Abstract
Plant cells contain a mixture of 26S and 20S proteasomes that mediate ubiquitin-dependent and ubiquitin-independent proteolysis, respectively. The 26S proteasome contains the 20S proteasome and one or two regulatory particles that are required for ubiquitin-dependent degradation. Comparative analyses of Arabidopsis proteasome mutants revealed that a decrease in 26S proteasome biogenesis causes heat shock hypersensitivity and reduced cell division rates that are compensated by increased cell expansion. Loss of 26S proteasome function also leads to an increased 20S proteasome biogenesis, which in turn enhances the cellular capacity to degrade oxidized proteins and thus increases oxidative stress tolerance. These findings suggest the intriguing possibility that 26S and 20S proteasome activities are regulated to control plant development and stress responses. This mini-review highlights some of the recent studies on proteasome regulation in plants.
Key words: proteasome, cell division, ubiquitin-dependent proteolysis, ubiquitin-independent proteolysis, stress responses
Introduction
The 26S proteasome (26SP), the proteolytic component of the ubiquitin (Ub)-dependent proteolytic system (UPS), is essential in eukaryotes.1,2 The 26SP degrades functional proteins that have been covalently linked to a polyUb chain, and thus negatively controls the abundance of numerous regulatory proteins involved in a myriad of signaling and metabolic pathways. In addition to this regulatory function, the 26SP is essential for protein quality control because it degrades misfolded and denatured proteins produced by translation errors or post-synthetic damage. These non-functional proteins are first recognized by chaperons, then ubiquitinated through the actions of chaperon-binding Ub ligases, and finally degraded by the 26SP.3–6
Most of the current research efforts in the UPS field are focused on the ubiquitination step, and because Ub ligases are key specificity factors of target ubiquitination, they are the topic of most of the published studies.7–9 This current focus has also been strengthened by the generally accepted belief that alterations in the specificity, processivity and abundance of the 26SP are not significant contributors to plant growth regulation. Recent reports however suggested that fluctuations in proteasomal activity impact plant cell proliferation and expansion rates and play a role in stress responses. Here we summarize these findings, and propose that developmentally and environmentally induced changes in proteasome abundance (and possibly, proteasome composition) are important for plant development and survival under adverse conditions.
The 26SP and Growth
The 26SP is composed of two subparticles, the cylindrically shaped 20S proteasome (20SP) that serves as the protein destruction chamber and regulatory particles (RPs) that are attached to both ends of the 20SP cylinder. The RPs provide the substrate specificity to the 26SP by interacting with and unfolding polyubiquitinated target proteins that are subsequently channeled into the 20SP for degradation.10 The 20SP is composed of 14 different subunits the majority of which are in Arabidopsis encoded by gene pairs. The RP is composed of at least 17 different subunits most of which are also encoded by two-membered gene families.10 The RP subunits form two different subparticles, the RP base that directly controls the opening into the 20SP and unfolds the target proteins, and the RP lid that is attached to the base and contains subunits of which the functions are largely unknown. Like in other eukaryotes, the 26SP is an essential cellular component of plant cells. For example, combining knock-out mutations in both copies of genes encoding Arabidopsis RP subunit isoforms led to the lethality of both male and female gametes.11,12
While the analyses of null mutants of RP subunits confirmed the essential function of the 26SP in plants, studies using weak loss-of-function mutants have allowed to determine the impact of fluctuations in proteasome activity on plant growth and stress responses.13–19 For example, comparative analyses of Arabidopsis 26SP mutant lines rpt2a-2, rpt2a-3, rpn10-1 and rpn12a-1 revealed that reductions in 26SP abundance invariably lead to increased cell expansion combined with decreased cell division rates.13 The decrease in cell numbers and compensative increase in cell sizes was already apparent in mutants in which 26SP activity was reduced by 40% compared to the wild type.13 It is easily imagined—but harder to prove in situ—that such a modest reduction in 26SP activity can be induced by developmental signals in a particular tissue or organ, thus tilting the balance of cellular development in favor of cell expansion. Furthermore, and following the same line of thinking, it would be expected that tissues with high cell division rates have a high 26SP activity. Analyses of the 26SP subunit gene set indeed revealed that the expression of 26SP genes is generally the highest in meristems and in young unexpanded organs and is consistently lower in mature tissues.10 Comparative analyses of weak 26SP loss-of-function mutants therefore suggested that the alteration of 26SP activity might be an important developmental mechanism for balancing cell division and expansion rates in plants.
The 26SP and Stress Responses
Since plant cell proliferation rates depend on an optimal 26SP activity, it is expected that stresses that directly impact 26SP activity will indirectly cause a reduction in cell proliferation. Several abiotic stress conditions are known to inhibit 26SP activity either by increasing the substrate load and thus slowing down the turnover rates of other 26SP targets or by directly inhibiting 26SP function. Heat shock and other stresses that cause protein misfolding lead to a substrate overload. Oxidative stress, on the other hand, directly leads to 26SP inhibition.20 In both cases, the inhibition of 26SP activity is predicted to decrease cell division rates.
In addition to these direct mechanisms that are likely to be relevant when the intensity of a stressor is high, plant cells may have evolved more elaborate molecular mechanisms that promote changes in 26SP activity in response to variations in environmental conditions. Recent studies have suggested that this type of mechanism exists and it depends on the UPS.21–23 The hot pepper (Capsicum annuum L.) U-box protein 1 (CaPUB1) and its Arabidopsis homologues AtPUB22 and AtPUB23 are Ub ligases, and the corresponding genes are rapidly induced by abiotic stresses such as desiccation, cold and mechanical wounding.21,22 Both the hot pepper and Arabidopsis PUBs seem to interfere with 26SP function by ubiquitinating specific subunits of the RP lid subcomplex. CaPUB1 ubiquitinates RPN6, and because the RPN6 level was reduced in CaPUB1 overexpressing plants, it has been suggested that the ubiquitination leads to the destabilization of this RP subunit.21 AtPUB22 and AtPUB23 ubiquitinate RPN12a, which leads to the relocation of a fraction of RPN12a into a cytosolic complex reminiscent of the proteasome related 500-kDa complex (PR500). PR500 contains subunits of the RP lid, and exists in unstressed plants as a separate particle, while it dissipates in response to heat shock and treatments with the amino acid analog canavanine.24 The dynamics of the PR500 complex during other stresses have not yet been investigated. Although the exact function of PR500 is still unknown, one hypothesis is that it functions as a reserve of RP subunits used for accelerated 26SP biogenesis that is needed to combat the effects of protein misfolding stresses.24 The current data however suggest that PR500 might be more than just a standby reserve of RP subunits; it appears to be a dynamic particle to which RP subunits are added during desiccation stress and from which RP subunits are recruited during protein misfolding stress.
The predicted effect of AtPUB22/23 action would therefore be to reduce 26SP levels in response to the drought stress by redirecting a portion of the RP subunits to the PR500 particle. It has been suggested that desiccation tolerance requires the UPS and therefore, a reduction in total 26SP activity should be detrimental for plant survival.25 Indeed, AtPUB22/23 overexpression caused hypersensitivity to drought stress, while their loss of function resulted in drought tolerance.21,22 This is somewhat paradoxical since the stress-induced expression of both ligases suggested that they are needed by plant cells to counteract the negative impact of the stress. The AtPUB22/23 overexpression studies did reveal two potentially beneficial traits for plants subjected to water stress: increased root elongation, which increases a plants' ability to maintain water uptake, and accelerated flowering that secures the production of progeny under adverse conditions. Thus, it is possible that both ligases provide a benefit to plants experiencing reduced water availability in the field. Loss of 26SP function is known to cause decreased root growth.14 Therefore, the increased root elongation in AtPUB22/23 overexpression plants might indicate that AtPUB22/23 have functions in addition to their effect on 26SP biogenesis. In summary, the exact mechanism and function of the 26SP regulation in the drought stress response remains unclear, and further studies are needed to understand the complexities of the proteasomal involvement in this prominent research area.
The 26SP to 20SP Ratio and Senescence
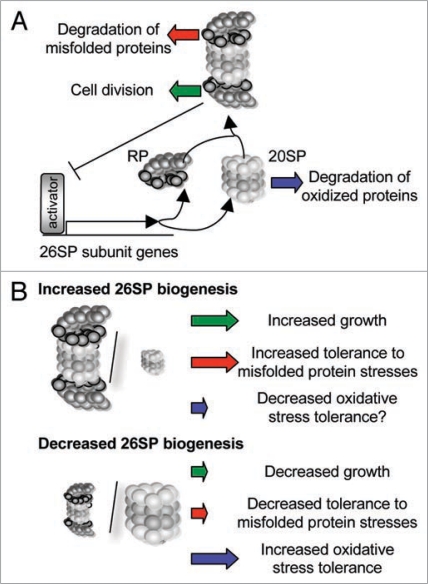
Proteins damaged by cellular stresses are degraded by proteasomes, and the current studies suggest that in all eukaryotes, the 26SP is responsible for the degradation of misfolded proteins and the free 20SP is needed for the removal of oxidized proteins.14,26–32 Plant cells contain a mixture of the 26SP and free 20SP, and comparative analyses of mutants that are defective in 26SP accumulation showed that optimal 26SP levels are needed to maintain tolerance to stresses, such as heat shock, that cause protein misfolding, while elevated levels of the free 20SP lead to increased tolerance to oxidative stress.14 These studies also revealed an inverse relationship between 26SP and 20SP abundance, with reductions in 26SP abundance invariably leading to increased 20SP biogenesis (Fig. 1), and suggested that regulation of the 26SP to 20SP ratio is important for plant development and stress responses.
Figure 1.
Model summarizing the effects of gain or loss of 26SP function on plant growth and abiotic stress tolerance. (A) 26SP-dependent feed-back regulation mechanism that controls 26SP and 20SP biogenesis. An unknown transcriptional activator, which is a UPS target itself, regulates the transcription of proteasome subunit genes.14 With help of a number of chaperones, proteaseome subunits assemble into the 20SP and rP complexes, and then join to form the 26SP.10,37 According to this model, sustained inhibition of 26SP assembly or function leads to the increased expression of proteasome subunit genes, resulting in increased 20SP but not 26SP activity. (B) Outcomes of increased and decreased 26SP activity on plant growth and stress tolerance levels.
The first evidence for a developmental regulation of the 26SP to 20SP ratio was obtained in Drosophila.33 This study shows that 26SP levels decline during the aging process while 20SP biogenesis increases. A similar trend, i.e., decline in 26SP levels combined with increase in 20SP levels, was suggested by a proteomics study of potato tuber aging.34 Combined, these studies imply that the regulation of 26SP to 20SP levels during aging and senescence may be a conserved mechanism in higher eukaryotes. Currently, this mechanism is poorly understood. The aging-dependent decrease in 26SP to 20SP ratio may be a causal factor in the aging process. A reduction in 26SP activity is expected to lead to a decreased capacity for removing misfolded proteins, resulting in the accumulation and aggregation of misfolded proteins which can accelerate cell death. In support of this explanation, a recent Drosophila study has shown that life span can be extended by preventing a decline in the 26S proteasome level during the ageing process.35 Alternatively, the change in 26SP to 20SP ratio could be induced by aging-related changes in the cell. Aging is typically accompanied by an increased production of free radicals and increased oxidative damage to cellular components.36 Under these conditions a shift in the proteasomes ratio in favor of the free 20SP would be beneficial, because it increases the cellular capacity to remove oxidized proteins that are potentially cytotoxic when allowed to accumulate.
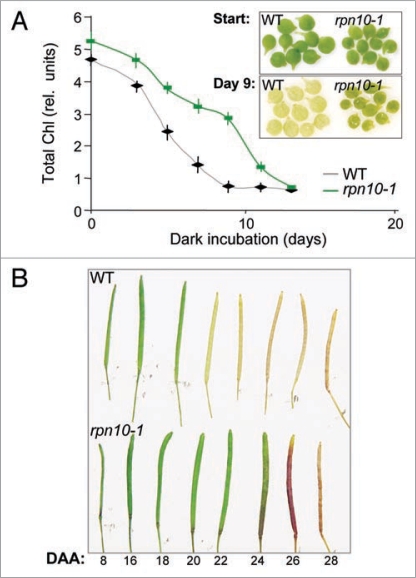
One way to answer the question whether the decreased 26SP to 20SP ratio is a cause or a consequence of the senescence process in plants is to test the timing of senescence in 26SP mutants that have decreased 26SP and increased 20SP activity. Accelerated senescence would indicate that optimal 26SP abundance needs to be maintained to prevent premature aging. Delayed senescence on the other hand would suggest that the increased 20SP activity counteracts the aging process. The Arabidopsis rpn10-1 proteasome mutant has a strong defect in 26SP-dependent proteolysis and a strong upregulation of the free 20SP.14 Analyses of this mutant reveal a significant delay in both natural and artificially induced senescence (Fig. 2). While further investigations are needed to determine if the delay is caused by the 26SP to 20SP shift, this observation however shows that contrary to animal species, reducing 26SP activity does not accelerate the aging process in plants. Furthermore, these results suggest that aging can be delayed by upregulating 20SP abundance.
Figure 2.
Delayed senescence in the 26SP mutant rpn10-1. (A) Dark-induced senescence. Cotyledons were detached from 5-days old wild type and rpn10-1 seedlings, and incubated in water in darkness for the designated time periods. Senescence was followed by measuring chlorophyll content of 10 cotyledons per time point. Total chlorophyll content was determined as in,38 and expressed as relative units (rel. units). Data are presented as mean ± SeM (n = 3). Insert shows cotyledons at the beginning of the experiment (start) and at 9 days into the treatment. (B) Senescence of siliques on inflorescences of wild type and rpn10-1. Individual flowers were tagged at anthesis along the primary inflorescences of wild type and mutant plants. A series of siliques were collected according to the denoted days after anthesis (DAA).
Conclusions and Perspectives
One of the conclusions of the Arabidopsis proteasome mutant studies was that a mild loss of 26SP significantly impacts plant growth, implying that plants need to maintain an optimal 26SP activity level to ensure developmentally programmed cell division and expansion rates. A second conclusion was that loss of 26SP function is not a priori detrimental for plant survival because it leads to increased 20SP biogenesis and thus, increased oxidative of interest to determine the 26SP to 20SP activity ratios in difstress tolerance and possibly also longevity. Given its potential ferent parts of a plant, at different stages in development, and in role in the development and stress responses of plants, it will be response to different biotic and abiotic stress conditions.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9469
References
- 1.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 2.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 3.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Höhfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 4.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chap. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan J, Wang J, Li Q, Hwang JR, Patterson C, Zhang H. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 2003;132:861–869. doi: 10.1104/pp.103.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotton SK, Callis J. Regulation of cullin RING ligases. Annu Rev Plant Biol. 2008;59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- 8.Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. F-box proteins everywhere. Curr Opin Plant Biol. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Kurepa J, Smalle J. Structure, function and regulation of plant proteasomes. Biochimie. 2008;90:324–335. doi: 10.1016/j.biochi.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 11.Book AJ, Smalle J, Lee KH, Yang P, Walker JM, Casper S, et al. The RPN5 subunit of the 26S proteasome Is essential for gametogenesis, sporophyte development, and complex assembly in Arabidopsis. Plant Cell. 2009;21:460–478. doi: 10.1105/tpc.108.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallois JL, Guyon-Debast A, Lécureuil A, Vezon D, Carpentier V, Bonhomme S, Guerche P. The Arabidopsis proteasome RPT5 subunits are essential for gametophyte development and show accession-dependent redundancy. Plant Cell. 2009;21:442–459. doi: 10.1105/tpc.108.062372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurepa J, Wang S, Li Y, Zaitlin D, Pierce AJ, Smalle JA. Loss of 26S proteasome function leads to increased cell size and decreased cell number in Arabidopsis shoot organs. Plant Physiol. 2009;150:178–189. doi: 10.1104/pp.109.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurepa J, Toh-e A, Smalle J. 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 2008;53:102–114. doi: 10.1111/j.1365-313X.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 15.Sung DY, Kim TH, Komives EA, Mendoza-Cozatl DG, Schroeder JI. ARS5 is a component of the 26S proteasome complex and negatively regulates thiol biosynthesis and arsenic tolerance in Arabidopsis. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03914.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda M, Matsui K, Ishiguro S, Sano R, Wada T, Paponov I, et al. The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development. 2004;131:2101–2111. doi: 10.1242/dev.01096. [DOI] [PubMed] [Google Scholar]
- 17.Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H. The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell. 2006;18:2479–2492. doi: 10.1105/tpc.106.045013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girod PA, Fu H, Zryd JP, Vierstra RD. Multiubiquitin chain binding subunit MCB1 (RPN10) of the 26S proteasome is essential for developmental progression in Physcomitrella patens. Plant Cell. 1999;11:1457–1472. doi: 10.1105/tpc.11.8.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahrami AR, Bastow R, Rolfe S, Price C, Gray JE. A role for nuclear localised proteasomes in mediating auxin action. Plant J. 2002;30:691–698. doi: 10.1046/j.1365-313x.2002.01320.x. [DOI] [PubMed] [Google Scholar]
- 20.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J. 1998;335:637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho SK, Chung HS, Ryu MY, Park MJ, Lee MM, Bahk YY, et al. Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-Box E3 ubiquitin ligase homolog. Plant Physiol. 2006;142:1664–1682. doi: 10.1104/pp.106.087965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell. 2008;20:1899–1914. doi: 10.1105/tpc.108.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yee D, Goring DR. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot. 2009;60:1109–1121. doi: 10.1093/jxb/ern369. [DOI] [PubMed] [Google Scholar]
- 24.Peng Z, Staub JM, Serino G, Kwok SF, Kurepa J, Bruce BD, et al. The cellular level of PR500, a protein complex related to the 19S regulatory particle of the proteasome, is regulated in response to stresses in plants. Mol Biol Cell. 2001;12:383–392. doi: 10.1091/mbc.12.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Q, Zhang J, Gao Q, Xing S, Li F, Wang W. Drought tolerance through overexpression of monoubiquitin in transgenic tobacco. J Plant Physiol. 2008;165:1745–1755. doi: 10.1016/j.jplph.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Kurepa J, Smalle J. To misfold or to lose structure? Detection and degradation of oxidized proteins by the 20S proteasome. Plant Signal Behav. 2008;3:386–388. doi: 10.4161/psb.3.6.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 28.Bader N, Grune T. Protein oxidation and proteolysis. Biol Chem. 2006;387:1351–1355. doi: 10.1515/BC.2006.169. [DOI] [PubMed] [Google Scholar]
- 29.Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 30.Grune T, Merker K, Sandig G, Davies KJ. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun. 2003;305:709–718. doi: 10.1016/s0006-291x(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 31.Jung T, Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 2008;60:743–752. doi: 10.1002/iub.114. [DOI] [PubMed] [Google Scholar]
- 32.Poppek D, Grune T. Proteasomal defense of oxidative protein modifications. Antioxid Redox Signal. 2006;8:173–184. doi: 10.1089/ars.2006.8.173. [DOI] [PubMed] [Google Scholar]
- 33.Vernace VA, Arnaud L, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J. 2007;21:2672–2682. doi: 10.1096/fj.06-6751com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaplace P, Fauconnier ML, Sergeant K, Dierick JF, Oufir M, van der Wal F, et al. Potato (Solanum tuberosum L.) tuber ageing induces changes in the proteome and antioxidants associated with the sprouting pattern. J Exp Bot. 2009;60:1273–1288. doi: 10.1093/jxb/erp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 37.Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 38.Graan T, Ort DR. Quantitation of the rapid electron donors to P700, the functional plastoquinone pool, and the ratio of the photosystems in spinach chloroplasts. J Biol Chem. 1984;259:14003–14010. [PubMed] [Google Scholar]