Abstract
Heterotrimeric G proteins (Gα, Gβ/Gγ subunits) constitute one of the most important components of cell signaling cascade. G Protein Coupled Receptors (GPCRs) perceive many extracellular signals and transduce them to heterotrimeric G proteins, which further transduce these signals intracellular to appropriate downstream effectors and thereby play an important role in various signaling pathways. GPCRs exist as a superfamily of integral membrane protein receptors that contain seven transmembrane α-helical regions, which bind to a wide range of ligands. Upon activation by a ligand, the GPCR undergoes a conformational change and then activate the G proteins by promoting the exchange of GDP/GTP associated with the Gα subunit. This leads to the dissociation of Gβ/Gγ dimer from Gα. Both these moieties then become free to act upon their downstream effectors and thereby initiate unique intracellular signaling responses. After the signal propagation, the GTP of Gα-GTP is hydrolyzed to GDP and Gα becomes inactive (Gα-GDP), which leads to its re-association with the Gβ/Gγ dimer to form the inactive heterotrimeric complex. The GPCR can also transduce the signal through G protein independent pathway. GPCRs also regulate cell cycle progression. Till to date thousands of GPCRs are known from animal kingdom with little homology among them, but only single GPCR has been identified in plant system. The Arabidopsis GPCR was reported to be cell cycle regulated and also involved in ABA and in stress signaling. Here I have described a general mechanism of signal transduction through GPCR/G proteins, structure of GPCRs, family of GPCRs and plant GPCR and its role.
Key words: heterotrimeric G proteins, GPCRs, seven-transmembrane receptors, signal transduction, stress signaling
Introduction
Cell signaling is one of the important processes required for the normal growth and development of cell. It is the basic process that helps the cells to sustain through various environmental cues and develop tolerance against stress conditions.1–3 The basic cell signaling machinery involves a receptor molecule that perceives the signal. The signal or primary stimulus could be light, hormone, odorant, antigen, neurotransmitter or the surface of another cell, which transport into the cell via membrane receptor, through signal transduction triad (receptor/transducer/effector).4 The second messenger could be Ca2+ (for ion channels) cAMP and cGMP (for adenylyl and guanlyl cyclases), inositol-1, 4,5-triphosphate (IP3), diacyl glycerol (DAG) and arachidonic acid (for phospholipases).2 The triad is responsible for converting the signal from first to second messenger, which could be further regulated by protein kinases or phosphatases in the cytoplasm. The target of the signal may be enzymes, intracellular receptors, special transport vehicles and finally transcription factors,5 which ultimately controls the gene expression. There are several signaling mechanisms present in the cell to carry out normal functions. One of an important signaling cascade is formed by GTP binding proteins or simply known as G proteins, so called because of their ability to bind to guanine nucleotide.6 One more molecule that is involved in this signaling cascade and forms an important part of the cascade is G Protein Coupled Receptor (GPCR). It is known that the signals are mostly perceived at the level of membrane and therefore transmembrane (TM) events are the likely routes for signal generation and transduction. In plants, the best-characterized plasma membrane-based receptors are of two kinds: (1) transmembrane receptor enzymes (usually kinase), (2) GPCRs. First step in the G protein mediated signaling cascade is binding of an agonist/ligand to GPCRs, so these molecules form an important part of this pathway. GPCRs are members of a large family of proteins found in eukaryotes and certain prokaryotes.8 A remarkable property of GPCRs which make it distinguishable from other classes of receptors is the presence of seven TM (7TM) α-helical region. Members of the GPCR superfamily share the same basic architecture i.e., 7TM α-helices, an extracellular amino-terminal segment and an intracellular carboxy-terminal tail. GPCRs form a large superfamily of membrane proteins that modulate sensory perception, chemotaxis, neurotransmission, cell communication, the senses of sight, smell and taste and many other vital physiological events. Purification of the first GPCR, the β-adrenergic receptor, was reported in 1981 by Shorr et al. and it was identified as one of the hormone-binding subunit.9 Characterized by their cell-surface localization and tissue-specificity, these protein receptors are the targets of 50–60% of all existing medicines including well-known blockers and anti-histamine therapeutics. These plasma membrane-bound receptors have evolved to recognize a diversity of extracellular physical and chemical signals, such as nucleotides, peptides, amines, Ca2+ and photons. On recognition of such signals, the GPCRs act as proximal event in signaling pathways that influence a wide variety of metabolic and differentiated functions.10,11 In this article a general mechanism of signal transduction through GPCR/G proteins, structure of GPCRs, family of GPCRs and plant GPCR and its role have been covered.
A General Mechanism of Signal Transduction through GPCR and G Proteins
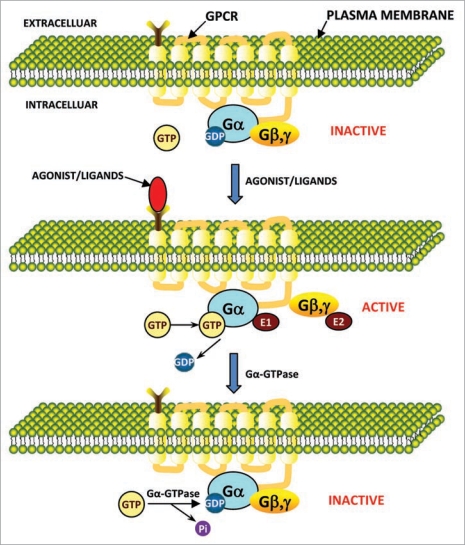
The regulatory cycle of G proteins i.e., activation/inactivation through GPCR is shown in Figure 1. In the inactive state, Gα is bound to Gβγ dimer and GDP. G protein mediated signaling starts by binding of an agonist molecule that leads to activation of GPCR. GPCR is also a guanine nucleotide exchange factor that promotes the exchange of guanosine disphosphate (GDP)/guanosine triphosphate (GTP) associated with the Gα subunit.12 Therefore, the activated GPCR catalyzes exchange of GTP for GDP on the Gα subunit, as a result conformational changes takes place in the GPCR, which leads to dissociation of Gβγ dimer from Gα and thus activates multiple molecules of G proteins (Fig. 1). The G proteins activated in this way constitute an amplified representation of the activated GPCR. Activated Gα and Gβγ proteins in turn binds to various effectors and thereby switches it either on or off in different systems, and effectors continue to pass the signal to different kinds of second messengers. Here intrinsic GTPase activity of Gα comes into play, that leads to conversion of bound GTP into GDP and hence the inactivation of G proteins cascade. GTPase activity of the Gα subunits may also be regulated by regulators of G proteins signaling (RGS proteins) as well as effectors. Moreover, effector enzymes such as adenylyl cyclases may also regulate the activation of G proteins by receptors (Fig. 1).
Figure 1.
Model for signal transduction by activation/inactivation of heterotrimeric G proteins through GPCR. The subunits of heterotrimeric G proteins (Gα and Gβγ) in their inactivated state are associated with each other. In inactivation state the GDP is bound to Gα (Gα-GDP). In signal transduction, first the GPCR gets activated by changing its conformation which resulted from binding of agonist/ligands to the extracellular region of GPCR. This activated GPCR further activate the inactive G protein to active G protein complex by dissociating the Gα from Gβγ. In active state the GTP is bound to Gα (Gα-GTP). Now free Gα and Gβγ have their own effectors (E1 and E2, respectively) to further transmit the signals and initiate unique intracellular signaling responses. Later, after the signal transduction, the Gα-GTPase activity hydrolyze the bound GTP (Gα-GTP) to GDP and Pi and inactivate the G protein complex by re-associating the Gα with Gβγ. In this state again GDP is bound to Gα (Gα-GDP) in the G protein complex. in this way the activation and inactivation cycle is completed.
The activated Gα interacts and regulates many effector molecules such as calcium, potassium channels, adenylyl cyclase, phospholipase C (PLC), PLD and protein kinases. Initially it was hypothesized that the βγ dimer acts as negative regulator and can block activation of adenylyl cyclase by this mechanism.13,14 But subsequently this hypothesis was changed with the discovery that the βγ subunit could activate the muscarinic K+ channel and βγ subunits positively regulate effectors.15,16 Finally, the βγ subunit was shown to be a positive regulator of a large number of effectors in addition to the K+ channel, including adenylyl cyclase, phospholipase C-β (PLC-β), phospholipase A2 (PLA2), phosphoinositide 3-kinase (PI3-kinase), and β-adrenergic receptor kinase. Also, Gβγ can activate Gα subunit.17 It is now clear that many effectors are regulated both by α and βγ subunits.
Structure of GPCRs
Members of the GPCR superfamily share the same basic architecture of 7TM α-helices, an extracellular amino-terminal segment and an intracellular carboxy-terminal tail. These plasma membrane-bound receptors have evolved to recognize a diversity of extracellular physical and chemical signals, such as nucleotides, peptides, amines, Ca2+ and photons. On recognition of such signals, the GPCRs act as proximal event in signaling pathways that influence a wide variety of metabolic and differentiated functions.10,11 An extensive analysis of about 200 GPCR sequences revealed that total length of GPCRs can vary between 311 and ∼1,490 amino acid residues. The largest variations in length are found in the N and C termini with size up to 879 and 371 amino acid residues, respectively.18 GPCRs are not only encoded by eukaryotic genes but also by viral genes. Human GPCRs genomic genes are predominantly intronless.18
The 7TM α-helices connected by three intracellular and three extracellular loops. The extracellular loops of the GPCR can be glycosylated and contain two highly conserved cysteine residues, which build disulfide bonds to stabilize the receptor structure. GPCRs contain extracellular N-terminus domains (ECL1, ECL2 and ECL3) of variable size, ranging from 154 residues (calcitonin receptor) to 36 residues (rhodopsin receptor). This domain contains asparagines residues and motifs for N-glycosylation, which influences intracellular trafficking of receptors to the plasma membrane, and cysteine residues in ECL1 and ECL2 loops that can influence protein folding critical for trafficking of a function receptor to the cell surface.19 The N terminus of some GPCRs is involved in ligand binding, activation and downregulation. The 7TM α-helices helices of GPCRs are arranged to form a tight, ring-shaped central core that is highly hydrophobic in nature. Similar to most TM proteins, the hydrophobic amino acid residues are presumably arranged to face the lipid bilayer, whereas the more hydrophilic amino acid residues face towards the core. Furthermore, helix-helix interaction contributes to the functional tertiary structure of the GPCRs necessary for receptor folding and stability, ligand binding and ligand-induced conformational changes for G protein coupling. Thus, mutations in the TM domain can have an array of deleterious effects.
GPCRs contains an intracellular carboxyl-terminal domains (ICL1, ICL2 and ICL3) which are involved in several aspects of GPCR signaling. This domain contains Ser and/or Tyr residues which serve as sites for G protein receptor kinase-mediated phosphorylation and receptor desensitization. Some GPCRs contain a cysteine residue in the C-terminal domain, which can serve as site for palmitoylation. This can create a fourth IL (intracellular loops) because of the ability of the palmitoylated cysteine to insert in the plasma membrane. Also, C terminus may be involved in the interactions with other proteins that mediate GPCR signaling, such as the calcyon, PDZ domain-containing proteins, and Homer/Vesl proteins.19 The GPCR vary not only in sequence, but also in length of amino acid and carboxy-termini (especially the C3 loop). The serine residue at carboxy terminus region of GPCRs gets phosphorylated by G protein-coupled receptor kinases (GRKs). GRKs constitute of six mammalian Ser/Thr protein kinase that phosphorylate agonist-bound, or activated, GPCR as their primary substrates, GRK-mediated receptor phosphorylation rapidly initiates profound impairment of receptor signaling, or desensitization.20 While the X-ray crystal structures of several GRKs have been solved, but the mechanism of GRK interaction with GPCRs was not known. Recently, Pao et al. (2009)21 proposed a mechanism whereby the N-terminus of GRK2 protein forms an intramolecular interaction that selectively enhances the catalytic activity of the kinase towards GPCR substrates.
Recently, Sheerer et al. (2008)22 reported the 3.2 angstrom (A°) crystal structure of the bovine GPCR (opsin) in its G-protein-interacting conformation. (Ops-GalphaCT peptide complex). Perk et al. (2008)23 reported the crystal structure of ligand-free native GPCR (opsin) from bovine retinal rod cells at 2.9 A° resolution. Compared to GPCR rhodopsin, opsin showed some structural changes in the conserved E(D)RY and NPxxY(x)5,6F regions and in TM5–TM7. At the cytoplasmic side, TM6 was found to be tilted outwards by 6–7 A°, whereas the helix structure of TM5 was more elongated and close to TM6. The authors have suggested that the opsin structure sheds new light on ligand binding to GPCRs and on GPCR activation.23
Family of GPCRs
These are the largest class of receptors, with more than one thousand GPCRs identified so far. The GPCRs family is the third most abundant family in Caenorhabditis elegans, comprising 5% of its genome with approximately 1,100 members. The Drosophila genome has at least 160 GPCRs. Based on the now entirely known human genome careful estimation suggest that about 3–4% of the human genes code for GPCRs, about 1,200–1,300 members of GPCR superfamily in the human genome,24,25 many of which are known to homo- and heterodimerize.26 However, in case of plants a single GPCR has been isolated from from pea27 and maize (Acc. No. NM_001153424), and computational analysis show their presence in Arabidopsis, Populus and rice.28 The GPCRs superfamily is divided into 6 major families which share little sequence homology among each other and some functional similarity.29 The 6 families of GPCRs are as follows:
Family A (rhodopsin receptor family): Family A is commonly known as rhodopsin family. It is the largest family of GPCRs and includes receptors for odorants and small ligands. This family is further divided into three groups. Group 1 contains GPCRs for small ligands including rhodopsin and β-adrenergic receptors. The binding site is localized within the seven TMs. Group 2 contains receptors for peptides whose binding site includes the N-terminal, the extracellular loops and the superior parts of TMs. Group 3 contains GPCRs for glycoprotein hormones. It is characterized by a large extracellular domain and a binding site which is mostly extracellular but at least with contact with extracellular loops e1 and e3.
Family B (secretin receptor family): Family B is commonly known as secretin family. It recruits about 60 members and is characterized not only by the lack of the structural signature present in family A but also by the presence of a large N-terminal ectodomain. Family B GPCRs have a similar morphology to group A3 GPCRs, but they do not share any sequence homology. Their ligands include high molecular weight hormones such as glucagon, secretine, calcitonin, growth hormone-releasing hormone, corticotropin-releasing factor, VIP-PACAP and the Black widow spider toxin, α-latrotoxin.
Family C (metabotropic glutamate receptors family): Family C recruits about two dozens GPCRs such as metabotropic glutamate receptors (mGluR) and the Ca2+ sensing receptors. This family also includes GABA-B receptors, taste receptors, olfactory receptors and a group of putative pheromone receptors coupled to the G protein Go (termed VRs and Go-VN). Like family B, these receptors possess large ectodomains responsible for ligand binding.
Family D (fungus pheromone receptor family): Family D comprises pheromone receptors (VNs) associated with Gi.
Family E (cAMP receptor family): cAMP receptors (cAR) have only seen found in D. discoideum but its possible expression in vertebrate has not yet been reported.
Family F (frizzled/smoothened receptor family): Family F includes the ‘frizzled’ and the ‘smoothened’ (Smo) receptors involved in embryonic development and in particular in cell polarity and segmentation.
These GPCR families are further classified into 64 different subfamilies based on the multiple receptor subtypes, each of which is encoded by separate genes. Each subfamily is further subdivided into different groups, based mainly on TiPS classification scheme that takes into account the native ligand(s) that binds to a particular GPCR. Therefore, GPCRs exist as a big family of receptors that binds to vast variety of ligands and are involved in various signaling pathways.
It is well known that the family of classical GPCRs is characterized by presence of 7TM domain and is also referred to as the heptahelical or serpentine receptors. However, recently it has become apparent that other receptors and proteins that are not heptahelical or serpentine also mediate some of their biological effects via activation of heterotrimeric G proteins. To date, the role of heterotrimeric G proteins in mediating the actions of these nonclassical GPCRs such as receptors for a variety of growth factors, atrial natriuretic hormone, extracellular matrix proteins, as well as zona pellucida glycoprotein ZP3 has not been elucidated.30 Papasaikas et al. (2004)31 suggested that GPCR recognition and classification at the family level can also be analyzed at PREDGPCR Internet service which is a free at http://bioinformatics.biol.uoa.gr/PRED-GPCR.
Gao and Wang (2006)32 introduced a nearest neighbor method to discriminate GPCRs from non-GPCRs and subsequently classify GPCRs at four levels on the basis of amino acid composition and dipeptide composition of proteins. They have classified 1,406 GPCRs into six families and for the subfamily prediction of 1,181 GPCRs of rhodopsin-like family. The sequence analysis of Family A and Family B of GPCRs indicated that there are functional sites on essentially all transmembrane helices, consistent with the parallel daisy chain model of GPCR oligomerization in which each GPCR makes interactions with a number of neighboring GPCRs.33 A simple JavaScript based web interface has been also developed to predict GPCR families and subfamilies (www.insilico-consulting.com/gpcrmotif.html). By using this method Gangal and Kumar, (2007)34 have annotated, 695 orphan receptors, and 121 were identified as belonging to Family A of GPCRs.
Plant GPCR and its Role
There are >1000s of GPCRs known in humans and animal till to date. GPCRs in plants are not well characterized as compared to GPCRs from animal system. Arabidopsis also contains a single regulator of G protein signaling protein (RGS1), which is shown to stimulate the intrinsic guanosine triphosphatase activity of Gα.35 The RGS1 also contains a heptahelical domain, RGS box domain and might be functioning as a receptor or co-receptor.36 Till to date there has been only one putative GPCR (GCR1) identified and experimentally investigated in Arabidopsis.37–40 Using a different bioinformatics tool it was shown that there might be as many as 394 divergent GPCR candidates in Arabidopsis.41 Through computational studies of Family A and Family B of GPCRs Vohra et al. (2007)33 reported that the plant GCR1 contained homology with Family A, Family B and Family E of GPCRs. The first plant GPCR was cloned from Arabidopsis by cDNA library screening and named as GCR1.42 It shows highest similarity to cAMP receptors receptors from the slime mold Dictyostelium discoideum.42 It was shown that GCR1 gene is cell cycle-regulated and its overexpression abolishes seed dormancy and shortens flowering time.38 It is shown that GPCR interacts with GPA1 in Arabidopsis and is involved in abscisic acid (ABA) signaling in guard cells.43 Also, it is shown that mutation in GCR1 leads to hypersensitivity to ABA, inhibition of seed germination, less sensitivity to GA and BR.44 Two putative cDNAs of gamma subunits of G protein complex were isolated from rice.45 PLD and heterotrimeric G proteins play important, diverse roles in cellular regulation and signal transduction. It is shown that Arabidopsis PLDa1 interacts with GPA1 through a motif analogous to the DRY motif in G protein-coupled receptors.46
Generally, the signal recognition by GPCRs is typically coupled by heterotrimeric G proteins to downstream effectors. However, some GPCRs in amoeboid cells and possibly in human cells can initiate downstream action independently of heterotrimeric G-proteins. It is reported that GCR1 can act independently of heterotrimeric G proteins in response to brassinosteroids and gibberellins in Arabidopsis seed germination.47 It has been indicated in loss-of-function gcr1 mutants that GCR1 also plays a positive role in gibberellin-(GA) and brassinosteroid-(BR) regulated seed germination. The null mutants of GCR1 were found to be less sensitive to GA and BR in seed germination. Chen et al. (2004)47 indicated that GCR1, unlike a typical 7TM receptor, apparently acts independently of the heterotrimeric G-protein in at least some aspects of seed germination, suggesting that this alternative mode of 7TM receptor action also functions in the plant kingdom.
It is known that GPCR genetically and physically interacts with the G protein alpha subunit (GPA1) to mediate all known ABA responses in Arabidopsis. Liu et al. (2007)48 observed an ABA-hypersensitive phenotype by overexpressing the GPCR in Arabidopsis. The GPCR also binds to ABA with high affinity at physiological concentration which leads to the dissociation of the GPCR-GPA1 complex in yeast. The authors suggested that the GPCR is a plasma membrane ABA receptor and is a new GPCR, GCR2, from Arabidopsis.48,49 Later, Gao et al. (2007)50 reported that this new GCR2 does contain the canonical 7TM topology structural (a hallmark of known GPCRs) in its amino acids sequence and also some discrepancies was reported regarding its purported plant hormone signaling function. The genetic analysis of gcr2 mutants failed to detect an ABA-related phenotype and some other studies suggested that GCR2 actually does not bind to ABA. These authors also reported that loss-of-function mutations in GCR2-LIKE 1 (GCL1) did not confer ABA insensitivity. All the findings of Gao et al. (2007)50 did not support the notion that GCR2 is an ABA-signaling GPCR in seed germination and early seedling development. Recently, Gao et al. (2008)51 further suggested through genetic evidence that GCR2 is unlikely to act as a receptor for ABA in the context of either seed germination or early seedling development. Still there is an unanswered question to know how many (yet undiscovered) candidates of GPCRs exist in plants? Various plant responses that are affected upon genetic knockout of alpha, beta and gamma subunits of G proteins44,52 suggested that there might be existence of more plant's GPCRs which may discover in future.
A novel GPCR containing a lipid kinase domain has also been reported in Dictyostelium that regulates cell density sensing.53 Earlier, we have reported the isolation of a pea GPCR gene (PsGPCR) and the characterization of the encoded GPCR protein.27 The full-length PsGPCR cDNA was found to be a 1.008 kb in size. The deduced amino acid sequence revealed a PsGPCR protein contained 335 amino acid residues with a predicted molecular mass of about 35 kDa and pI 10.60. The PsGPCR protein was reported to form oligomers by self-interacting, and also found to interacted with all three subunits of G-proteins but not with PLC-delta or the C2 domain of PLC-delta.27 The Arabidopsis GCR1 was also reported to interact with Ga (GPA1) and thereby regulate abscisic acid signaling.43 The oligomeric nature of PsGPCR probably allow for a more complex ligandreceptor relationship.54
Due to low sequence conservation GPCRs it difficult to identify novel candidates from plants. Recently, Gookin et al. (2008)28 by whole-proteome analyses reported the identification of candidate GPCRs within the Arabidopsis, Oryza and Populus proteomes. Most of them were found to be interacting with Galpha of Arabidopsis (GPA1).
Recently, from the laboratory of Assmann another pair of GPCRs have been implicated in ABA response. These are novel GPCR-type G-proteins (GTG1 and GTG2) from Arabidopsis which show homology to an orphan vertebrate GPCR (GPR89) and interact with Arabidopsis G protein alpha subunit, GPA1.55 Interestingly, these proteins also have intrinsic GTP-binding and GTPase activity. These authors also showed that Arabidopsis mutants lacking both GTG1 and GTG2 exhibit ABA hyposensitivity which indicated that GTG1 and GTG2 may mediate ABA responses during germination, flowering, stomatal closure and root elongation. Furthermore, GTG1 and GTG2 were also shown to bind ABA. Finally it has been suggested that GTG proteins function both as a new type of G protein and as a class of membrane-localized ABA receptors.49,55
Acknowledgements
Work on signal transduction and plant stress signaling in NT's laboratory is partially supported by Department of Science and Technology (DST), Government of India.
Abbreviations
- ABA
abscisic acid
- ECL
extracellular amino-terminal domain
- GCR1
GPCR from Arabidopsis
- GDP
guanosine disphosphate
- GPCRs
G protein coupled receptors
- GRK
GPCR receptor kinase
- GTG
GPCR-type G-protein
- GTP
guanosine triphosphate
- ICL
intracellular carboxyl-terminal domain
- 7TM
seven transmembrane α-helical regions
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9530
References
- 1.Redhead CR, Palme K. The genes of plant signal transduction. Crit Rev Plant Sci. 1996;15:425–454. [Google Scholar]
- 2.Mahajan S, Tuteja N. Cold, salinity and drought stresses: An overview. Arch Biochem Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Tuteja N. Mechanisms of high salinity tolerance in plants. Meth Enzymol. 2007;428:419–438. doi: 10.1016/S0076-6879(07)28024-3. [DOI] [PubMed] [Google Scholar]
- 4.Trewavas AJ, Malho R. Signal perception and Transduction: The origin of the phenotype. Plant Cell. 1997;9:1181–1195. doi: 10.1105/tpc.9.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hucho F, Buchner K. Signal transduction and protein kinases: the long way from the plasma membrane into the nucleus. Naturwissenschaften. 1997;84:281–290. doi: 10.1007/s001140050396. [DOI] [PubMed] [Google Scholar]
- 6.Temple BR, Jones AM, Juliano RL. The plant heterotrimeric G-protein complex. Annu Rev Plant Biol. 2007;58:249–266. doi: 10.1146/annurev.arplant.58.032806.103827. [DOI] [PubMed] [Google Scholar]
- 7.Tuteja N, Sopory SK. Plant signaling in stress: G-protein coupled receptors, heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signal Behav. 2008;3:79–86. doi: 10.4161/psb.3.2.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strader CD, Fong TM, Tota MR, Underwood D, Dixon RA. Structure and function of G protein-coupled receptors. Annu Rev Biochem. 1994;63:101–132. doi: 10.1146/annurev.bi.63.070194.000533. [DOI] [PubMed] [Google Scholar]
- 9.Shorr RGL, Lefkowitz RJ, Caron MG. Purification of the β-adrenergic receptor. Identification of the hormone-binding subunit. J Biol Chem. 1981;256:5820–5826. [PubMed] [Google Scholar]
- 10.Bockaert J, Pin JP. Molecular tinkering of G-protein-coupled receptors: An evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamm HE. How activated receptors coupled to G proteins. Proc Natl Acad Sci USA. 2001;98:4819–4821. doi: 10.1073/pnas.011099798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamm H. The many faces of G-protein signaling. J Biol Chem. 1966;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 13.Gilman AG. G proteins and dual control of adenylate cyclase. Cell. 1984;36:577–579. doi: 10.1016/0092-8674(84)90336-2. [DOI] [PubMed] [Google Scholar]
- 14.Gilman AG. G proteins: transducer of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 15.Logothetis DE, Kurachi Y, Galper J, Neer EJ, Clapham DE. The βγ subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature. 1987;325:321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- 16.Ford CE, Skiba NP, Bae H, Daaka Y, Reuveny E, Shekter LR, et al. Molecular basis for interaction of G protein βγ subunits with effectors. Science. 1998;280:1271–1273. doi: 10.1126/science.280.5367.1271. [DOI] [PubMed] [Google Scholar]
- 17.Rondard P, Iiri T, Srinivasan S, Meng E, Fujita T, Bourne HR. Mutant G protein alpha subunit activated by Gbetagamma: a model for receptor activation. Proc Natl Acad Sci USA. 2001;98:6150–6155. doi: 10.1073/pnas.101136198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentles A, Karlin S. Why are human G-protein-coupled receptors predominantly intronless? Trends in Gen. 1999;15:47–48. doi: 10.1016/s0168-9525(98)01648-5. [DOI] [PubMed] [Google Scholar]
- 19.Baldwin JM. Structure and function of receptors coupled to G proteins. Curr Opin Cell Biol. 1994;6:180–190. doi: 10.1016/0955-0674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 20.Pingret J, Journet E, Barker DG. Rhizobium Nod factor signaling: evidence for a G protein-mediated transduction mechanism. Plant Cell. 1998;10:659–671. doi: 10.1105/tpc.10.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pao CS, Barker BL, Benovic JL. Role of the amino terminus of G protein-coupled receptor kinase 2 in receptor phosphorylation. Biochemistry. 2009 doi: 10.1021/bi900408g. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 23.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008 doi: 10.1038/nature07063. www.nature.com/doifinder/10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 24.Schoneberg T, Schulz A, Gudermann T. The structural basis of G-protein-coupled receptor function and dysfunction in human diseases. Rev Physiol Biochem Pharmacol. 2002;144:143–227. [PubMed] [Google Scholar]
- 25.Fredriksson R, Schioth HB. The repertoire of G-protein coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 26.Bai M. Dimerization of G-protein-coupled receptors: roles in signal transduction. Cellular Sig. 2004;16:175–186. doi: 10.1016/s0898-6568(03)00128-1. [DOI] [PubMed] [Google Scholar]
- 27.Misra S, Wu Y, Venkataraman G, Sopory S, Tuteja N. Heterotrimeric G-protein complex and G-protein-coupled receptor from a legume (Pisum sativum): role in salinity and heat stress and cross-talk with Phospholipase C. Plant J. 2007;51:656–669. doi: 10.1111/j.1365-313X.2007.03169.x. [DOI] [PubMed] [Google Scholar]
- 28.Gookin TE, Kim J, Assmann SM. Whole proteome identification of plant candidate G-protein coupled receptors in Arabidopsis, rice and poplar: computational prediction and in-vivo protein coupling. Genome Biol. 2008;9:120. doi: 10.1186/gb-2008-97-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolakowski LF., Jr “GCRDb: a G-protein-coupled receptor database”. Receptor Channel. 1994;2:1–7. [PubMed] [Google Scholar]
- 30.Patel TB. Single transmembrane spanning heterotrimeric G protein-coupled receptors and their signaling cascades. Pharmacol Rev. 2004;56:371–385. doi: 10.1124/pr.56.3.4. [DOI] [PubMed] [Google Scholar]
- 31.Papasaikas PK, Bagos PG, Litou ZI, Promponas VJ, Hamodrakas SJ. PRED-GPCR: GPCR recognition and family classification server. Nucleic Acids Res. 2004;32:380–382. doi: 10.1093/nar/gkh431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao QB, Wang ZZ. Classification of G-protein coupled receptors at four levels. Protein Eng Des Sel. 2006;19:511–516. doi: 10.1093/protein/gzl038. [DOI] [PubMed] [Google Scholar]
- 33.Vohra S, Chintapalli SV, Illingworth CJ, Reeves PJ, Mullineaux PM, Clark HS, et al. Computational studies of Family A and Family B GPCRs. Biochem Soc Trans. 2007;35:749–754. doi: 10.1042/BST0350749. [DOI] [PubMed] [Google Scholar]
- 34.Gangal R, Kumar KK. Reduced alphabet motif methodology for GPCR annotation. J Biomol Struct Dyn. 2007;25:299–310. doi: 10.1080/07391102.2007.10507178. [DOI] [PubMed] [Google Scholar]
- 35.Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- 36.Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, et al. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plakidou-Dymock S, Dymock D, Hooley R. A higher plant seven transmembrane receptor that influences sensitivity to cytokinins. Curr Biol. 1998;8:315–324. doi: 10.1016/s0960-9822(98)70131-9. [DOI] [PubMed] [Google Scholar]
- 38.Colucci G, Apone F, Alyeshmerni N, Chalmers D, Chrispeels MJ. GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc Natl Acad Sci USA. 2002;99:4736–4741. doi: 10.1073/pnas.072087699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apone F, Alyeshmerni N, Wiens K, Chalmers D, Chrispeels MJ, Colucci G. The G-protein-coupled receptor GCR1 regulates DNA synthesis through activation of phosphatidylinositol-specific phospholipase C. Plant Physiol. 2003;133:571–579. doi: 10.1104/pp.103.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, et al. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007;143:1590–1600. doi: 10.1104/pp.106.089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriyama EN, Strope PK, Opiya SO, Chen Z, Jones AM. Mining the Arabidopsis thaliana genome for highly-divergent seven transmembrane receptors. Genome Biol. 2006;7:96. doi: 10.1186/gb-2006-7-10-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Josefsson LG, Rask L. Cloning of a putative G-protein-coupled receptor from Arabidopsis thaliana. Eur J Biochem. 1997;249:415–420. doi: 10.1111/j.1432-1033.1997.t01-1-00415.x. [DOI] [PubMed] [Google Scholar]
- 43.Pandey S, Assmann SA. The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell. 2004;16:1616–1632. doi: 10.1105/tpc.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr Opin Plant Biol. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 45.Kato C, Mizutani T, Tamaki H, Kumagai H, Kamiya T, Hirobe A, et al. Characterization of heterotrimeric G protein complexes in rice plasma membrane. Plant J. 2004;38:320–331. doi: 10.1111/j.1365-313X.2004.02046.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhao J, Wang X. Arabidopsis Phospholipase Dα1 interacts with the heterotrimeric G protein α-subunit through a motif analogous to the DRY motif in G protein-coupled receptors. J Biol Chem. 2004;279:1794–1800. doi: 10.1074/jbc.M309529200. [DOI] [PubMed] [Google Scholar]
- 47.Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 2004;135:907–915. doi: 10.1104/pp.104.038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Yue Y, Li B, Nie Y, Li W, Wu WH, Ma L. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1676–1677. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- 49.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signaling. Nature. 2009;450:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 50.Gao Y, Zeng Q, Guo J, Cheng J, Ellis BE, Chen JG. Genetic characterization reveals no role for the reported ABA receptor, GCR2, in ABA control of seed germination and early seedling development in Arabidopsis. Plant J. 2007;52:1001–1013. doi: 10.1111/j.1365-313X.2007.03291.x. [DOI] [PubMed] [Google Scholar]
- 51.Guo J, Zeng Q, Emami M, Ellis BE, Chen JG. The GCR2 gene family is not required for ABA control of seed germination and early seedling development in Arabidopsis. PLoS One. 2008;3:2982. doi: 10.1371/journal.pone.0002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, et al. Heterotrimeric G protein gamma subunits provide functional selectivity in Gbetagamma dimer signaling in Arabidopsis. Plant Cell. 2007;19:1235–1250. doi: 10.1105/tpc.107.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bakthavatsalam D, Brazill D, Gomer RH, Eichinger L, Rivero F, Noegel AA. “A G protein-coupled receptor with a lipid kinase domain is involved in cell-density sensing”. Curr Biol. 2007;17:892–897. doi: 10.1016/j.cub.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 54.Park PS-H, Palczewski K. Diversifying the repertoire of G protein-coupled receptors through oligomerization. Proc Natl Acad Sci USA. 2005;102:8793–8794. doi: 10.1073/pnas.0504016102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:21–23. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]