Abstract
In flowering plants, gravity perception appears to involve the sedimentation of starch-filled plastids, called amyloplasts, within specialized cells (the statocytes) of shoots (endodermal cells) and roots (columella cells). Unfortunately, how the physical information derived from amyloplast sedimentation is converted into a biochemical signal that promotes organ gravitropic curvature remains largely unknown. Recent results suggest an involvement of the Translocon of the Outer Envelope of (Chloro) plastids (TOC) in early phases of gravity signal transduction within the statocytes. This review summarizes our current knowledge of the molecular mechanisms that govern gravity signal transduction in flowering plants and summarizes models that attempt to explain the contribution of TOC proteins in this important behavioral plant growth response to its mechanical environment.
Key words: gravitropism, root, amyloplast, TOC complex, TOC132, TOC75
Introduction
Gravity provides a directional cue that enables plants to coordinate growth patterns. Using gravity as a guide, plants drive their roots into the soil and send their shoots skyward. Plants continue to coordinate their growth relative to the gravity vector not only throughout development, but also in response to their dynamic environments, which confront them with changes in light, humidity, oxygen, ions, nutrients, temperature and mechanical forces. To coordinate these various growth responses, a plant must convert the mechanical force of gravity into a chemical signal. This chemically-transduced signal will ultimately influence the growth pattern of the responding organ(s). While there are several reviews that discuss in great detail the extensive investigations into this process,1–3 there are still many outstanding issues. In particular, it has long been a goal to understand the earliest mechanisms of gravity perception and signal transduction. Recently, we identified new genetic mutants that lend important insight into this enigmatic process. Additionally, these mutants could provide useful material for investigating protein import into plastids, especially amyloplasts.
To produce a gravitropic response, a plant must first be able to perceive gravity, then convert the mechanical force into a chemical signal. This chemical signal will ultimately influence the growth pattern of the responding organ. In Arabidopsis shoots, gravity perception and response happen in overlapping sites. In Arabidopsis roots, there is a spatial separation between the primary site of gravity perception and early signal transduction, the root cap, and the responding site that develops a curvature, the distal elongation zone (DEZ). This separation makes Arabidopsis root gravitropism an ideal model to dissect the earlier phases from the later ones.1
Gravity sensing occurs primarily in the statocytes, which are the columella cells of the root and lie in the endodermal layer of the shoot. The statocytes are characterized by the presence of large, starch-filled amyloplasts. These plastids are denser than the surrounding cytoplasm, and tend to sediment to the bottom of statocytes. The movement and/or position of the amyloplasts contribute to the formation of a chemical signal, which is then transmitted basipetally to the DEZ, where it elicits a curvature response (Fig. 1A and B).
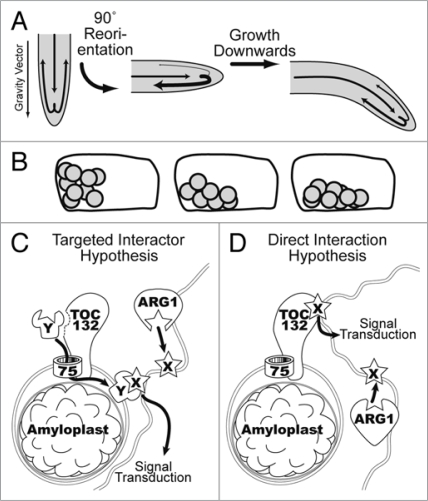
Figure 1.
Schematic representation of the various phases of root gravitropism. (A) Fountain model of auxin transport in roots.The direction of auxin transport is represented by arrows whose widths reflect relative flow rates. In vertical roots auxin, which is mainly synthesized in young shoot tissues, is transported through the vasculature into the root tip where it is redistributed symmetrically toward more peripheral tissues of the lateral cap. There, auxin is transported basipetally toward the elongation zone where it inhibits cell elongation. Upon 90° reorientation within the gravity field, the lateral transport of auxin across the cap is re-directed downward, leading to a lateral gradient that, upon transmission to the elongation zone, promotes differential cellular elongation between upper and lower flanks, responsible for tip curvature. (B) The starch-statolith hypothesis proposes that the sedimentation of amyloplasts toward the new bottomside of gravistimulated root-cap columella cells triggers a transduction pathway that leads to cell polarization and lateral auxin transport toward to lower flank of the root (see A). (C and D) Models of TOC action in gravitropism. TOC132 may mediate the insertion of a gravity-signal transducer (Y) into the outer-envelope of amyloplasts. Upon amyloplast sedimentation within the statocytes,Y may interact with a plasma-membrane or ER-associated signal transducer (X), regulating its activity (C). Alternatively, TOC132 may interact directly with X upon gravistimulation thereby triggering signal transduction (D). In either case,ARG1 or ARL2 (not shown here) may modulate the location and/or activity of transducer X within the sensitive membrane. These figure panels were modified from figures published in Stanga et al.59
Gravity Signal Transmission and Curvature Response
The phytohormone auxin is extremely important for the transmission of a gravitropic signal from the root cap to the DEZ. In a vertically-growing root, auxin is transmitted from its main site of synthesis in the shoot apex, through the vasculature to the root cap, where it is laterally distributed and then basipetally transmitted to the growing cells of the elongation zone.4 According to the Cholodny-Went theory, a gravistimulated root will establish a lateral auxin gradient across the root cap, with the new bottom flank accumulating greater concentrations of auxin than the new top flank.5,6 The auxin gradient in the root cap is then transmitted to the distal elongation zone, where it promotes differential cell elongation between the top and bottom sides, responsible for downward curvature (Fig. 1A and B).
Abundant experimental and genetic evidence supports the Cholodny-Went theory. For instance, mutations in genes that affect auxin transport or response cause gravitropic defects (reviewed in ref. 7). Also, transgenic reporters of auxin levels in plants, such as the GUS or GFP genes under the control of the auxin-responsive artificial DR5 promoter, are differentially activated in cells of the peripheral-cap and epidermis on the bottom side of gravistimulated roots.8–12 Analysis of the auxin efflux facilitator, PIN3, and the corresponding mutant also lends support for the Cholodny-Went theory. pin3 is an auxin transport mutant that affects gravitropism.13 The PIN3 protein is expressed in statocytes where it localizes uniformly to the plasma membrane of vertically growing roots. In the root-cap statocytes, gravistimulation causes PIN3 to relocalize to the bottom side of the plasma membrane within about 2 minutes. This process is believed to promote the establishment of the lateral auxin gradient discussed above. It should however be cautioned that it is also possible that the gravity-induced PIN3 relocalization in the root statocytes results from a differential promotion of auxin efflux activity at the bottom flank of the responding cells.14 In fact, while pin3 mutants are defective in both root and hypocotyl gravitropism, genetic analyses show that PIN3 is regulated by different mechanisms in the different organs.12
While auxin plays a crucial role in gravitropism, other phytohormones are also involved. Ethylene, brassinosteroids, abscisic acid, gibberellins, salicylic acid and jasmonic acid are all known to affect gravitropism.7 However, their contribution is primarily due to their effects on auxin-related processes. Another phytohormone, cytokinin, is capable of inducing curvature when applied exogenously, although comprehensive genetic evidence for the involvement of cytokinin is currently lacking.15
Perceiving Gravity
According to a long-standing idea, the starch-statolith hypothesis, it is the location and/or motion of the amyloplasts that provides the directional cue necessary for gravitropism.16 Physical and genetic ablation experiments indicate the importance of statocytes to gravitropism.17,18 While ablation of peripheral root cap cells did not alter root curvature, ablation of the innermost columella cells significantly altered root curvature without affecting growth rates. Of these inner columns, the second story columella cells (S2) contribute most to gravitropism.19 Importantly, these S2 cells exhibit the largest amyloplast sedimentation velocities.
Artificial displacement of amyloplasts illustrates the importance of their movement to gravitropism. A natural property of the starch molecules that accumulate in these plastids is their diamagnetism, which induces them to move away from high-gradient magnetic fields (HGMF). When a HGMF is applied near the tip of a vertically oriented root, amyloplasts within the statocytes move away from the field. This lateral movement of the amyloplasts, away from the gravity vector, promotes a root-tip curvature in the direction of amyloplast displacement, as predicted by the starch statolith hypothesis.20 Importantly, phosphoglucomutase 1 mutants (pgm1), which contain starchless amyloplasts, did not curve when exposed to a lateral HGMF, suggesting that the curvature response was not due to other magnetic effects on the plant.
Studies on the developmental timing of the inception of gravisensitivity during germination and of starchless and starch-deficient mutants demonstrate the importance of sedimenting amyloplasts to gravitropism. For instance, in flax the onset on gravicompetence was established to correlate with the appearance of mature, sedimentable amyloplasts during germination, 11 h before root emergence.21 Similarly, Arabidopsis mutants with varying amounts of starch show varying levels of gravicompetence that correlate with the ability of amyloplasts to sediment. Starch excess mutant (sex1) hypocotyls contain increased levels of starch and display enhanced sensitivity to gravistimulation.22 On the other hand, mutants with intermediate levels of starch display little or moderately decreased sensitivity to gravistimulation. Similarly, pgm1 mutant plants contain amyloplasts that lack starch and do not appear to settle to the bottom of statocytes upon gravistimulation. They show a severely attenuated gravity response.
The shoot statocytes of wild-type Arabidopsis plants contain large vacuoles that seem to impede amyloplast movement. Consequently, amyloplasts seem to sediment through transvacuolar strands in shoot statocytes upon gravistimulation.2 This is quite distinct from what is seen in the columella cells of the root cap, which lack large vacuoles, thus allowing easy amyloplast sedimentation upon gravistimulation. Interestingly, several shoot gravity response (sgr) mutants of Arabidopsis were shown to be defective in genes that contribute to the biogenesis and/or function of their vacuoles, a phenotype that is accompanied by altered amyloplast sedimentation and gravitropic deficiency.22–27
While the starch-statolith hypothesis may be sufficient to explain most gravitropic sensing ability of flowering plant organs, it should be cautioned that several lines of evidence also suggest the existence of alternative mechanisms. One such line of evidence comes from analyses of plant gravitropism that involve a clever apparatus designed to keep a single region of a root at a constant angle from the gravity vector.28 This apparatus, named the “ROTATO”, comprises a rotating stage controlled by a computer and microscope that monitor a selected region of the root and maintain it at a selected angle from the gravity vector. When the DEZ was maintained at a constant gravistimulating angle from vertical, the root continued to develop curvature long after the root cap had reached a vertical orientation, indicating that a secondary site of gravity sensing, distinct from the root cap, might exist in the DEZ. Because cells in this region of the root contain no sedimentable amyloplasts, it was suggested that gravity sensing in that region might involve a mechanism that detects the total weight of the protoplast on its cell wall. This hydrostatic-pressure model of gravity sensing was previously proposed to function in the large internodal cells of the alga Chara,29 and some evidence also supports its involvement in gravity sensing by rice roots.30 An alternative mechanism of gravity sensing in the root DEZ may help explain several unexpected observations on root gravitropism: (1) decapped maize roots display an actin turnover-dependent gravitropic response;31 (2) a bidirectional movement of curvature can be detected by high-resolution time-lapse imaging approaches during the first 2–3 hours of gravistimulation, which includes an unexpected acropetal component;32 and (3) the starchless pgm1 mutants of Arabidopsis still show some response to gravistimulation despite displaying no evidence of amyloplast sedimentation.33,34
Transducing Mechanical Forces into Chemical Signals
Although there is a bounty of experimental evidence supporting the starch-statolith hypothesis as well as more recent evidence suggesting the possibility of additional mechanisms, the means by which the first signal transduction events are triggered remain elusive. A few models have been proposed, involving mechanosensitive ion channels, receptor-ligand interactions, actin tensegrity and protoplast pressure.
Mechanosensitive ion channels are appealing candidates for gravity signal transduction because experimental evidence indicates the involvement of Ca2+ flux in gravitropism. Gravitropism can be inhibited by chemically interfering with calcium channels, calmodulin, Ca2+ ATPases, or calcium itself.35–37 Aequorin, a Ca2+ reporter, shows a biphasic cytosolic Ca2+ transient following gravistimulation of Arabidopsis seedlings.38 This transient consists of an initial spike followed by a sustained secondary peak. The first peak seems to correlate with rotational stimulation of the seedlings whereas the latter appears associated with the signaling events that accompany gravity perception in shoots.39,40 Unfortunately, the aequorin signal was so low that the experiment required observing large numbers of gravistimulated seedlings simultaneously, rendering efforts to identify subcellular patterns of Ca2+ difficult. Subsequent work showed that these signals derive from hypocotyls and petioles, but not roots, although the possibility that there are Ca2+ fluxes in roots, below the threshold of detection in this experimental system, could not be eliminated.39
Despite the evidence tying Ca2+ to gravitropism, it has not yet been shown that mechanosensitive ion channels are responsible for Ca2+ fluxes. Bioinformatic approaches have failed to identify orthologs to known eukaryotic mechanosensitive channels. However, ten Arabidopsis genes encoding orthologs to bacterial mechanosensitive channels (MSL genes) have been identified recently, and some members of this family may be able to contribute to gravitropism.41,42 In particular, MSL9 and MSL10 are found in the plasma membrane of root cells. Future experiments may address a possible role for these channels in gravitropism or other forms of mechanotransduction. Additionally, an Arabidopsis Ca2+ channel recently implicated in mechanotransduction has been shown to rescue a stretch-activated Ca2+ channel in yeast.43 This protein defines an additional class of potential mechanosensitive channels that warrant further study.
Further support for the role of Ca2+ in gravity signal transduction comes from experiments investigating the contribution of inositol-1,4,5-triphosphate (IP3) to that pathway. IP3 is involved in releasing Ca2+ from intracellular stores in both animals and plants.44 Gravistimulation of oat and maize pulvini (graviresponsive organs at the base of monocot leaves) showed a biphasic IP3 response. Within the first few minutes after gravistimulation, IP3 levels fluctuated between upper and lower flanks of the pulvini. This response was followed by sustained increases of IP3 along the lower flank of the stimulated organ.45–47 A similar response was observed in Arabidopsis inflorescence stems. Furthermore, hydrolysis of Arabidopsis IP3 by transgenic expression of human inositol polyphosphate 5-phosphatase coincided with a perturbation of inflorescence stem gravitropism.48
Within 1–2 minutes following gravistimulation, a pH spike can be observed in the cytoplasm of Arabidopsis columella cells, concomitant with an apoplastic acidification.49,50 This is a transient flux that returns to the unstimulated state within 8–12 minutes. When caged protons are released in columella cells, the gravitropic response is delayed.49 Chemicals that alter the pH balance of the apoplast also alter gravitropism: apoplastic alkalinization delays gravitropism, apoplastic acidification enhances gravitropism.50 The importance of starch to this process is emphasized by the observation that pgm1 mutant roots show little to no pH spike following gravistimulation.49
The involvement of mechanosensitive ion channels fits well with the starch-statolith hypothesis whereby sedimenting amyloplasts would act as activators of the channels. The ability of the amyloplasts to sediment, which is correlated with starch content, would likewise be correlated with the ability to activate these mechanosensitive ion channels.51 A similar, alternative mechanism could account for gravity perception. In this model, the weight of the protoplast produces tension differences between the top and bottom sides of a cell, which may differentially activate mechanosensitive ion channels.52 As discussed above, this hydrostatic-pressure model derives from experimentation with the large Chara internodal cells, and as of yet has not been demonstrated to be functioning in Arabidopsis.
It has also been proposed that sedimenting amyloplasts might interact with the dynamic actin cytoskeleton network that is attached to the plasma membrane of the statocytes, thereby influencing membrane tension with potential impact on the gating of mechanosensitive ion channels embedded in those membranes. Interestingly, repeated gravistimulation of shorter duration than the time required for sedimenting amyloplasts to reach the bottom of the statocyte, but sufficient to alter interactions between amyloplasts and the dynamic cytoskeleton, is effective at inducing a gravitropic response. This observation supports a role for the actin cytoskeleton in transducing the mechanical information of amyloplasts movement to the peripheral membranes.53 On the other hand, sedimenting plastids have also been proposed to locally disrupt the actin cytoskeleton in the statocytes, thereby altering tensions between the cytoskeleton and connected membranes with potential impact on membrane-associated mechanosensitive ion channels.54 Unfortunately, these models of actin-mediated gravity signal transduction seem contradicted by the results of pharmacological experiments showing that disruption of the actin cytoskeleton increases, rather than inhibit, root55 and shoot gravitropic curvature.56 Actin is involved in many subcellular processes likely to function in gravitropism.57 For instance, the actin cytoskeleton network has been implicated in amyloplast saltation, a process that constantly repositions amyloplasts within the statocytes, thereby potentially resetting the gravity sensing machinery.55 It has also been proposed to mediate the subcellular localization of the PIN3 auxin efflux facilitator in the statocytes.13 Therefore, precisely how it is involved in gravity signaling remains elusive.
There are also models that do not involve mechanosensitive ion channels. Again, studies of Chara outline a mechanism that could possibly have an Arabidopsis analog. It should be cautioned that while it may be possible to draw parallels between the gravitropic mechanisms of Chara and Arabidopsis, these organisms share a relatively distant common ancestor approximately 420–480 million years ago, plenty of time for many important features to diverge.58 One such important difference is that statoliths in Chara are BaSO4-filled vacuoles, not amyloplasts. Despite these cytological differences, Chara is a useful system to distinguish between the possibilities of the protoplast-pressure model of gravity perception and a ligand-receptor model that postulates that ligands on the surface of sedimenting statoliths have to interact with receptors embedded in sensitive membranes on the side of the cells in order to trigger gravity signal transduction. In a microgravity environment provided by parabolic flights, the weight of the protoplast would not be sufficient to cause the opening of mechanosensitive ion channels, as suggested by the protoplast-pressure model. Yet, Chara did exhibit gravitropic curvature under these conditions as long as the statoliths contacted a specialized surface of the plasma membrane.59 These experiments suggest the possibility of a receptor-ligand interaction between two proteins, one displayed on the surface of the statolith, and the other on the sensitive region of the plasma membrane. We recently described two new mutants, mar1 and mar2 (discussed later), that suggest the exciting possibility that a similar mechanism might also exist in Arabidopsis.60
ARG1: An Important Player in Gravity Signal Transduction
An important participant of early gravity signal transduction was identified in a screen designed to isolate Arabidopsis mutants with root gravitropic abberations. altered response to gravity1 (arg1) roots, when reoriented 90°, are slow to resume downward growth, though they retain the ability to respond to gravistimulation.61 A similar phenotype is observed in arg1 hypotocyls. This phenotype is not accompanied by any alterations in sensitivity to phytohormones or polar auxin transport inhibitors, growth rate or starch accumulation. Furthermore, phototropism is normal in arg1, demonstrating that the curvature defect is specific to gravitropism.
Transgenic rescue of the gravitropic defect with ARG1 fused to promoters that drive expression in the statocytes suggest that ARG1 participates in early gravity signal transduction.9 Because only the earliest phases occur within the root cap, rescuing the arg1 defect by expressing ARG1 with the RCP1 promoter showed that ARG1 is indeed involved in the early stages of gravitropism. Likewise, expressing ARG1 with the SCR1 promoter, which drives expression within the endodermis, rescued the hypocotyl gravitropism defect of arg1.
Two paralogs of ARG1 exist in Arabidopsis, named ARG1-Like 1 and ARG1-Like 2 (ARL1 and ARL2). arl2 mutants display similar phenotypes to arg1, while arl1 does not show any defects. The arl2 arg1 double mutant, the arl1 arl2 arg1 triple mutant, and the arg1 and arl2 single mutants all display similar reorientation defects, indicating that arl2 and arg1 operate in the same genetic pathway.62 However, arg1 and arl2 operate in a different genetic pathway than pgm1, because the pgm1 arg1 and pgm1 arl2 double mutants are more severely deficient in their gravitropic response than either single mutant. This distinction led to the identification of the mar mutations, which also enhance the gravitropic defect of arg1, and are discussed in the next section.
The ARG1 and ARL2 proteins are characterized primarily by two domains: a J-domain and a predicted coiled-coil region.61 J-domains are widely conserved and constitute a broad family in Arabidopsis. Via interactions with heat shock cognate 70 (hsc70) chaperones, J-domain proteins modulate the folding, activity, targeting and abundance of different substrates.63 Coiled-coil domains are involved in protein-protein interactions, and may help to determine the protein substrates of ARG1 and ARL2.64
ARG1 is expressed ubiquitously in plants whereas ARL2 is expressed specifically in the statocytes.12,61 Biochemical fractionation experiments showed association of ARG1 with a variety of different cellular membranes of the endomembrane system, along with the plasma membrane.9 Subcellular localization using GFP- and myc-tagged ARG1 transgenes showed signals in discrete puncta throughout the cell, particularly in areas of high vesicle trafficking activity, such as the nascent phragmoplast. Localization to plastids was conspicuously absent. Similar studies demonstrated ARL2 association with the plasma membrane and some endomembranes of the root statocytes. Furthermore, when ectopicallyexpressed in other regions of the plant, ARL2-GFP was also found to associate with cellular regions undergoing intense vesicular trafficking activity, such as the phragmoplast.12
In addition to the evidence discussed already, two important observations support the conclusion that ARG1 and ARL2 participate in gravity signal transduction events. First, the cytoplasmic alkalinization required for normal gravitropism is absent in arg1–2 mutants.9 This observation, coupled with the localization data discussed above, suggest that ARG1 may be modulating the localization and/or function of a proton pump. Secondly, the arg1 and arl2 mutants eliminate the contribution of the auxin efflux facilitator PIN3 to the gravitropic response by altering its accumulation at the bottom side of gravistimulated root statocytes.12 As a consequence of this alteration, gravistimulated arg1 and arl2 roots are defective in their ability to establish a lateral auxin gradient, resulting in delayed gravitropic curvature. These data may reflect the role in the statocytes of cytoplasmic H+ as a modulator of PIN3 localization and/or activity. Indeed, a similar relationship was established between a proton pyrophosphatase (AVP1) and a different auxin efflux facilitator, PIN1.65 Interestingly, gravistimulated arg1 pin3 and arl2 pin3 hypocotyls have slower gravitropic bending than any single mutant.128 However, during the first 6 hours after gravistimulation, the double mutants resemble the single mutants. Therefore, while ARG1 and ARL2 act in the same genetic pathway as PIN3 in the roots, they seem to act independently of PIN3 in the hypocotyls.
Genetic Modifiers of arg1
We recently reported the identification of two new mutants, modifiers of arg1 1 and 2 (mar1; mar2).60 These genetic enhancers show little or no gravitropic defects on their own, yet roots and hypocotyls grow in random directions when in an arg1 background. While the double mutants are agravitropic, their defect appears to affect the early stages of gravitropism only, as they respond normally to lateral light stimulation and exogenous application of phytohormones and auxin transport inhibitors. Importantly, these agravitropic double mutants have amyloplasts that appear normal in their overall morphology, starch content and sedimentation in response to gravity stimulation. Amazingly, though the mar mutants do not appear to have altered amyloplasts, the genes encode plastid-localized proteins, previously characterized as components of the Translocon of the Outer envelope of Chloroplasts (TOC), TOC75 and TOC132. Compared with other plastid types, chloroplasts are abundant and relatively simple to isolate. Therefore, much of the initial characterization of the TOC complex was done with isolated pea chloroplasts, and the complex earned its chloroplastic namesake.66–68 It is important to note that additional plastid types require these translocons, and TOC-complex subunits are expressed in tissues devoid of chloroplasts.69 To hypothesize how the TOC complex contributes to gravitropic signaling, we must first review some key observations in the broadly expanding field of plastidic protein import. Table 1 will help the reader through this section of our review. It summarizes the characteristics of the three main components of the TOC complex in Arabidopsis (TOC159, TOC75 and TOC34), the genes encoding them, the corresponding mutants and their phenotypes.
Table 1.
Characteristics of the main components of the TOC complex in arabidopsis
| Protein Family | Family Members | Locus | Mutants of Note | Comments | References |
| TOC75 Pores | TOC75-III | At3g46740 | toc75-III; mar1 | null lethal; genetic interaction with arg1 | Stanga et al 2009; Baldwin et al 2005 |
| TOC75-IV | At4g09080 | toc75-IV | abnormal etioplasts | Baldwin et al. 2005 | |
| TOC159 Receptors | TOC159 | At4g02510 | ppi2 | Pale leaves; abnormal chloroplast development | Bauer et al. 2000; Kubis et al. 2004; Ivanova et al. 2004 |
| TOC132 | At2g16640 | mar2; attoc132-1; toc132-2 | Genetic interaction with arg1; some functional redundancy with TOC120 | Stanga et al 2009; Kubis et al. 2004; Ivanova et al. 2004 | |
| TOC120 | At3g16620 | attoc120-1; toc120-2 | Defective root plastids in toc120 toc132 mutant | Kubis et al. 2004; Ivanova et al. 2004 | |
| TOC90 | At5g20300 | toc90-1 | Role unknown | Kubis et al. 2004 | |
| TOC34 Receptors | TOC33 | At1g02280 | ppil | Photosynthesis-related protein import | Jarvis et al. 1998; Kubis et al. 2003 |
| TOC34 | At5g05000 | ppi3 | Abnormal root growth | Constan et al 2004 |
For each component, the names of the family members are provided, along with the loci encoding them, the mutations cited in this review and their phenotypes.
The TOC Complex
Although plastids possess their own genetic machinery, most plastid proteins are encoded in the nuclear genome and translated in the cytosol.70 In order for these plastidic proteins to function where they are needed, they must first translocate across two membranes, an outer envelope membrane and an inner envelope membrane. Chaperones maintain the proteins that are destined to be translocated, or preproteins, in an unfolded state and target them to the TOC complex.71 The TOC complex is situated on the outer envelope of plastids. There, it interacts with the preproteins and translocates them across the outer envelope in a GTP-dependent process.72 Once this step has happened, preproteins are fed into an analogous protein complex of the inner envelope, the TIC complex, possibly by interacting with proteins in the inner membrane space.73,74 The TIC complex completes the translocation, and preproteins adopt their final conformation or continue on to subplastidic destinations. As an important alternative to translocation, plastid-localized proteins may be targeted to the outer envelope via interactions with the TOC complex,75 which could possibly reflect the mechanism of the TOC complex's involvement in gravitropism.
TOC75: The Central Pore
The TOC complex comprises three primary subunits, named for their size (kDa) in pea: TOC159, TOC34 and TOC75. These are found with a stoichiometry of 1:4–5:4.76,77 Each of the genes encoding these proteins is a member of a small gene family. TOC75 is the central pore through which all translocated pre-proteins pass. The central pore of TOC75 bears significant structural similarity to bacterial porin proteins. The key topological feature of these proteins is an array of short β-strands that form a β-barrel.78,79 The Arabidopsis genome has two TOC75-like genes (in addition to a third pseudogene): TOC75-III and TOC75-IV.80 A reverse genetic approach identified only subtle phenotypes associated with mutant forms of TOC75-IV. Structural abnormalities in etioplasts coupled with the TOC75-IV expression pattern suggest a role in dark-grown seedlings. In stark contrast to this subtle phenotype, an insertional allele of TOC75-III is lethal: embryos arrest at the 2-cell stage.80 mar1-1 is a hypomorphic allele of TOC75-III: while there is a mutation in a β-strand, mar1-1 plants survive, though they are pale and stressed.60 This partly-functional mutant may prove to be a valuable tool for dissecting the features of the TOC75 pores. Although the mar1-1 mutants enhance the gravitropic phenotype of arg1–2, they also have pleiotropic defects even as single mutants, likely due to alterations in import of many critical proteins. In contrast, the mar2-1 mutants appear very similar to wild type as single mutants, while producing the same agravitropic phenotype in the arg1–2 background. mar2-1 has a mutation in TOC132, one of the members of the TOC159 gene family.
TOC159: A 4-Member Family of GTPase Receptors
TOC159 interacts with preproteins during early stages of protein import, and is proposed to mediate preprotein recognition.67 TOC159 has three paralogs: TOC132, TOC120 and TOC90.67,81,82 These four proteins have highly conserved C-terminal membrane anchor domains (M-domains) and central GTP-binding domains (G-domain). Their N-terminal acidic domains (A-domains) vary considerably in length and are the major source of diversity amongst the four. Removal of the cytosolic domains, the A- and G-domains, from TOC159 nearly eliminates detectable binding of preproteins at the chloroplast surface.83 However, preprotein translocation can still occur. A forward genetic screen for mutants defective in plastid protein import identified a mutant version of TOC159 (ppi2;84). ppi2 plants have an albino phenotype due to the arrested development of chloroplasts. Non-photosynthetic plastids of the root, however, appear to develop normally. Furthermore, transcription of photosynthetic genes was repressed, and photosynthetic proteins were found not to accumulate. Consistent with these findings, expression analysis of the TOC159 family members identified higher relative expression of TOC159 in leaves compared to roots, while TOC132 and TOC120 are more abundant in roots than above-ground tissues.69,70,81
Analysis of RNA-null insertional alleles of these other paralogs supports a role for them in translocation of non-photosynthetic proteins.81 No visible phenotypes were seen in toc120 and toc90 mutants. When two separate groups analyzed toc132 mutants, one group detected no phenotype, while the other saw a very slight paleness in young plants, which developed into a reticulate pattern in older leaves.81,82 Quantification of chlorophyll content detected a slight decrease. Ecotype differences may account for these subtle differences. Genetic analysis of these mutants revealed phenotypes not found in any single mutant.81 toc132 toc159 double mutants are lethal at an early stage of development, due to either a reduction in import of a broad range of proteins, or complete abrogation of a single import pathway that is only disrupted in either single mutant. Double mutant analysis also shows redundancy between toc132 and toc120. The double mutants resemble the ppi2 mutant, in that they are small and very pale. Unlike the ppi2 mutant, toc132 toc120 root plastids have structural abnormalities, notably a high proportion of large inclusion bodies. These experiments support the model that the TOC receptors derive functional specialization by interacting with different classes of preproteins. Further support comes from our interesting observation that while TOC132 and TOC120 share some functional redundancy, they retain functional specialization, as arg1 toc120 mutants do not display the same random-growth phenotype we observe in arg1 toc132 mutants.60 It should however be cautioned here that the lower level of TOC120 expression relative to TOC132 in roots may be partially responsible for the latter result.
TOC34: A 2-Member Family of GTPase Receptors
Like members of the TOC159 family, the third component of the TOC complex, TOC34, is also a membrane-anchored protein with a cytosolic GTPase-domain.85 Unlike TOC159, the GTPase domain composes nearly the entirety of the cytosolic domain. TOC34 is also thought to have activity as a receptor, as it has been shown to interact directly with preproteins prior to translocation. The Arabidopsis TOC34 gene family has two members: TOC33 and TOC34 (note that Arabidopsis TOC33 is orthologous to pea TOC34:86). Like the TOC159 receptors, TOC33 and TOC34 have different affinities for different preproteins.87,88 Additional ppi mutants may help to identify the different classes of preproteins transduced through TOC33 and TOC34. ppi1 is a mutant form of the Arabidopsis TOC33 gene, ppi3 is a mutant form of the TOC34 gene. Like ppi2, ppi1 is specifically defective in expression, chloroplast import and accumulation of photosynthetic proteins.89 On the other hand, ppi3 mutants have shorter roots and aberrant root plastids, while their aerial tissues appear normal.90 These phenotypes are consistent with the relative abundance of TOC34 in the root and TOC33 in leaves.69 As such, ppi3 is an interesting candidate for possible involvement in root gravitropism; its contributions are being investigated currently.
Targeting to the TOC Complex
The TOC complex is necessary for classical signal sequence-based targeting to the plastid.91 Most nuclear-encoded plastid proteins possess a cleavable N-terminal transit peptide that is both necessary and sufficient for plastidic targeting.92 TOC complex targeting sequences are diverse in both size and composition, which has so far precluded a thorough understanding of structural elements that determine targeting specificity. The diversity of transit peptide sequences may provide a means for the diverse types of TOC complexes to recognize subsets of preproteins.
Ample evidence supports the classical cleavable signal-sequence mechanism of plastidic targeting. However, recent evidence suggests that this is not the only process that can target proteins to the plastids. First, proteomic studies of isolated plastids have identified proteins lacking a cleavable signal sequence, suggesting an alternate mechanism of recognition or possibly even an alternative mechanism of translocation for some proteins.93 Second, multiple targeting pathways for both intermembrane-space-localized and outer envelope proteins have been proposed that would not require a cleavable signal.94–96 This raises an interesting possibility for the role of the TOC complex in gravitropic signaling: that the TOC complex mediates the insertion into the outer amyloplast envelope of a particular protein(s) that acts as a ligand (or receptor) that modulates gravity signal transduction.
Model and Concluding Remarks
Other observations support the possiblity of a ligand-receptor model of gravitropism: the wild-type-like sedimentation and saltation behaviors of the mar2 mutant amyloplasts suggest not only that actin-related gravity signal transduction processes are unlikely to be affected in the mar mutants, but also that models requiring the mass of the plastids to trigger mechanosensitive ion channels are insufficient to explain the gravitropic defect of mar2 arg1 mutants.60
As discussed above, the ARG1 and ARL2 proteins associate with both the plasma membrane and components of vesicle trafficking pathway, suggesting a role in mediating the targeting or activity of gravity signal transducers at the plasma membrane or at organelles of the secretory pathway.9 Importantly, these J-domain proteins are conspicuously absent from plastids, suggesting that the genetic interactions between arg1/arl2 and toc132 do not represent a continuous physical interaction between the corresponding proteins. It is however possible that both ARG1/ARL2 and the TOC complex mediate the proper localization and/or activity of gravity signal transducers at distinct compartments within the cell that come together upon gravistimulation (plasma membrane or endoplasmic reticulum for ARG1, and plastid envelope for the TOC complex). In this context, the genetic interactions we observe between arg1–2/arl2–3 and the mar mutations would reflect a decreased ability for these transducers to interact and promote gravity signal transduction when sedimenting plastids hit the peripheral ER or plasma membrane of the mutant statocytes. Accordingly, removal of TOC132 would reduce the amount of plastid-associated transducer to a level that remains sufficient to trigger a normal gravitropic response in ARG1 ARL2 plants, but is insufficient in the context of the attenuated pool of functional interacting ligand in arg1–2 or arl2–3 membranes. Similarly, removal of ARG1 or ARL2 may reduce the amount of peripheral membrane-associated signal transducer to a level that still permits a significant, though partially altered, gravitropic response in a wild-type TOC132 background, but becomes insufficient in a TOC132-deficient background.
In the context of the ligand-interaction model of gravity sensing, it is an important reminder that TOC75 also contributes to the proper targeting of plastid outer-envelope proteins, including members of the TOC159 family.75 Therefore, the missense mutation in TOC75 associated with mar1-1 may affect the proper targeting of the proposed plastid-associated transducer, which could potentially be TOC132 itself or another outer-envelope-associated protein.
While the ligand-receptor model of gravity sensing is complelling, it is also possible that the TOC complex affects gravitropism through an unidentified regulatory molecule. Indeed, plastids contribute to the manufacture and/or storage of important biological products.97 However, the strong genetic interaction observed between toc132 and arg1–2 or arl2–3 would require that the corresponding regulatory molecule be needed only in the absence of ARG1 or ARL2, a less likely scenario. Alternatively, internal plastid proteins may act in a signal transduction pathway without altering amyloplast structure or behavior. For example, a thylakoid-localized calcium-binding protein has recently been implicated in the transduction of stomatal-closing signals.98 However, here again, such internal plastid protein would have to be needed for gravity signal transduction only in the absence of ARG1 or ARL2. Metabolic and/or proteomic profiling of mar2-1 arg1–2 or mar2-1 arl2–3 amyloplasts, along with the genetic identification of new participants of this genetic pathway, will be needed to further elucidate the molecular mechanisms that allow components of the TOC complex to contribute to gravity signal transduction.
To the best of our knowledge, the results discussed above constitute so far the strongest evidence consistent with a ligandreceptor model of gravity signal transduction in flowering plants. This model could be similar to the ligand-receptor model of gravity sensing that has been proposed to function in Chara rhizoids, based on careful evaluations of gravity sensing in the context of parabolic-flight microgravity environments.59 Research is under way to test this exciting alternative scenario of gravity sensing and signal transduction.
Acknowledgements
This review was made possible by grants from the National Science Foundation (grants nos. MCB-0240084 and IOS-0642865), the National Aeronautics and Space Administration (grant no. NAG2-1602), HATCH funds from the College of Agriculture and Life Sciences, University of Wisconsin-Madison, to P.H.M., and a fellowship from the National Institutes of Health Training Grant in Genetics at the University of Wisconsin-Madison to J.P.S. This is manuscript no. 3644 of the Laboratory of Genetics.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9470
References
- 1.Perrin R, et al. Gravity signal transduction in primary roots. Ann Bot. 2005;96:737–743. doi: 10.1093/aob/mci227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison B, et al. Signal transduction in gravitropism. In: Gilroy S, Masson P, editors. Plant Tropisms. Vol. 2. Blackwell Publishing; 2008. pp. 21–45. [Google Scholar]
- 3.Boonsirichai K, et al. Root gravitropism: An experimental tool to investigate basic cellular and molecular processes underlying mechanosensing and signal transmission in plants. Ann Rev Plant Biol. 2002;53:421–447. doi: 10.1146/annurev.arplant.53.100301.135158. [DOI] [PubMed] [Google Scholar]
- 4.Trewavas A. What remains of the Cholodny-Went theory? Plant Cell Environ. 1992;15:759–794. [PubMed] [Google Scholar]
- 5.Went F. Wuchstoff und Wachstum. Rec Trav Bot Neerl. 1928;25:1–116. (Ger). [Google Scholar]
- 6.Cholodny H. Beiträge zur hormonalen theorie von tropismen. Planta. 1928;6:118–134. (Ger). [Google Scholar]
- 7.Muday G, Rahman A. Auxin transport and the integration of gravitropic growth. In: Gilroy S, Masson P, editors. Plant Tropisms. Vol. 3. Blakwell Publishing; 2008. pp. 47–78. [Google Scholar]
- 8.Rashotte A, et al. Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 2000;122:481–490. doi: 10.1104/pp.122.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boonsirichai K, et al. ARG1 is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes. Plant Cell. 2003;15:2612–2625. doi: 10.1105/tpc.015560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ottenschlager I, et al. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA. 2003;100:2987–2991. doi: 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis D, et al. Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell. 2007;19:1838–1850. doi: 10.1105/tpc.107.051599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison B, Masson P. ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. Plant J. 2008;53:380–392. doi: 10.1111/j.1365-313X.2007.03351.x. [DOI] [PubMed] [Google Scholar]
- 13.Friml J, et al. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2005;415:806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- 14.Paciorek T, et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435:1251–1256. doi: 10.1038/nature03633. [DOI] [PubMed] [Google Scholar]
- 15.Aloni R, et al. Role of cytokinin in the regulation of root gravitropism. Planta. 2004;220:177–182. doi: 10.1007/s00425-004-1381-8. [DOI] [PubMed] [Google Scholar]
- 16.Sack F. Plastids and gravitropic sensing. Planta. 1997;203:63–68. doi: 10.1007/pl00008116. [DOI] [PubMed] [Google Scholar]
- 17.Tsugeki R, Fedoroff N. Genetic ablation of root cap cells in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:12941–12946. doi: 10.1073/pnas.96.22.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka A, et al. Positional effect of cell inactivation on root gravitropism using heavy-ion microbeams. J Exp Bot. 2002;53:683–687. doi: 10.1093/jexbot/53.369.683. [DOI] [PubMed] [Google Scholar]
- 19.Blancaflor E, et al. Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol. 1998;116:213–222. doi: 10.1104/pp.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuznetsov O, Hasenstein K. Intracellular magnetophoresis of amyloplasts and induction of root curvature. Planta. 1996;198:87–94. doi: 10.1007/BF00197590. [DOI] [PubMed] [Google Scholar]
- 21.Ma Z, Hasenstein K. The onset of gravisensitivity in the embryonic root of flax. Plant Physiol. 2006;140:159–166. doi: 10.1104/pp.105.073296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vitha S, et al. Gravitropism in starch-excess mutant of Arabidopsis thaliana. Am J Bot. 2007;94:590–598. doi: 10.3732/ajb.94.4.590. [DOI] [PubMed] [Google Scholar]
- 23.Kato T, et al. SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell. 2002;14:33–46. doi: 10.1105/tpc.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yano D, et al. A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc Natl Acad Sci USA. 2003;100:8589–8594. doi: 10.1073/pnas.1430749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita M, et al. Inovolvement of the vacuoles of the endodermis in the early process of shoot gravitropism in Arabidopsis. Plant Cell. 2002;14:47–56. doi: 10.1105/tpc.010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tasaka M, et al. Genetic regulation of gravitropism in higher plants. Int Rev Cytol. 2001;206:135–154. doi: 10.1016/s0074-7696(01)06021-1. [DOI] [PubMed] [Google Scholar]
- 27.Silady R, et al. The GRV2/RME-8 protein of Arabidopsis functions in the late endocytic pathway and is required for vacuolar membrane flow. Plant J. 2007;53:29–41. doi: 10.1111/j.1365-313X.2007.03314.x. [DOI] [PubMed] [Google Scholar]
- 28.Wolverton C, et al. Root gravitropism in response to a signal originating outside of the cap. Planta. 2002;215:153–157. doi: 10.1007/s00425-001-0726-9. [DOI] [PubMed] [Google Scholar]
- 29.Staves M, et al. Hydrostatic pressure mimics gravitational pressure in characean cells. Protoplasma. 1992;168:141–152. doi: 10.1007/BF01666260. [DOI] [PubMed] [Google Scholar]
- 30.Staves M, et al. The effect of external medium on the gravitropic curvature of rice (Oryza sativa, Poaceae) roots. Am J Bot. 1997;84:1522–1529. [PubMed] [Google Scholar]
- 31.Mancuso S, et al. Actin turnover-mediated gravity response in maize root apices. Gravitropism of decapped roots implicates gravisensing outside of the root cap. Plant Signal Behav. 2006;1:52–58. doi: 10.4161/psb.1.2.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller N, et al. Computer-vision analysis of seedling responses to light and gravity. Plant J. 2007;52:374–381. doi: 10.1111/j.1365-313X.2007.03237.x. [DOI] [PubMed] [Google Scholar]
- 33.Caspar T, Pickard B. Gravitropism in a starchless mutant of Arabidopsis: implications for the starchstatolith theory of gravity sensing. Planta. 1989;177:185–197. [PubMed] [Google Scholar]
- 34.Kiss J, et al. Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta. 1989;177:198–206. [PubMed] [Google Scholar]
- 35.Sinclair W, Trewavas A. Calcium in gravitropism: A re-examination. Planta. 1997;203:85–90. doi: 10.1007/pl00008120. [DOI] [PubMed] [Google Scholar]
- 36.Fasano J, et al. Ionic signaling in plant responses to gravity and touch. J Plant Growth Reg. 2002;21:71–88. doi: 10.1007/s003440010049. [DOI] [PubMed] [Google Scholar]
- 37.Monshausen G, et al. Touch sensing and thigmotropism. In: Gilroy S, Masson P, editors. Plant Tropisms. Vol. 5. Blackwell Publishing; 2008. pp. 91–122. [Google Scholar]
- 38.Plieth C, Trewavas A. Reorientation of seedlings in the Earth's gravitational field induces cytosolic calcium transients. Plant Physiol. 2002;129:786–796. doi: 10.1104/pp.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toyota M, et al. Cytoplasmic calcium increases in response to changes in the gravity vector in hypocotyls and petioles of Arabidopsis seedlings. Plant Physiol. 2008;146:505–514. doi: 10.1104/pp.107.106450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toyota M, et al. Critical consideration on the relationship between auxin transport and calcium transients in gravity perception of Arabidopsis seedlings. Plant Signal Behav. 2009;3:521–524. doi: 10.4161/psb.3.8.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haswell E, Meyerowitz E. MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr Biol. 2006;16:1–11. doi: 10.1016/j.cub.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 42.Haswell E, et al. Two MscS homologs provide mechanosensitive channel activities in the Arabidopsis root. Curr Biol. 2008;18:730–734. doi: 10.1016/j.cub.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa Y, et al. Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc Natl Acad Sci USA. 2007;104:3639–3644. doi: 10.1073/pnas.0607703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mikoshiba K. The IP3 receptor/Ca2+ channel an its cellular function. Biochem Soc Symp. 2007;74:9–22. doi: 10.1042/BSS0740009. [DOI] [PubMed] [Google Scholar]
- 45.Perera I, et al. Transient and sustained increases in inositol-1,4,5-trisphosphate precede the differential growth response in gravistimulated maize pulvini. Proc Natl Acad Sci USA. 1999;96:5838–5843. doi: 10.1073/pnas.96.10.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perera I, et al. A role for inositol 1,4,5-trisphosphate in gravitropic signaling and the retention of cold-perceived gravistimulation of oat shoot pulvini. Plant Physiol. 2001;125:1499–1507. doi: 10.1104/pp.125.3.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perera I, et al. Phosphoinositide signaling and plant gravitropism. Grav Space Biol Bull. 2003;17:102. [Google Scholar]
- 48.Perera I, et al. A universal role for inositol 1,4,5-trisphosphate-mediated signaling in plant gravitropism. Plant Physiol. 2006;140:746–760. doi: 10.1104/pp.105.075119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fasano J, et al. Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell. 2001;13:907–921. doi: 10.1105/tpc.13.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott A, Allen N. Changes in cytosolic pH within Arabidposis root columella cells play a key role in the early signaling pathway for root gravitropism. Plant Physiol. 1999;121:1291–1298. doi: 10.1104/pp.121.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leitz G, et al. Statolith sedimentation kinetics and force transduction to the cortical endoplasmic reticulum in gravity-sensing Arabidopsis columella cells. Plant Cell. 2009;21:843–860. doi: 10.1105/tpc.108.065052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staves M. Cytoplasmic streaming and gravity sensing in Chara internodal cells. Planta. 1997;203:79–84. doi: 10.1007/pl00008119. [DOI] [PubMed] [Google Scholar]
- 53.Perbal G, et al. The dose-response curve of the gravitropic reaction: a re-analysis. Physiol Plant. 2002;114:336–342. doi: 10.1034/j.1399-3054.2002.1140302.x. [DOI] [PubMed] [Google Scholar]
- 54.Yoder T, et al. Amyloplast sedimentation dynamics in maize columella cells support a new model for the gravity-sensing apparatus of roots. Plant Physiol. 2001;125:1045–1060. doi: 10.1104/pp.125.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou G, et al. The promotion of gravitropism in Arabidopsis roots upon actin disruption is coupled with the extended alkalinization of the columella cytoplasm and a persistent lateral auxin gradient. Plant J. 2004;39:113–125. doi: 10.1111/j.1365-313X.2004.02114.x. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto K, Kiss J. Disruption of the actin cytoskeleton results in the promotion of gravitropism in inflorescence stems and hypocotyls of Arabidopsis. Plant Physiol. 2002;128:669–681. doi: 10.1104/pp.010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hou G, et al. Enhanced gravitropism of roots with a disrupted cap actin cytoskeleton. Plant Physiol. 2003;131:1360–1373. doi: 10.1104/pp.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bowman J, et al. Green genes—comparative genomics of the green branch of life. Cell. 2007;129:229–234. doi: 10.1016/j.cell.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 59.Limbach C, et al. How to activate a plant gravireceptor. Early mechanisms of gravity sensing studied in Characean rhizoids during parabolic flights. Plant Physiol. 2005;139:1030–1040. doi: 10.1104/pp.105.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanga J, et al. A role for the TOC complex in Arabidopsis root gravitropism. Plant Physiol. 2009;149:1896–1905. doi: 10.1104/pp.109.135301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sedbrook J, et al. ARG1 (Altered Response to Gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc Natl Acad Sci USA. 1999;96:1140–1145. doi: 10.1073/pnas.96.3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guan C, et al. The ARG1-LIKE2 (ARL2) gene of Arabidopsis thaliana functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway. Plant Physiol. 2003;133:100–112. doi: 10.1104/pp.103.023358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mayer M, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanisms. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 65.Li J, et al. Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science. 2005;310:121–125. doi: 10.1126/science.1115711. [DOI] [PubMed] [Google Scholar]
- 66.Waegemann K, Söll J. Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1991;191:149–158. [Google Scholar]
- 67.Perry L, Keegstra K. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 1994;6:93–105. doi: 10.1105/tpc.6.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kessler F, et al. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- 69.Vojta A, et al. The protein translocon of the plastid envelopes. J Biol Chem. 2004;279:21401–21405. doi: 10.1074/jbc.M401968200. [DOI] [PubMed] [Google Scholar]
- 70.Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.May T, Soll J. 14-3-3-proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12:53–64. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soll J, Schleiff E. Protein import into chloroplasts. Nature Rev Mol Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- 73.Kouranov A, et al. Tic22 is targeted to intermembrane space of chloroplasts by a novel pathway. J Biol Chem. 1999;274:25181–25186. doi: 10.1074/jbc.274.35.25181. [DOI] [PubMed] [Google Scholar]
- 74.Becker T, et al. Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts. Mol Cell Biol. 2004;15:5130–5144. doi: 10.1091/mbc.E04-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tu S, et al. Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell. 2004;16:2078–2088. doi: 10.1105/tpc.104.023952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hust B, et al. Analysis of the subunit composition of TOC protein import complexes from chloroplasts in Arabidopsis thaliana reveals distinct complex subtypes. 2008 Submitted. [Google Scholar]
- 77.Schleiff E, et al. Characterization of the translocon of the outer envelope of chloroplasts. J Cell Biol. 2003;160:541–551. doi: 10.1083/jcb.200210060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schleiff E, et al. Prediction of the plant β-barrel proteome: A case study of the chloroplast outer envelope. Protein Sci. 2003;12:748–759. doi: 10.1110/ps.0237503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sveshnikova N, et al. Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proc Natl Acad Sci USA. 2000;97:4973–4978. doi: 10.1073/pnas.080491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baldwin A, et al. A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol. 2005;138:715–733. doi: 10.1104/pp.105.063289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kubis S, et al. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell. 2004;16:2059–2077. doi: 10.1105/tpc.104.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ivanova Y, et al. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol Biol Cell. 2004;15:3379–3392. doi: 10.1091/mbc.E03-12-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen K, et al. Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol. 2000;122:813–822. doi: 10.1104/pp.122.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bauer J, et al. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- 85.Ma Y, et al. Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation or precursor proteins at the outer envelope. J Cell Biol. 1996;134:315–327. doi: 10.1083/jcb.134.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jackson-Constan D, Keegstra K. Arabidopsis genes encoding components of chloroplastic protein import apparatus. Plant Physiol. 2001;125:1567–1576. doi: 10.1104/pp.125.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gutensohn M, et al. Functional analysis of the two Arabidopsis homologues of Toc34, a component of the chloroplast protein import apparatus. Plant J. 2000;23:771–783. doi: 10.1046/j.1365-313x.2000.00849.x. [DOI] [PubMed] [Google Scholar]
- 88.Jelic M, et al. Two Toc34 homologues with different properties. Biochemistry. 2003;42:5906–5916. doi: 10.1021/bi034001q. [DOI] [PubMed] [Google Scholar]
- 89.Kubis S, et al. The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import and accumulation of photosynthetic proteins. Plant Cell. 2003;15:1859–1871. doi: 10.1105/tpc.012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Constan D, et al. An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 2004;38:93–106. doi: 10.1111/j.1365-313X.2004.02024.x. [DOI] [PubMed] [Google Scholar]
- 91.Kessler F, Schnell D. The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic. 2006;7:248–257. doi: 10.1111/j.1600-0854.2005.00382.x. [DOI] [PubMed] [Google Scholar]
- 92.Li H, Chen L. Protein targeting and integration signal for the chloroplastic outer envelope membrane. Plant Cell. 1996;8:2117–2126. doi: 10.1105/tpc.8.11.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jarvis P. Organellar proteomics: chloroplasts in the spotlight. Curr Biol. 2004;14:317–319. doi: 10.1016/j.cub.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 94.Miras S, et al. Toc159-and Toc75-independent import of a transit sequence-less precursor into the inner envelope of chloroplasts. J Biol Chem. 2007;282:29482–29492. doi: 10.1074/jbc.M611112200. [DOI] [PubMed] [Google Scholar]
- 95.Nada A, Soll J. Inner envelope protein 32 is imported into chloroplasts by a novel pathway. J Cell Sci. 2004;117:3975–3982. doi: 10.1242/jcs.01265. [DOI] [PubMed] [Google Scholar]
- 96.Radhamony R, Theg S. Evidence for an ER to Golgi to chloroplast protein transport pathway. Trends Cell Biol. 2006;16:385–387. doi: 10.1016/j.tcb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 97.Neuhaus H, Emes M. Nonphotosynthetic metabolism in plastids. Ann Rev Plant Physiol Plant Mol Biol. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- 98.Nomura H, et al. Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. Plant J. 2008;53:988–998. doi: 10.1111/j.1365-313X.2007.03390.x. [DOI] [PubMed] [Google Scholar]