Abstract
Colletotrichum higginsianum causes typical anthracnose lesions on the leaves, petioles, and stems of cruciferous plants. Inoculation of Arabidopsis thaliana ecotype Columbia leaves with C. higginsianum results in fungal growth and disease symptoms reminiscent of those induced in other cruciferous plants. We performed map-based cloning and natural variation analysis of 19 A. thaliana ecotypes to identify a dominant resistance locus against C. higginsianum. We found that the A. thaliana RCH2 (for recognition of C. higginsianum) locus encodes two NB-LRR proteins, both of which are required for resistance to C. higginsianum in the A. thaliana ecotype Ws-0. Both proteins are well-characterized R proteins involved in resistance against bacterial pathogens; RRS1 (resistance to Ralstonia solanacearum 1) confers resistance to strain Rs1000 of R. solanacearum and RPS4 to Pseudomonas syringae pv. tomato strain DC3000 expressing avrRps4 (Pst-avrRps4). Furthermore, we found that both RRS1-Ws and RPS4-Ws genes are required for resistance to Pst-avrRps4 and to Rs1002 R. solanacearum. We therefore demonstrate that a pair of neighboring genes, RRS1-Ws and RPS4-Ws, function cooperatively as a dual R-gene system against at least three distinct pathogens.
Key words: R gene, RPS4, RRS1, Colletotrichum higginsianum, Pseudomonas syringae, Ralstonia solanacearum
Plants are exposed to various types of potentially invasive organisms, including viruses, bacteria, fungi, nematodes and protozoa, but are able to defend themselves by activating multiple defense mechanisms. The gene-for-gene hypothesis1 provides a mechanism for specific recognition of the pathogen by the plant. This recognition is mediated by direct or indirect interactions between the product of a plant resistance (R) gene and the corresponding effectors encoded by avirulence genes in the pathogen.2 Most R-genes encode non-membrane proteins that contain a conserved nucleotide-binding (NB) site and a carboxy-terminal leucine-rich repeat (LRR) domain.
The A. thaliana genome contains about 150 genes coding for NB-LRR-containing proteins.3 This is far less than the number of genes that would be required to respond individually and specifically to all of its potential pathogens. However, plants may have been able to limit the number of required NB-LRR-encoding genes if host proteins perceive sets of distinct pathogens.4
Colletotrichum species cause devastating anthracnose diseases in a large number of agronomically important crops. These diseases can often be controlled by introduction of genetic resistance traits, but the molecular components of resistance remain unknown. Inoculation of A. thaliana ecotype Columbia (Col-0) leaves with Colletotrichum higginsianum results in fungal growth and disease symptoms reminiscent of those induced in other cruciferous plants.5,6 Inoculation of a large number of ecotypes with isolates of C. higginsianum showed that A. thaliana has at least two dominant resistance gene loci, designated RCH1 and RCH2 (for recognition of C. higginsianum), indicating that A. thaliana resistance to C. higginsianum is controlled by a “gene-for-gene” interaction.5 In a previous study, we identified a single putative R locus, RCH1 on the top of chromosome 4, in the C. higginsianum-resistant A. thaliana ecotype Eil-0.5
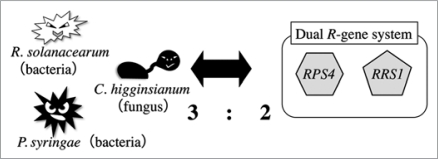
In the present study, the locus named RCH2 maps in an extensive cluster of disease-resistance loci known as MRC-J in the A. thaliana ecotype Ws-0. By analyzing natural variations within the MRC-J region, we found that alleles of RRS1 (resistance to Ralstonia solanacearum 1) from susceptible ecotypes contain single nucleotide polymorphisms that may affect the encoded protein. Consistent with this finding, two susceptible mutants, rrs1-1 and rrs1–2, were identified by screening a T-DNA-tagged mutant library for the loss of resistance to C. higginsianum. The screening identified an additional susceptible mutant (rps4-21), which has a 5-bp deletion in the neighboring gene, RPS4-Ws, a well-characterized R gene that provides resistance to Pseudomonas syringae pv. tomato strain DC3000 expressing avrRps4 (Pst-avrRps4). To assess if RRS1-Ws and RPS4-Ws function in concert, we generated an rps4-21/rrs1-1 double mutant by crossing rps4-21 and rrs1-1 mutants. The susceptibility levels of rps4-21/rrs1-1 double mutant to C. higginsianum were similar to that exhibited by the single mutants, suggesting that RRS1-Ws and RPS-4-Ws function cooperatively. We also found that both RRS1 and RPS4 are required for resistance to R. solanacearum and Pst-avrRps4. Thus, these two adjacent R genes confer resistance, in tandem or individually, to three distinct pathogens with very different infection strategies and virulence mechanisms (Fig. 1).
Figure 1.
RPS4 and RRS1 function as a dual resistance gene system that prevents infection by three distinct pathogens (Colletotrichum higginsianum, Ralstonia solanacearum and Pseudomonas syringae pv. tomato strain DC3000 expressing avrRps4).
Several examples of two NB-LRR genes acting cooperatively to confer resistance against a pathogen have been reported. For example, A. thaliana RPP2A and RPP2B reside adjacently in the RPP2 locus.7 Blast resistance in Pikm-containing rice is conferred by a combination of two NB-LRR encoding genes, Pikm1-TS and Pikm2-TS.8 Pi5-mediated resistance against rice blast requires two NB-LRR-encoding genes.9 It is not known whether these NB-LRR genes function cooperatively or independently. Because of structural similarity with RRS1/RPS4 genes, it is possible that resistance to the pathogens is conferred by cooperation between the two NB-LRR genes.
Several reports have shown that a single R gene/locus can confer resistance to multiple pathogens. For instance, tomato Mi mediates resistance against three distinct types of pests, including root-knot nematodes, potato aphids and sweet potato whitefly.10 In the present study, we suggest that two distinct R-genes located in a conserved head-to-head organization confer resistance to three distinct pathogen species by acting cooperatively.
The tandem function of RRS1-Ws and its neighboring gene RPS4-Ws is also supported by the evolutionary conservation of the gene pair. Close homologs of RPS4 are often physically paired with homologs of RRS1 in a head-to-head (inverted) tandem arrangement.11 The evolutionary conservation of homologous gene pairs in a head-to-head arrangement also supports the idea that cooperative function of two R genes could be a common mechanism of defense against pathogens. Since the two open reading frames are only 264 bp apart, the promoter regions of the gene pairs possibly overlap, leading to co-regulation of the genes. The head-to-head configuration may assure balanced levels of the protein pair to meet a strict stoichiometric requirement to act together, possibly in a complex. As a practical application, this finding may provide a new strategy for creating transgenic plants that express R genes from other plants. Introduction of two R genes in a head-to-head orientation may be necessary for effective pathogen resistance.
Acknowledgements
This work was supported in part by an Industrial Technology Research Grant Program from the New Energy and Industrial Technology Development Organization (NEDO) of Japan, by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) to Y.N., by KAKENHI (19580053 to Y.N., and 18780028 to M.N.), and by The Sumitomo Foundation to M.N.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9640
References
- 1.Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 2.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van der Biezen EA, Jones JDG. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 5.Narusaka Y, Narusaka M, Park P, Kubo Y, Hirayama T, Seki M, et al. RCH1, a locus in Arabidopsis that confers resistance to the hemibiotrophic fungal pathogen Colletotrichum higginsianum. Mol Plant-Microbe Interact. 2004;17:749–762. doi: 10.1094/MPMI.2004.17.7.749. [DOI] [PubMed] [Google Scholar]
- 6.O'Connell R, Herbert C, Sreenivasaprasad S, Khatib M, Esquerré-Tugayé MT, Dumas B. A novel Arabidopsis-Colletotrichum pathosystem for the molecular dissection of plant-fungal interactions. Mol Plant-Microbe Interact. 2004;17:272–282. doi: 10.1094/MPMI.2004.17.3.272. [DOI] [PubMed] [Google Scholar]
- 7.Sinapidou E, Williams K, Nott L, Bahkt S, Tör M, Crute I, et al. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 2004;38:898–909. doi: 10.1111/j.1365-313X.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- 8.Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumono T, et al. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics. 2008;180:2267–2276. doi: 10.1534/genetics.108.095034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SK, Song MY, Seo YS, Kim HK, Ko S, Cao PJ, et al. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics. 2009;181:1627–1638. doi: 10.1534/genetics.108.099226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nombela G, Williamson VM, Muñiz M. The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol Plant-Microbe Interact. 2003;16:645–649. doi: 10.1094/MPMI.2003.16.7.645. [DOI] [PubMed] [Google Scholar]
- 11.Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]