetylases (Evert et al., 2006).
The normal role of ataxin-3 appears to be within the ubiquitin signalling system. The protein has two ubiquitin interacting motifs (or three in the shorter splice variant) (Albrecht et al., 2004) and additionally there are two ubiquitin binding sites in the Josephin domain (Nicastro et al., 2009). Ataxin-3 has been demonstrated to possess ubiquitin protease activity (Burnett et al., 2003) and it may be involved in delivery of ubiquitinated substrates to the proteasome for degradation (Doss-Pepe et al., 2003). In particular, ataxin-3 may be important in the regulation of endoplasmic reticulum associated protein degradation, removing mis-folded proteins from the endoplasmic reticulum (Wang et al., 2006; Zhong and Pittman, 2006). Overexpression of wild-type ataxin-3 suppresses toxicity in Drosophila models of polyglutamine mediated neurodegeneration (Warrick et al., 2005) and this was hypothesized to result from an increased turnover of the mutant polyglutamine proteins.
The ubiquitin protease activity of ataxin-3 is not altered by the presence of the expanded polyglutamine repeat in the mutant protein (Burnett et al., 2003; Chai et al., 2004; Todi et al., 2009). In fact, polyglutamine mutations appear to cause disease predominantly by toxic gain-of-function effects (Imarisio et al., 2008). Furthermore, two copies of the mutant ataxin-3 gene have been reported to result in more severe disease in spinocerebellar ataxia type 3 (Carvalho et al., 2008). Thus, we have considered that reducing the levels of toxic proteins by enhancing the degradation of the mutant allele relative to the wild-type allele may be a possible therapeutic strategy for this group of diseases. This may be possible by inducing macroautophagy (which we will call autophagy). Along with degradation by the proteasome, autophagy forms the major route for degradation of cellular proteins. Autophagy begins with the formation of double-membraned structures called autophagosomes in the cytosol, which ultimately fuse with lysosomes to degrade their contents. Previously, we and others have shown in tissue culture and in Drosophila that a range of aggregate-prone, polyglutamine-expanded constructs (including mutant huntingtin fragments, full-length mutant huntingtin and full-length mutant ataxin-3) are highly dependent on autophagy for their clearance (Ravikumar et al., 2002; Berger et al., 2006; Shibata et al., 2006). When autophagy is blocked either chemically or genetically, these proteins accumulate. Interestingly, the clearance of wild-type forms of mutant huntingtin has little or no dependence on autophagy (Ravikumar et al., 2002; Shibata et al., 2006).
Autophagy can be up-regulated by inhibiting the mammalian target of rapamycin (mTOR) or by mTOR-independent pathways (Noda and Ohsumi, 1998; Sarkar et al., 2005; Boland et al., 2008; Williams et al., 2008). Both rapamycin (a specific mTOR inhibitor) and drugs acting on mTOR-independent pathways enhance the clearance of mutant polyglutamine proteins (including full-length mutant ataxin-3) in cell culture (Sarkar et al., 2005; Berger et al., 2006; Williams et al., 2008). We have shown in cells, Drosophila and zebrafish, that autophagy up-regulation decreases soluble levels of aggregate-prone proteins, decreases the proportion of cells with inclusions and attenuates toxicity (Ravikumar et al., 2004; Sarkar et al., 2005; Williams et al., 2008). We have also shown that the rapamycin analogue temsirolimus (cell cycle inhibitor-779) reduces aggregate number and behavioural abnormalities in a transgenic mouse expressing a mutant huntingtin fragment when administered ∼6 weeks before the onset of signs (Ravikumar et al., 2004). Temsirolimus is a rapamycin ester designed for injection and shows the same activity as rapamycin (sirolimus, Rapamune®) (reviewed in Huang and Houghton, 2001). This finding raised the possibility that up-regulation of autophagy may be an effective therapeutic strategy for a variety of diseases caused by aggregate-prone intracytosolic proteins. Here we have tested the efficacy of temsirolimus in an expanded full-length spinocerebellar ataxia type 3 mouse model administered soon after the onset of disease signs.
Materials and methods
Mice and behavioural tests
All procedures were carried out under the jurisdiction of appropriate UK Home Office Project and Personal animal licences and with local ethical committee approval. The generation of the mouse model (spinocerebellar ataxia type 3 line 70.61) used here has been previously described (Bichelmeier et al., 2007). These mice overexpress full-length human ataxin-3 with an expanded polyglutamine repeat region of 70 amino acids under the control of the prion protein promoter on a C57/BL6 background. Mice were genotyped at 3 weeks of age by PCR using published primers (Bichelmeier et al., 2007). Male mice were selected for use in behavioural studies as they demonstrated less variable performance in rotarod trials and hence a more consistent phenotype than females (data not shown). Transgenic and non-transgenic or drug and placebo treated animals were matched within litters and housed in the same cage. Temsirolimus (kind gift from Wyeth) was administered by intraperitoneal injections three times a week at a dose of 20 mg/kg (Ravikumar et al., 2004). Control mice were given a placebo of injection buffer only with the same frequency. Rotarod testing was carried out exactly as previously described (Vacher et al., 2005). Placebo and drug treated mice within each litter were tested on the rotarod simultaneously and at the time of testing were not identified as to their treatment status.
The original generation and genotyping of the Huntington’s disease mouse line, HD-N171-N82Q [B6C3F1/J-Tg(HD82Gln)81Dbo/J, Jackson Laboratory, Bar Harbour, ME] has been previously described (Schilling et al., 1999; Vacher et al., 2005). These mice express exon 1 of Huntingtin with an expanded polyglutamine region of 82 amino acids under the control of the prion protein promoter.
Culturing and treatment of primary cortical neurons
For direct assessment of autophagy following rapamycin treatment primary cortical neurons were isolated from embryonic day 16.5 Sprague Dawley rat pups (Charles River). Briefly, pup brains were harvested and placed in ice-cold Dulbecco’s modified eagle medium where the meninges were removed; the cerebral cortices were dissected and then incubated in Dulbecco's; modified eagle medium containing Accutase (Innovative Cell Technologies, San Diego, CA) for 10 min at 37°C. After mechanical dissociation using sterile micropipette tips, dissociated neurons were resuspended in Dulbecco's; modified eagle medium and centrifuged. Cell count and viability assay was performed using trypan blue. Viable cells were seeded on a poly-d-lysine and laminin coated 6 multiwell (7.5 × 105 cells per well). Cells were maintained in Dulbecco’s modified eagle medium containing 2 mM glutamine, 2% B27 supplement and 1% penicillin/streptomycin/fungizone (Invitrogen). One-half of the plating medium was changed every third day until treatment. After 7 days in vitro, cultured neurons were treated for 8 h with 0.2 µM rapamycin (Sigma-Aldrich). Where used, a saturating concentration (400 nM) of bafilomycinA1 (Calbiochem) was added to the cells in the last 4 h before harvesting.
Immunohistochemistry
For aggregate analysis, brains of 3 month old mice were perfusion fixed with 4% paraformaldehyde. Brains were then frozen and 12 µm sections cut. Sections were post-fixed for 10 min with 4% paraformaldehyde, blocked for 1 h in block buffer (5% normal goat serum, 1% bovine serum albumin 0.1% triton X-100 in phosphate buffered saline) and incubated with primary antibody, mouse anti-ataxin-3 (MAB5360, Millipore, Watford, Hertfordshire) diluted 1 in 1000 in block buffer overnight at 4°C. Primary antibody binding was visualized by peroxidase labelling using vectastain elite avidin and biotinylated enzyme complex kit with 3,3′-diaminobenzidine substrate following the manufacturers protocol (both from Vector Laboratories, Burlingame, CA). Aggregates were counted in the motor cortex, as this region showed clear accumulation of aggregates at a level that could be accurately counted. Three fields (diameter of 1 mm) were counted on different sections of brains from five animals in each treatment group (n = 5). Statistical analysis was carried out by unpaired t-test.
For S6 staining, tissue was treated as above; primary antibody was either anti-phospho-S6 or anti-total-S6 (both Cell Signaling Technologies, Danvers, MA). Sections were then incubated in secondary antibody, goat-anti rabbit conjugated to alexa 488 (Invitrogen) for 1 h at room temperature and then mounted in citifluor (Citifluor, London) with 4′,6-diamidino-2-phenylindole.
Protein extraction and western blotting
For autophagy analysis, cell pellets were lysed on ice in Laemmli buffer (6.5 mM Tris–HCl pH 6.8, 2% sodium dodecyl sulphate, 5% β-mercaptoethanol, 10% glycerol and 0.01% bromophenol blue) for 30 min in presence of protease inhibitors (Roche Diagnostics). Nuclear and cytoplasmic extracts were prepared from fresh brain tissue using a nuclear extract kit (Active motif, Carlsbad, CA), following the manufacturer’s protocol. Total brain lysates were prepared by homogenising in buffer B (50 mM Tris pH 7.5, 10% glycerol, 5 mM magnesium acetate, 0.2 mM ethylenediaminetetraacetic acid, 0.5 mM dithiothreitol and protease inhibitor) at 4°C. The homogenate was centrifuged at 13 000g at 4°C, the supernatant was removed and used for western blot analysis.
Samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, using a standard protocol and transferred to polyvinylidene fluoride membrane (Millipore, Watford, Hertfordshire). Membranes were incubated overnight with primary antibody followed by peroxidase conjugated secondary antibody and detection using Amersham electro or enhanced chemiluminescence substrate kit (GE Healthcare, Little Chalfont, Buckinghamshire). Membranes were then stripped using Re-Blot plus (Millipore) and reprobed with loading control antibody, either anti α-tubulin clone DM1A or rabbit actin (both Sigma, Poole, Dorset) or anti-histone H3 (Cell Signaling Technology, Danvers, MA). Densitometric analysis was carried out using Image J software. Primary antibodies used were mouse anti-ataxin-3 (Millipore, Watford, Hertfordshire), rabbit anti-light-chain-3 (LC3: Novus Biologicals), rabbit anti-phospho and total p70S6Kinase, rabbit anti-phospho and total S6 ribosomal protein, rabbit anti-phospho and total eukaryotic initiation factor 4E-binding-protein 1 (Cell Signaling Technology), goat anti-ubiquitin specific peptidase (Usp)-15, rabbit anti-Grik2, mouse anti-splicing factor proline/glutamine-rich and rabbit anti-RORa (all Abcam, Cambridge) (see Table 1 for expansions).
Table 1.
Genes identified to be down-regulated in ataxin-3 transgenic mice versus non-transgenic mice and normalized by temsirolimus treatment
| Probe set ID | Gene title | Gene symbol | Ataxin-3 placebo versus wild-type placebo |
Ataxin-3 temsirolimus versus SCA3 placebo |
||
|---|---|---|---|---|---|---|
| P-value | Fold change | P-value | Fold change | |||
| 1417472_at | Myosin, heavy polypeptide 9, non-muscle | Myh9 | 0.036 | 1.50 | 0.006 | 2.71 |
| 1436326_at | RAR-related orphan receptor alpha | RORa | 0.019 | 1.53 | 0.016 | 1.54 |
| 1436891_at | Ubiquitin specific peptidase 15 | Usp15 | 0.009 | 1.50 | 0.006 | 1.91 |
| 1436898_at | Splicing factor proline/glutamine rich | Sfpq | 0.002 | 1.86 | 0.001 | 1.85 |
| 1437728_at | AlkB, alkylation repair homolog 5 (E. coli) | Alkbh5 | 0.004 | 1.62 | 0.001 | 1.93 |
| 1438268_at | Membrane associated DNA binding protein | Mnab | 0.003 | 2.19 | 0.026 | 1.53 |
| 1438403_s_at | Metastasis associated lung adenocarcinoma transcript 1 | malat1 | 4.49E-04 | 1.70 | 0.001 | 1.72 |
| 1439940_at | RIKEN cDNA 2900019G14 | 0.001 | 1.87 | 0.002 | 1.77 | |
| 1439946_at | Myocyte enhancer factor 2C | Mef2c | 0.003 | 2.23 | 0.015 | 1.61 |
| 1440602_at | Glutamate receptor, ionotropic, kainate 2 (beta 2) | Grik2 | 0.003 | 1.62 | 0.004 | 1.69 |
| 1442813_at | Diacylglycerol kinase, iota | Dgki | 0.032 | 1.64 | 0.007 | 1.83 |
| 1443086_at | Activated leukocyte cell adhesion molecule | Alcam | 0.014 | 1.54 | 0.002 | 1.60 |
| 1444811_at | Translocation protein 1 | Tloc1 | 0.036 | 1.62 | 0.045 | 1.51 |
| 1445676_at | RIKEN clone C230043A04 | 0.007 | 1.59 | 0.004 | 1.55 | |
| 1456005_a_at | BCL2-like 11 (apoptosis facilitator) | Bcl2l11 | 0.038 | 1.65 | 0.024 | 1.68 |
| 1457817_at | Breast carcinoma amplified sequence 3 | Bcas3 | 0.004 | 1.52 | 6.51E-04 | 1.60 |
SCA3 = Spinocerebellar ataxia type 3.
Microarray methods
Mice were adminsistered temsirolimus or placebo as for behavioural studies, from 5 weeks of age for a period of 6 weeks. Mice were then sacrificed and brains dissected and snap frozen on dry ice. Microarray experiments were performed on an Affymetrix platform. Fragmented and labelled cRNA of the different groups (four wild-type—placebo mice, five wild-type—temsirolimus mice; ataxin-3-placebo and ataxin-3-temsirolimus) were hybridized on Affymetrix mouse genome 430 2.0 arrays. A probe level summary was determined using the Affymetrix GeneChip Operating Software using the Microarray Analysis Software 5 algorithm. Normalization of raw data was performed by the Array Assist®Software 4.0 (Stratagene, La Jolla, Canada), applying a robust multichip average algorithm with correction for guanine and cytosine content. Significance was calculated using a t-test without multiple testing correction (Array Assist® Software), selecting all transcripts with a minimum change in expression level of 1.5-fold together with a P-value < 0.05. When identifying transcriptional changes between multiple treatment groups, genes were selected based on changes in the same probe set and changes in different probe sets corresponding to the same gene were not included.
Real-time PCR
Initial validation was carried out on RNA samples isolated for microarray analysis. Quantitative real-time PCR was performed with the QuantiTect SYBR green PCR kit (Qiagen, Hilden, Germany) in the LightCycler 2.0 (Roche). Standard curves of each amplified gene were created to calculate the PCR efficiency. Pyruvate dehydrogenase (PDH), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ) and succinate dehydrogenase complex, subunit A (SDHA) were analysed as the reference (housekeeping) genes and were used for normalization. The Cp-values of the reference genes and the target genes were obtained with the LightCycler Software 4.0 using the ‘Fit Points’ method. The relative expression levels of all genes were calculated using the mathematical model of Pfaffl (2001). Primers used were as follows: Grik2 (Glutamate receptor, ionotropic, kainate 2 (beta 2)) forward 5′ AGTGCCACCATACCATCCAG 3′, reverse 5′ GCTGGCACTTCAGAGACATTC 3′, PDH forward 5′ GTAGAGGACACGGGCAAGAT 3′, reverse 5′ TGAAAACGCCTCTTCAGCA 3′, SDHA forward 5′ GCAGCACAGGGAGGTATCA 3′, reverse 5′ CTCAACCACAGAGGCAGGA 3′, Tloc (translocation protein 1) forward 5′ CATCGGGTTGACTATTTTATTGC 3′, reverse 5′ CCACAGACTCCCTGGTTGTAA 3′, Usp15 forward, 5′ GAAGCTGGGTGCTGAGGA 3′, reverse 5′ CCACAGACTCCCTGGTTGTAA 3′, YWHAZ forward 5′ AGACGGAAGGTGCTGAGAAA 3′, reverse 5′ TCAAGAACTTTTCCAAAAGAGACA 3′.
For further analysis RNA from 3.5–4 month old mouse brains was extracted using Trizol (Invitrogen). One micro gram RNA was used for reverse transcription, performed with Supercript III kit (Invitrogen) according to the manufacturer’s instructions. The quantitative PCR reaction for green fluorescent protein was carried out with SYBR PCR mix (Applied Biosystems) and the following primer pairs, RORa (Retinoic acid receptor-related orphan receptor alpha) forward 5′ GAACACCTTGCCCAGAACAT 3′, reverse 5′ AGCTGCCACATCACCTCTCT 3′, Sfpq (splicing factor proline/glutamine rich) 5′ ACGATGGAAGTCCTTGGATG 3′, reverse 5′ TCCATGCGCCTTAATTCTTC 3′, Mnab1 (membrane associated DNA binding protein) forward 5′ CGAATGACCCAAGGACAACT 3′, reverse 5′ AGGAGAAGGTGGTGTTGGTG 3′, Mef2c (Myocyte enhancer factor 2C) forward 5′ CCATTGGACTCACCAGACCT 3′, reverse 5′ AGCACACACACACACTGCAA 3′, Dgki (Diacylglycerol kinase, iota) forward 5′ ATGACATCCATCAGGTGCAA 3′, reverse 5′ CCGGAGTTTGTCCTTGTCAT 3′, Alcam2 (Breast carcinoma amplified sequence 3) forward 5′ CTCGTTGCTGGTGTCGTCTA 3′, reverse 5′ ATCCGCTCCTCTCTTAGGC 3′, Bcas3 (Breast carcinoma amplified sequence 3) forward 5′ GGAGAGCGTCGTGACTTTTC 3′, reverse 5′ CAGAGGAGGCTCATTTCCAG 3′, Alkbh5 (alkylation repair homolog 5) forward 5′ GGAACCTGTGCTTTCTCTGC 3′, reverse 5′ TGCTCAGGGATTTTGTTTCC 3′, Bcl2l11 (B cell lymphoma protein 2 (Bcl-2) like 11) forward 5′ GCCCCTACCTCCCTACAGAC 3′, reverse 5′ TGGGGATCTGGTAGCAAAAG 3′, Myh9 (myosin, heavy polypeptide 9, non-muscle) forward 5′ AGGAGACAAAGGCGCTATCA 3′, reverse 5′ GACTTCTCCAGCTCGTGGAC 3′, Glyceraldehyde 3-phosphate dehydrogenase-2 forward 5′ TGTGTCCGTCGTGGATCTGA 3′, reverse 5′ CCTGCTTCACCACCTTCTTGAT 3′.
Data were captured on a 7900 HT fast real time PCR System (Applied Biosystems) and analysed as a function of glyceraldehyde 3-phosphate dehydrogenase expression.
Aggregation and toxicity of polyglutamine constructs
SK-N-SH and HeLa cells were grown in Dulbecco’s modified eagle medium supplemented with 5% fetal calf serum, penicillin/streptomycin and 2 mM l-glutamine at 37°C in 5% CO2. Transfections were performed in 6 well plates using Lipofectamine or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Plasmids were enhanced green fluorescent protein (pEGFP-C1, Clontech) fused at its C terminus with an Huntington’s disease gene exon 1 fragment with 74 polyQ repeats (EGFP-httQ74) (Narain et al., 1999), wild-type Usp15 and mutant Usp15 (Usp15C783A) were a kind gift of Bettina Hetfeld (Hetfeld et al., 2005). SK-N-SH cells were co-transfected with 1.5 µg of Usp15 construct or empty vector and 0.5 µg EGFP-httQ74 and incubated for 24 h. Small interfering RNA transfections in HeLa cells were carried out using 50 nM control (non-targeting SMARTpool), or specific ON-TARGETplus SMARTpool (Dharmacon). After 24 h cells were further transfected with 0.5 µg EGFP-httQ74 and incubated for 48 h. Cells were fixed for 20 min in 4% formaldehyde at room temperature. Slides were mounted in Citifluor (Citifluor Ltd.) containing 4′,6-diamidino-2-phenylindole (3 µg/ml). Transfected cells were scored for aggregation and cell death using an Eclipse E600 fluorescence microscope (plan-apo 60×/1.4 oil immersion lens) (Nikon). Cell death was measured as the number of nuclei demonstrating apoptotic morphology (fragmentation or pyknosis). At least 200 transfected cells per slide were scored in triplicate with the scorer blinded to treatment and each experiment was carried out on three independent occasions.
Results
Temsirolimus enhances rotarod performance in a mouse model of spinocerebellar ataxia type 3
Transgenic mice overexpressing full-length human ataxin-3 with an expanded polyglutamine repeat of 70 glutamines have previously been shown to possess a neuropathological phenotype (Bichelmeier et al., 2007). In our laboratory, the mice did not demonstrate as severe a phenotype as previously reported. Transgene expression was not seen to decrease lifespan and no overt cellular phenotype was noted by staining of cerebellum with calbindin or phosphorylated neurofilament antibodies (data not shown), although clear mutant ataxin-3 aggregation was observed (see below). We examined the motor coordination of these mice using rotarod performance. A decreased ability to walk on the rotarod was observed in mice expressing expanded polyglutamine ataxin-3 relative to non-transgenic littermate controls (Fig. 1A). This decrease was most pronounced in young mice as the rotarod performance of wild-type mice declined with age. Rotarod assessment was the only motor phenotype that gave quantifiable readouts in the first 6 months of life; we saw no clear effects with tremor, fore- and hind-limb grip strength, wire manoeuvre or hind limb grasping (data not shown).
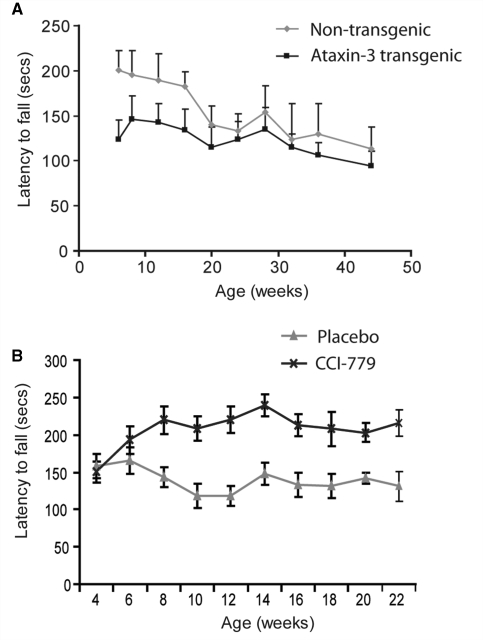
Figure 1.
Temsirolimus administration improves rotarod performance in ataxin-3 transgenic mice. (A) Mice expressing mutant ataxin-3 (black line) demonstrate a decreased latency to fall on the accelerating rotarod test compared with non-transgenic littermate controls (grey line). Data shown represent mean values and standard error. P < 0.001 by ANOVA. (B) Mice administered temsirolimus (CCI-779; black line) show an increase in performance on the rotarod over placebo treated control animals (grey line) throughout the period of the study. P < 0.001 by ANOVA. For placebo treated mice n = 17 at each time point. For temsirolimus treated mice n = 17 up to and including 14 weeks, n = 15 at 16 weeks and n = 14 thereafter.
We treated spinocerebellar ataxia type 3 transgenic mice with temsirolimus, a water-soluble rapamycin analogue, or vehicle control administered by intraperitoneal injection exactly as described previously (Ravikumar et al., 2004). Temsirolimus had no effect on rotarod performance in wild-type mice (data not shown). The polyQ repeat length of all animals in the trial was confirmed as being 70 (data not shown). Mice were tested for rotarod performance at 5 weeks of age, before beginning temsirolimus treatment. Animals were split into placebo and treatment groups (within littermate groups) ensuring both groups had similar initial rotarod performance (placebo 157 ± 14 s, treatment 150 ± 16 s; not significant by t-test). Treatment was started at 6 weeks of age by which time transgenic mice already show decreased rotarod performance (Fig. 1A). Performance was monitored every second week until the mice reached 21 weeks of age. Throughout this time mice given temsirolimus were able to walk for longer on the accelerating rotarod test than placebo treated mice (Fig. 1B).
Ataxin-3 transgenic mice did not show premature lethality in our animal house, at least until 18 months. We lost two mice in the study as a result of injection complications, one mouse was culled due to peritonitis and another died acutely during injection (presumably due to injection into a vessel or organ). In addition, two mice died unexpectedly of unknown causes during the period of the trial.
Rapamycin up-regulates autophagy in cortical neurons
Rapamycins, including temsirolimus, up-regulate autophagy by inhibiting the kinase mTOR. We therefore confirmed that the mTOR pathway was inhibited in the brains of treated mice. As we previously observed in a mouse model of Huntington’s disease (Ravikumar et al., 2004), mice treated with temsirolimus showed a decrease in immunoreactivity for phosphorylated S6 ribosomal protein, a downstream target of the mTOR pathway, whilst immunoreactivity for total S6 ribosomal protein did not change (Fig. 2A).
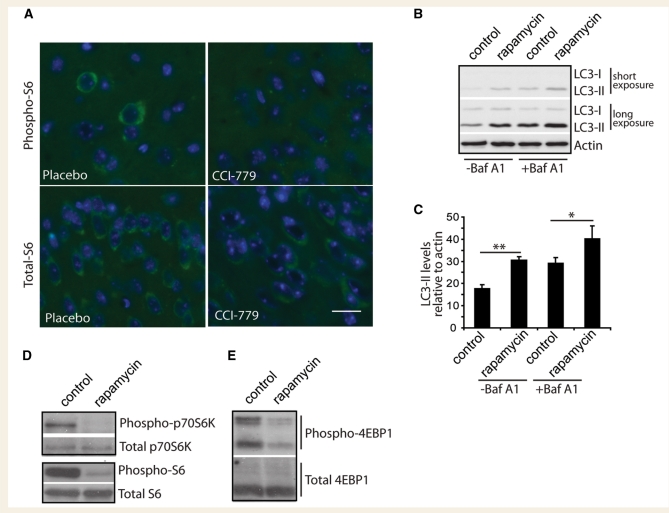
Figure 2.
Temsirolimus inhibits the mTOR pathway in vivo, as rapamycin does in cultured primary neurons. (A) Sections from mouse brains treated with either temsirolimus (CCI-779) or placebo were immunostained for phosphorylated S6 protein (Phospho-S6, top panels) or total S6 protein (bottom panels), nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Total S6 immunoreactivity was seen in the perinuclear region of all cells. In placebo treated animals some cells stained positive for phosphorylated S6, however, this staining was not seen in sections from animals treated with temsirolimus. Scale bar represents 5 µm and is valid for all panels. In cultured primary neurons, LC3-II levels were assessed by western blot (B). Two different exposures are shown to allow comparison of weaker bands in untreated lanes (−Baf A1) and stronger bands in Bafilomycin A1 (+Baf A1) lanes without saturation. Densitometric quantification of LC3-II levels relative to actin in triplicate experiments is shown in (C). **P < 0.01 and *P < 0.05 by t-test. Effect of rapamycin treatment of phosphorylation of downstream mTOR targets was investigated by western blotting, (D) phosphorylated p70 S6 kinase levels and phosphorylated S6 ribosomal protein and (E) phosphorylated eukaryotic initiation factor 4E-binding-protein (EBP)-1.
Inhibition of this pathway in vivo strongly suggests the up-regulation of autophagy. A more direct assessment of autophagy levels can be achieved by measuring LC3-II levels, as this protein is specifically associated with autophagosomes. However, demonstration of increased LC3-II levels in vivo is not possible as neurons appear to clear autophagosomes very efficiently (Boland et al., 2008). We therefore used primary neuronal culture to show that treatment with rapamycin increases LC3-II levels (Fig. 2B and C). This also allowed us to demonstrate that this increase in LC3-II levels results from an increase in synthesis rather than a decrease in degradation of autophagosomes, as increased levels were also seen in the presence of bafilomycin-A1, an autophagy inhibitor that blocks autophagosome lysosome fusion and therefore LC3-II degradation (Fig. 2B and C). As demonstrated in vivo, we observed that treatment of cortical neurons with rapamycin inhibited the mTOR pathway. By western blotting we could observe a decrease in phosphorylation of p70S6 kinase and eukaryotic initiation factor 4E-binding-protein-1, both substrates for mTOR, as well as decreased levels of phosphorylated S6 protein, a substrate of p70S6 kinase (Fig. 2D and E).
Temsirolimus treatment decreases aggregate number in the brains of ataxin-3 transgenic mice
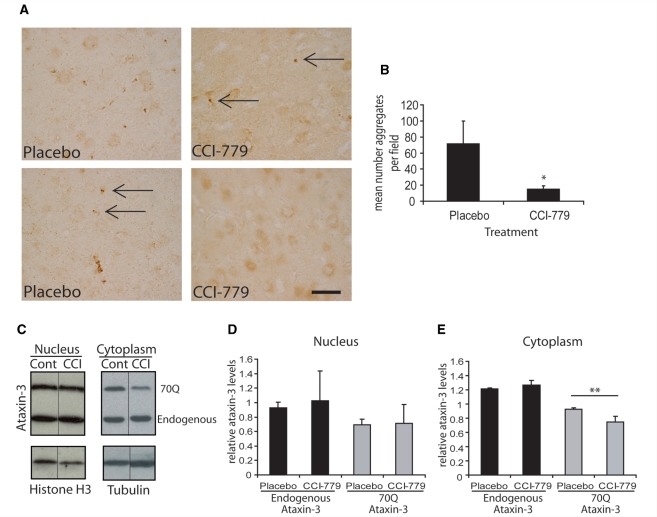
To assess the cellular effects associated with the improvement in behavioural testing seen in temsirolimus treated mice we looked at ataxin-3 positive aggregates in the brains of treated and control mice. Mice treated with placebo show a large number of aggregates throughout the brain. We scored aggregates in the motor cortex as this region demonstrated a high level of aggregates, while aggregate number could still be counted accurately, making it the most suitable region for this assay. Mice were treated for two months with temsirolimus or placebo beginning at 5 weeks of age (as for rotarod performance). Mice were then sacrificed and the number of aggregates in the motor cortex quantified. Aggregate number was significantly reduced in mice treated with temsirolimus compared to placebo treated mice (Fig. 3A and B).
Figure 3.
Aggregation and clearance of ataxin-3 with temsirolimus treatment. (A) Ataxin-3 staining in the motor cortex reveals the presence of aggregates in transgenic mice. Control (placebo) mice show more aggregates in the motor cortex than treated (temsirolimus, CCI-779) animals. Examples of aggregates are indicated by arrows in both treated and untreated brains. Scale bar represents 20 µm and is valid for all pictures. Quantification of the mean number of aggregates is shown in (B). Aggregates were counted in the motor cortex on three sections each for five mice in each treatment group (n = 5). *P < 0.05 placebo versus temsirolimus treated mice by t-test. Levels of endogenous, and transgenic mutant ataxin-3 in placebo (cont) and drug treated (CCI) mice were measured by western blotting (C). The expanded polyglutamine stretch of the mutant protein results in its slower migration in the gel (upper band). Tubulin is used as a control to ensure equal loading of cytoplasmic samples and Histone H3 for nuclear samples. The black line marks where lanes of the western have been omitted for the sake of clarity. Quantification by densitometry of ataxin-3 levels in nuclear protein extracts (D) and cytosolic extracts (E) from three mice corrected for the level of tubulin (n = 3). **P < 0.01 placebo versus temsirolimus treated mice by t-test.
Temsirolimus reduces cytoplasmic levels of expanded ataxin-3
In spinocerebellar ataxia type 3, ataxin-3 aggregates show predominantly nuclear localization, and this nuclear localization is required for toxicity (Bichelmeier et al., 2007). Autophagy is a cytoplasmic process and we were therefore interested to increase our understanding of the cellular localization of the effect of temsirolimus. We measured soluble protein levels in the cytoplasmic and nuclear fractions from mouse brains. The levels of soluble 70Q ataxin-3 were reduced in the cytoplasm of treated mouse brains, but not in the nuclear fraction (Fig. 3C–E). We also observed that, while the levels of the expanded polyglutamine ataxin-3 protein are reduced in the cytoplasmic fraction, there was no change in the levels of the endogenous wild-type ataxin-3 protein in either the cytoplasm or the nucleus. This suggests the mutant, aggregate-prone protein is more efficiently degraded by autophagy than the wild-type, endogenous protein. This is entirely consistent with our previous data from cell culture models of different aggregate-prone proteins, where the mutant form is much more dependent on autophagy for clearance (Williams et al., 2008).
Modest effects on transcription are seen with the protective effect of temsirolimus
Whilst the pathogenisis of spinocerebellar ataxia type 3 is unknown it has been hypothesized that transcriptional dysregulation may play a role (reviewed in Riley and Orr, 2006). In order to investigate the importance of transcriptional changes in disease pathogenesis we used a microarray approach to look at changes in gene expression in ataxin-3 transgenic mice and the reversal of these changes associated with the improved phenotype in temsirolimus treated mice. As our ataxin-3 transgenic mice do not have a severe phenotype, transcriptional changes identified are likely to be associated with early disease. We hypothesized that investigating gene expression changes that are associated with the improvement in the phenotype seen in temsirolimus treated ataxin-3 transgenic mice would allow us to identify gene expression changes that are important in the progression of the disease, since temsirolimus administration reduces the levels of the primary ‘toxin’, mutant ataxin-3.
RNA was extracted from the brains of four groups of mice, wild-type or ataxin-3 transgenic mice administered either placebo or temsirolimus. From these datasets we identified transcripts that were altered by more than 1.5-fold in transgenic, control (placebo-treated) animals compared to wild-type, control animals. This analysis identified 91 transcripts. The overall magnitude of the changes in transcription were small with 77 genes showing changes of between 1.5- and 2-fold, 13 genes showing changes between 2- and 3-fold and one gene, CUG triplet repeat, RNA binding protein 2 showing a 3.16-fold down-regulation in transgenic versus non-transgenic placebo treated animals. This is of interest as another CUG repeat binding protein, muscleblind (Miller et al., 2000), has been shown to be a modulator of ataxin-3 toxicity in Drosophila (Li et al., 2008). Overexpression of muscleblind led to an increase in toxicity of mutant ataxin-3 by a mechanism involving toxicity of the mutant RNA (Li et al., 2008).
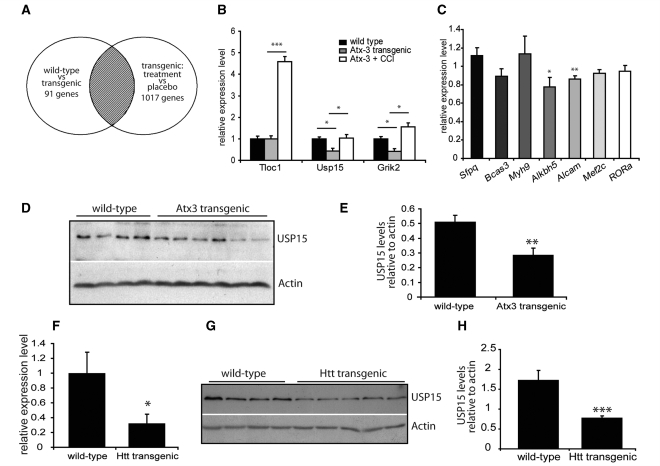
From the 91 transcripts altered in wild-type versus transgenic animals we selected those that were also changed in the opposite direction (i.e. returned to normal levels) in transgenic animals treated with temsirolimus (Fig. 4A). This analysis identified only 16 genes (Table 1) whose expression was decreased in transgenic ataxin-3 mice and increased in transgenic mice treated with temsirolimus. We did not identify any genes whose expression increased in transgenic ataxin-3 mice and decreased in temsirolimus treated mice.
Figure 4.
Microarray analysis of transgenic ataxin-3 mice. Microarray analysis of four experimental groups of mice was carried out and changes between pairs of experimental groups were identified. (A) Shows a representation of these comparisons as a Venn diagram. The number of genes with altered regulation are given for each pair of groups. The shaded region identifies the transcripts selected i.e. those altered in wild-type versus transgenic mice that were normalized following the treatment of mice with temsirolimus (see Table 1). Microarray changes were verified by quantitative PCR. (B) Shows mRNA relative expression levels of Tloc1, Usp15 and Grik2, expression levels are normalized to wild-type, placebo treated mice (black bars). Grey bars represent placebo treated, ataxin-3 transgenic mice and white bars temsirolimus treated ataxin-3 transgenic mice. *P < 0.05 and ***P < 0.001 by t-test. (C) Additional verification of transcript alterations in wild-type versus ataxin-3 transgenic mice. mRNA was extracted from brains of untreated mice and expression levels of measured relative to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase 2. Expression was normalized to levels in wild-type mice and the relative levels in ataxin-3 transgenic mice are shown. *P < 0.05 and **P < 0.01 by t-test, n = 3, other comparisons are not statistically significant. Protein levels of Usp15 were analysed in 3.5–4 month old wild-type (n = 4) and transgenic ataxin-3 (n = 6) mouse brain by western blotting (D). Protein levels were quantified by densitometry relative to the level of the loading control, actin (E). **P < 0.01 by t-test. (F) mRNA expression of Usp15 was measured in brains of 6 month old wild-type and huntingtin (Htt) transgenic mice. Expression levels are corrected for the housekeeping gene SDHA and are shown relative to wild- type mice. *P < 0.05 by t-test. (G) Protein levels of Usp15 were investigated by western blot with actin as a loading control. Protein levels were quantified by densitometry relative to the level of actin (H). ***P < 0.001 by t-test.
Expression changes seen by microarray were verified by quantitative PCR. Of the first three genes we looked at, we confirmed a decrease in expression of Usp15 and Grik2 between wild-type and ataxin-3 transgenic mice, which were reversed in mice treated with temsirolimus (Fig. 4B). However, no decrease was seen in Tloc1 levels in ataxin-3 transgenic mice compared to wild-type mice, although an increase in Tloc1 levels was seen following temsirolimus administration. Of the remaining 10 known genes identified by our microarray experiments we were able to successfully amplify mRNA transcripts from seven (Bcl2l11, Dgki and Mnab PCRs were unsuccessful) and we confirmed decreases in Alkbh5 and Alcam mRNA levels in ataxin-3 transgenic mice compared to wild-type (Fig. 4C). Whilst mRNA levels were clearly altered for both Usp15 and Grik2 changes at the protein level could only be observed with Usp15 (Fig. 4D and E). This may be because western blotting is a less sensitive method of detecting changes and small changes in Grik2 level may be missed, or because the rate of transcription of Grik2 is not an important step in the control of protein expression level. Additionally, in agreement with the quantitative PCR results we did not observe any changes in the level of protein expression of Sfpq and RORa (Supplementary Fig. 1).
Decreases in ubiquitin specific peptidase-15 levels are seen in mice expressing mutant ataxin-3 and mutant huntingtin
The identification and verification of changes in Usp15, a deubiquitinating protein, was of great interest. Ataxin-3 has been previously shown to possess deubiquitinating activity (Burnett et al., 2003) the N-terminal Josephin domain bears homology to the UBP (ubiquitin specific protease) family of deubiquitinating enzymes, which includes Usp15 (Scheel et al., 2003). Additionally, the deubiquitinating activity of ataxin-3 has been shown to be regulated by ubiquitination of ataxin-3 itself (Todi et al., 2009). We were therefore interested to investigate further the potential role of changes in expression level of Usp15 in spinocerebellar ataxia type 3.
To establish if this decrease was specific for spinocerebellar ataxia type 3 and associated with the function of ataxin-3, we investigated the levels of both Usp15 mRNA and protein in a mouse model of another polyglutamine disorder, Huntington’s disease (Schilling et al., 1999). These mice express an N-terminal fragment of huntingtin (171 amino acids) with an expanded polyglutamine repeat region of 82 glutamines. As for mutant ataxin-3 transgenic mice, mutant huntingtin transgenic mice also showed a decrease in mRNA and protein levels of Usp15 (Fig. 4F–H).
Overexpression of ubiquitin specific peptidase-15 results in an increase in polyglutamine aggregation
To investigate further the relationship between Usp15 and polyglutamine containing proteins we looked at the effect of altering levels of Usp15 expression in cells. As mutant ataxin-3 has minimal toxicity and does not form aggregates in cell culture systems, exon 1 of huntingtin with an expanded polyglutamine repeat region of 74 glutamines was used in toxicity and aggregation studies. To mimic the alteration in Usp15 levels seen in mutant ataxin-3 and huntingtin transgenic mice, cells were treated with small interfering RNA against Usp15. This resulted in a decrease in Usp15 protein levels (Supplementary Fig. 2A) although no changes in aggregation or toxicity of mutant huntingtin were observed (Supplementary Fig. 2B and C). Whilst decreasing Usp15 levels showed no effect, overexpression of Usp15 resulted in an increase in aggregation levels (Supplementary Fig. 2D). Usp15 is able to cleave both polyubiquitin chains and monoubiquitinated substrates (Hetfeld et al., 2005). The effect of Usp15 overexpression on polyglutamine aggregation is independent of its polyubiquitin protease activity as overexpression of a mutant form of the protein, Usp15C783A, which is unable to degrade polyubiquitin substrates (Hetfeld et al., 2005), showed the same effect as the wild-type protein. This mutant Usp15 is still able to cleave monoubiquitinated substrates. Despite increasing aggregation, overexpression of Usp15 did not increase toxicity of the expanded polyglutamine protein (Supplementary Fig. 2E).
Discussion
We have shown that administration of an mTOR inhibitor, temsirolimus, ameliorates the effects of ataxin-3 overexpression in a mouse model of spinocerebellar ataxia type 3. mTOR inhibition induces autophagy in all cells tested, from S. cerevisiae to mammalian neurons (Noda and Ohsumi, 1998; Boland et al., 2008). While rapamycin induces autophagy in primary neurons, this will be difficult to demonstrate definitively in vivo as neurons appear to clear autophagosomes very efficiently (Boland et al., 2008). However, consistent with mTOR inhibition inducing autophagy, ataxin-3 transgenic mice treated with temsirolimus showed decreased levels of phosphorylated S6 ribosomal protein (a downstream target of the mTOR pathway), and levels the mutant protein in the cytosol were decreased, as were the number of aggregates.
Temsirolimus treatment also resulted in an improvement in motor coordination as measured by rotarod performance. As the phenotype of the ataxin-3 overexpressing mice used in this study is mild, we were unable to assess a broader range of phenotypes. Ideally, it would be helpful to test this therapeutic strategy in other rodent models of spinocerebellar ataxia type 3 in future, if they show stronger and more robust phenotypes. Despite this, our study demonstrates that temsirolimus may have additional therapeutic values beyond the effect previously demonstrated by pre-symptomatic treatment in a mouse model of Huntington’s disease (Ravikumar et al., 2004). Indeed, autophagy induction may be effective even if initiated after disease onset. However, treatment strategies aiming to enhance protein clearance may be most effective prior to overt disease onset or early in disease course. In an elegant study, Orr and colleagues developed an inducible mouse model of the polyglutamine disease spinocerebellar ataxia type 1 (Zu et al., 2004). They demonstrated that, if transgene expression was turned off early in the disease course, there was excellent recovery, but switching off the mutant transgene later in the disease did not result in the same extent of recovery. However, as human diseases do not progress as rapidly as most of the mouse models used for preclinical compound testing, there may be an increased window of opportunity for treatment of patients.
In our study we observe a reduced level of soluble 70Q ataxin-3 in the cytoplasm of treated mouse brains, but not in the nuclear fraction (Fig. 3C–E). We believe that autophagy predominantly clears the soluble species of the mutant protein, rather than the large aggregates that are visible by light microscopy (discussed in Rubinsztein, 2006). However, the number of cells with aggregates is directly proportional to expression of the mutant protein (Narain et al., 1999). The aggregate load in the nucleus is likely to change when cytosolic ataxin-3 is depleted, since the protein shuttles between the nucleus and cytoplasm (Pozzi et al., 2008). It is possible that changes in the nuclear soluble pool assessed by western blotting are more difficult to quantify than changes in aggregate number.
While the major contributing factor to the loss of toxicity of ataxin-3 by temsirolimus is likely to be the up-regulation of autophagy, there are also other factors that could play a protective role. Firstly, as well as increasing protein clearance, up-regulation of autophagy has been demonstrated to have cytoprotective effects against apoptotic death via the intrinsic, mitochondria-dependant pathway (Ravikumar et al., 2006). Secondly, in addition to inducing autophagy, rapamycin has been suggested to act to reduce toxicity of aggregate-prone proteins by decreasing protein translation (King et al., 2008). However, it is important to point out that the clearance of these aggregate-prone proteins is impaired when one blocks autophagy via a wide range of mTOR-independent chemical and genetic pathways (Ravikumar et al., 2002; Berger et al., 2006). Likewise, induction of autophagy via mTOR-independent pathways enhances the clearance of aggregate-prone proteins and is protective in different polyglutamine models in cells, flies and zebrafish (Williams et al., 2008). Finally, the beneficial effects of rapamycin in fly models of these diseases appear to be autophagy-dependent, as rapamycin had no effects on toxicity in flies expressing different aggregate-prone mutant proteins on a background of reduced activity of different autophagy genes (Berger et al., 2006; Pandey et al., 2007). While these data suggest that rapamycin treatment is dependent on autophagy in Drosophila, we cannot exclude an additional effect of temsirolimus via mild translational inhibition in the spinocerebellar ataxia type 3 transgenic mice. However, any such effect could not be a consequence of global translational inhibition, as we observed no changes in the levels of wild-type ataxin-3, consistent with previous data suggesting that the mutant forms of these proteins are much more dependent on autophagy for their clearance compared to their wild-type forms (discussed in Rubinsztein, 2006).
Treatment of proteinopathies with drugs that up-regulate autophagy has shown tantalizing results in a small trial of another human disease associated with intracytosolic inclusion formation, amyotrophic lateral sclerosis. Lithium induces autophagy by lowering intracellular inositol or inositol 1,4,5-trisphosphate levels in an mTOR independent manner (Sarkar et al., 2005) and delays disease progression in patients with amyotrophic lateral sclerosis and in a mouse model of amyotrophic lateral sclerosis overexpressing a mutant form of Superoxide dismutase 1 (Fornai et al., 2008). While lithium did induce autophagy in neurons in the mouse models and facilitated Superoxide dismutase 1 clearance, it is possible that this drug had additional autophagy-independent protective effects. Nevertheless, the mTOR-independent nature of the action of lithium and its cytoprotective and autophagy- inducing properties offer the possibility for the development of treatment rationales involving simultaneous lithium and rapamycin administration (Sarkar et al., 2008). Therefore there may be much potential for the use of drugs upregulating autophagy in the treatment of a diverse range of proteinopathies.
The mouse model described here and the rescue seen with the administration of temsirolimus offered the possibility to investigate the role of transcriptional dysregulation in spinocerebellar ataxia type 3. Polyglutamine disorders have been suggested to result from an altered transcription, (reviewed in Riley and Orr, 2006). The evidence for this comes largely from studies of Huntington’s disease, but has also been suggested in spinocerebellar ataxia type 3 due to the observations that ataxin-3 may affect histone acetylation (Li et al., 2002; Evert et al., 2006). In this study we attempted to identify changes in transcription that may be important in the early disease progression of spinocerebellar ataxia type 3. Ataxin-3 transgenic mice show a relatively mild phenotype, this is advantageous as secondary transcriptional changes that are not causative of disease phenotype are likely to be reduced. Additionally, focusing on transcripts that were reverted to normal expression levels with a reversal of the disease phenotype by temsirolimus allowed us to look more directly for transcriptional changes specific to disease pathogenesis. Surprisingly we observed only a few such changes in gene expression. It is also of note that the magnitude of expression changes seen was small, with most transcripts showing changes of 1.5–2-fold.
We were able to confirm the changes in expression level of Usp15 seen by microarray at both the mRNA and protein level. Furthermore, its expression was also decreased in mutant huntingtin transgenic mice. However, changes in expression of Usp15 did not modulate toxicity of a model polyglutamine protein in cell culture. Whilst this appears to suggest that changes in Usp15 expression are not a major contributing factor to the pathogenesis of spinocerebellar ataxia type 3, it is also possible that changes in this gene may be important in the slowly progressing phenotype seen in mouse models and in the human disease, but not in the acute cell system used here.
In conclusion, we have demonstrated that treatment of transgenic ataxin-3 mice with temsirolimus, a drug which up-regulates autophagy, leads to a decrease in protein levels of the mutant polyglutamine protein and that this is associated with improved behavioural outcomes in the mice. Transgenic ataxin-3 mice also demonstrate a mild perturbation in transcription which is partially relieved by treatment with temsirolimus. However, the importance of these transcriptional alterations in the pathogenesis of spinocerebellar ataxia type 3 remains unclear.
Funding
Wellcome Trust Senior Research Fellowship awarded to DCR, by Wyeth and EUROSCA.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Acknowledgements
The authors would like to thank Wyeth for providing temsirolimus.
Glossary
Abbreviations
- Alcam
activated leukocyte cell adhesion molecule
- Alkbh5
alkylation repair homolog 5
- Bcas3
Breast carcinoma amplified sequence 3
- Bcl2l11
B cell lymphoma protein 2 like 11
- Dgki
Diacylglycerol kinase, iota
- EGFP
enhanced green fluorescent protein
- Grik2
Glutamate receptor, ionotropic, kainate 2 (beta 2)
- LC
light chain
- Mef2c
Myocyte enhancer factor 2C
- Mnab1
membrane associated DNA binding protein
- mTOR
mammalian target of rapamycin
- Myh9
myosin, heavy polypeptide 9, non-muscle
- RORa
retinoic acid receptor-related orphan receptor alpha
- Sfpq
splicing factor proline/glutamine rich
- Usp
ubiquitin specific peptidase
References
- Albrecht M, Golatta M, Wullner U, Lengauer T. Structural and functional analysis of ataxin-2 and ataxin-3. Eur J Biochem. 2004;271:3155–70. doi: 10.1111/j.1432-1033.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–42. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- Bichelmeier U, Schmidt T, Hubener J, Boy J, Ruttiger L, Habig K, et al. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J Neurosci. 2007;27:7418–28. doi: 10.1523/JNEUROSCI.4540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's; disease. J Neurosci. 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett B, Li F, Pittman RN. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet. 2003;12:3195–205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- Carvalho DR, La Rocque-Ferreira A, Rizzo IM, Imamura EU, Speck-Martins CE. Homozygosity enhances severity in spinocerebellar ataxia type 3. Pediatr Neurol. 2008;38:296–9. doi: 10.1016/j.pediatrneurol.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Chai Y, Berke SS, Cohen RE, Paulson HL. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J Biol Chem. 2004;279:3605–11. doi: 10.1074/jbc.M310939200. [DOI] [PubMed] [Google Scholar]
- Doss-Pepe EW, Stenroos ES, Johnson WG, Madura K. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol Cell Biol. 2003;23:6469–83. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evert BO, Araujo J, Vieira-Saecker AM, de Vos RA, Harendza S, Klockgether T, et al. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. J Neurosci. 2006;26:11474–86. doi: 10.1523/JNEUROSCI.2053-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornai F, Longone P, Cafaro L, Kastsiuchenka O, Ferrucci M, Manca ML, et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2008;105:2052–7. doi: 10.1073/pnas.0708022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, et al. The zinc finger of the CSN-associated deubiquitinating enzyme Usp15 is essential to rescue the E3 ligase Rbx1. Curr Biol. 2005;15:1217–21. doi: 10.1016/j.cub.2005.05.059. [DOI] [PubMed] [Google Scholar]
- Huang S, Houghton PJ. Mechanisms of resistance to rapamycins. Drug Resist Updat. 2001;4:378–91. doi: 10.1054/drup.2002.0227. [DOI] [PubMed] [Google Scholar]
- Imarisio S, Carmichael J, Korolchuk, Chen CW, Saiki S, Rose C, et al. Huntington's; disease: from pathology and genetics to potential therapies. Biochem J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- King MA, Hands S, Hafiz F, Mizushima N, Tolkovsky AM, Wyttenbach A. Rapamycin inhibits polyglutamine aggregation independently of autophagy by reducing protein synthesis. Mol Pharmacol. 2008;73:1052–63. doi: 10.1124/mol.107.043398. [DOI] [PubMed] [Google Scholar]
- Li F, Macfarlan T, Pittman RN, Chakravarti D. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J Biol Chem. 2002;277:45004–12. doi: 10.1074/jbc.M205259200. [DOI] [PubMed] [Google Scholar]
- Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–11. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–48. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narain Y, Wyttenbach A, Rankin J, Furlong RA, Rubinsztein DC. A molecular investigation of true dominance in Huntington's; disease. J Med Genet. 1999;36:739–46. doi: 10.1136/jmg.36.10.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro G, Masino L, Esposito, Menon RP, De Simone A, Fraternali F, et al. The josephin domain of ataxin-3 contains two distinct ubiquitin binding sites. Biopolymers. 2009;91:1203–14. doi: 10.1002/bip.21210. [DOI] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Perez MK, Paulson HL, Pendse, Saionz, Bonini NM, Pittman RN. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J Cell Biol. 1998;143:1457–70. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C, Valtorta M, Tedeschi G, Galbusera E, Pastori, Bigi A, et al. Study of subcellular localization and proteolysis of ataxin-3. Neurobiol Dis. 2008;30:190–200. doi: 10.1016/j.nbd.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Berger Z, Vacher C, O'K;ane CJ, Rubinsztein DC. Rapamycin pre-treatment protects against apoptosis. Hum Mol Genet. 2006;15:1209–16. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–17. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Riley BE, Orr HT. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–92. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–11. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Krishna G, Imarisio S, Saiki S, O'K;ane CJ, Rubinsztein DC. A rational mechanism for combination treatment of Huntington's; disease using lithium and rapamycin. Hum Mol Genet. 2008;17:170–8. doi: 10.1093/hmg/ddm294. [DOI] [PubMed] [Google Scholar]
- Scheel H, Tomiuk S, Hofmann K. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum Mol Genet. 2003;12:2845–52. doi: 10.1093/hmg/ddg297. [DOI] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- Schols L, Bauer P, Schmidt T, Schulte T, Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3:291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–85. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–8. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- Todi SV, Winborn BJ, Scaglione KM, Blount JR, Travis SM, Paulson HL. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 2009;28:372–82. doi: 10.1038/emboj.2008.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's; disease. Hum Mol Genet. 2005;14:3425–33. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Cell Biol. 2006;174:963–71. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, et al. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington's; disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet. 2006;15:2409–20. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- Zu T, Duvick LA, Kaytor MD, Berlinger MS, Zoghbi HY, Clark HB, et al. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J Neurosci. 2004;24:8853–61. doi: 10.1523/JNEUROSCI.2978-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.