Abstract
Since the initial biochemical study of a putative invertase inhibitor half a century ago, it has remained as a puzzle as whether such an inhibitory protein indeed limits invertase activity in vivo and, if it does, what is the developmental or physiological significance of such an interaction? Recently, we demonstrated that an invertase inhibitor, INVINH1, specifically inhibited cell wall invertase activity in tomato and Arabidopsis. Silencing INVINH1 expression in tomato released a significant amount of extra cell wall invertase activity. This posttranslational elevation of invertase activity resulted in a blockage of ABA-induced leaf senescence and an increase in fruit sugar levels and seed weight. Here, we discuss the implication of the findings and propose a model that the invertse inhibitor may act as a key modulator in controlling leaf longevity and seed development to ensure success during plant evolution. This may be achieved by optimizing carbon and nitrogen allocation and sugar signaling via interaction between invertase and inhibitor. The discoveries open up exciting new areas for exploring fundamental questions in sugar signaling, carbon allocation and plant development as well as avenues for improving crop productivity.
Key words: carbon allocation, evolution, invertase, invertase inhibitor, leaf senescence, seed development, sugar signaling
Introduction
In most of the higher plants, sucrose is the major sugar transported through phloem from photosynthetic leaves (source) to non-photosynthetic tissues (sink). Upon delivery to the recipient sinks, sucrose must be either degraded by sucrose synthase (Sus) into UDP-glucose and fructose or hydrolyzed by invertase (Inv) into glucose and fructose for further use.1 Invs are categorized into apoplasmic, vacuolar and cytoplasmic subgroups. While Sus appears to be predominately involved in energy (ATP) generation and starch and cell wall biosynthesis,2 Inv has been shown to play multiple pivotal roles in plant development and in response to stress via controlling carbon allocation and sugar signaling.3
Research on the roles of Invs has been focused primarily at the transcriptional level. However, it has been speculated, largely based on in vitro studies, that Inv activity may be limited by a small group of inhibitory proteins with molecular masses (Mr) ranging from 15 to 23 kD.4 Given that apoplamic and vacuolar Invs are intrinsically stable due to their glycan decoration, control of their activity may be highly dependent on posttranslational regulation.5 Thus, elucidation of the potential control of Inv activity by its inhibitor in vivo is critically required for advancing our understanding of carbon allocation and plant development.5,6
Interaction between Invertase and its Inhibitor Plays Key Roles in Plant Development
By using a combination of molecular genetic, biochemical and cell biology approaches, we recently identified an invertase inhibitor, INVINH1, from tomato that specifically inhibits cell wall Inv activity in planta.6 RNAi-mediated silencing of INHINV1 expression increased cell wall Inv activity by ∼50% and ten times in mature and old leaves, respectively, and two-fold in developing seeds and fruits, leading to delayed leaf senescence and an increase in seed weight and fruit hexose level.6 The data demonstrate, for the first time, that cell wall Inv activity is highly elastic and subject to posttranslational control by its inhibitory protein in vivo. This functional protein-protein interaction could have profound implications in sugar signaling, plant senescence and evolution (see below).
Sugar Signaling Mediated by Cell Wall Invertase and its Inhibitor Acts Upstream of Hormone-Regulated Leaf Senescence
It is well known that abscisic acid (ABA) induces leaf senescence in many plant species including tomato7 and rice.8 The underlying mechanism, however, remains elusive. We found that silencing INVINH1 blocked ABA-induced leaf senescence and expression of senescence-associated genes, SENU2 and SENU3 without impacting on endogenous cytokinin levels.6 These observations show expression of INVINH1, hence the decrease of cell wall Inv activity, is required for the senescence process. The reduction of cell wall inv activity by its inhibitor likely decreases the apoplasmic hexose level. The low hexose concentration in the leaf apoplasm may signify to (1) induce observed increase in the senescence-related Cys protease genes, SENU2 and SENU3 and (2) promote phloem loading of sucrose from storage carbohydrate in the leaves, which together leads to leaf senescence. Consistently, cytokinin-mediated delay in leaf senescence is dependent upon high cell wall Inv activity.9 It is thus logical to conclude that sugar signaling from apoplasm likely acts upstream of ABA- and cytokinin-mediated leaf senescence.
Invertase Inhibitor as a Key PlayerModulating Resource Allocation during Evolution
Given the essential role the apoplasmic Inv plays in carbon partitioning, it is intriguing as why there are inhibitory proteins such as INVINH1 that limit Inv activity? Here, we propose a resource allocation model where modulation by inv inhibitor may function to control and optimize nutrient distribution during plant evolution. During senescence, leaves remobilize carbon and nitrogen back to the remaining bodies such as stem.10 This nutrient-recycling strategy ensures efficient utilization of limited resources for the survival of plant species through evolution. The remobilization of carbon and nitrogen is achieved by breaking down stored protein into amino acids and starch into soluble sugars.7,10 The resultant amino acids and sugars (mainly in the form of sucrose) are then loaded into the phloem for translocation to the parental bodies. In this context, the dramatic increase in the expression of Inv inhibitor, INVINH1, as leaf ages, upon which expression of two Cys proteases SENU2 and SENU3 was induced and ABA-induced leaf aging proceeded,6 may dictate the timing of senescence, thereby guaranteeing nutrient recycling.
A similar evolutionary role may be played by INVINH1 in reproductive organs. Here, INVINH1 interacted with a cell wall Inv, Lin 5, in the phloem parenchyma cells of fruit placenta connecting young seeds, where sucrose unloading follows an apoplasmic pathway.6 This cell-specific limitation of the Inv activity by the inhibitor could ensure that each seed receives a small proportion of carbon just sufficient for its survival from a limited amount of resource available. This control could maximize the number of viable seeds from a given plant, a critical feature of plant evolution. Like offspring in mammals, being born at the right weight is the best strategy for survival and living.
Sugar Sensing from Apoplasm: A Link between Cell Wall Invertase and G-Protein?
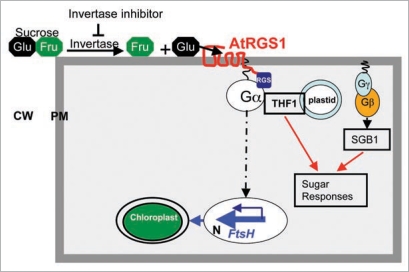
One of the intriguing questions arisen from the analyses above is how plant cells sense changes of sugar levels in the apoplasm, a critical but unresolved issue faced by modern plant biology. The notion that plasma membrane sucrose or hexose carriers may function as ‘receptors’ for sugar sensing has not been backed by experimental evidence thus far. Based on recent work in Arabidopsis, we propose that glucose generated by invertase in apoplasm may be sensed by RGS1 (regulator of G-protein signaling) protein that consists of an N-terminal transmembrane domain characteristic of G-protein-coupled receptors and a C-terminal RGS box in Arabidopsis. The latter domain acts as a GTPase-accelerating protein (GAP) for the heterotrimeric G-protein α subunit (AtGPA1).11 The fact that glucose can promote in vivo interaction between AtRGS1 and AtGPA1 suggests that AtRGS1 probably functions as a glucose-activated GAP to switch off G-protein signal transduction.12 However, it remains unknown whether glucose can directly bind to AtRGS1. In addition, AtGPA1 is physically interacted with thylakoid formation 1 (THF1) that is localized both on the out envelope of plastids and stroma.13,14 The physiological significance of this interaction between AtGPA1 and THF1 is also associated with sugar sensing.13,14 Thus, it is likely that apoplasmic-localized glucose may be recognized by RGS1 which transmits the extracellular sugar signal into the cell through protein partners including GPA1 and THF1 (Fig. 1). These observations, together with the finding that cell wall Inv activity is highly regulated by its inhibitor,6 provide tools and opportunities to dissect the relationship between cell wall invertase, its inhibitor and G-proteins in sugar sensing and signaling from extra- to intra-cellular compartments.
Figure 1.
A model on sugar sensing and signaling from extra- to intra-cellular compartments mediated by cell wall invertase, its inhibitor and G-proteins. Glucose generated by interaction between invertase and its inhibitor in apoplasm may be sensed by AtRGS1. The GTP-bound active form of Gα can transmit glucose signal into the cytoplasm via direct interaction with AtRGS1 and THF1, and induce sugar responses. The active form of Gα can also regulate chloroplast development and photosynthesis in a THF1-independent pathway via controlling nuclear gene expression. A Golgi-localized hexose transporter (SGB1) has been genetically identified to mediate Gβγ signaling. CW; cell wall. PM; plasma membrane. N; nucleus.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9667
References
- 1.Ruan Y-L, Llewellyn DJ, Furbank RT. Suppression of sucrose synthase expression represses cotton fibre cell initiation, elongation and seed development. Plant Cell. 2003;15:952–964. doi: 10.1105/tpc.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruan Y-L. Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fiber. Funct Plant Biol. 2007;34:1–10. doi: 10.1071/FP06234. [DOI] [PubMed] [Google Scholar]
- 3.Koch K. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Krausgrill S, Greiner S, Koster U, Vogel R, Rausch T. In transformed tobacco cells the apoplasmic invertase inhibitor operates as a regulatory switch of cell wall invertase. Plant J. 1998;13:275–280. [Google Scholar]
- 5.Rausch T, Greiner S. Plant protein inhibitors of invertases. Biochim Biophy Acta. 2004;1696:253–261. doi: 10.1016/j.bbapap.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, Ni D-A, Ruan Y-L. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell. 2009;21:2072–2089. doi: 10.1105/tpc.108.063719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghanem ME, Albacete F, Martínez-Andújar C, Manuel AM, Romero-Aranda R, Dodd I, et al. Hormonal changes during salinity-induced leaf senescence in tomato (Solanum lycopersicum L.) J Exp Bot. 2008;59:3039–3050. doi: 10.1093/jxb/ern153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Zhang J, Wang Z, Zhu Q, Liu L. Abscisic acid and cytokinins in the root exudates and leaves and their relationship to senescence and remobilization of carbon reserves in rice subjected to water stress during grain filling. Planta. 2002;215:645–652. doi: 10.1007/s00425-002-0789-2. [DOI] [PubMed] [Google Scholar]
- 9.Lara MEB, Garcia M-CG, Fatima T, Ehneß E, Lee TK, Proels R, et al. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell. 2004;16:1276–1287. doi: 10.1105/tpc.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drake R, Isaac J, Farrell A, Cooper W, Schuch W, Grierson D. Isolation and analysis of cDNAs encoding tomato cysteine proteases expressed during leaf senescence. Plant Mol Biol. 30:755–767. doi: 10.1007/BF00019009. [DOI] [PubMed] [Google Scholar]
- 11.Chen JG, Willard FS, Huang J, Liang SA, Chasse AM, Jones AM, Siderovski DP. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- 12.Johnston CA, Taylor JP, Gao Y, Kimple AJ, Chen JG, Siderovski DP, et al. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, et al. The plastid protein thylakoid formation1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell. 2006;18:1226–1238. doi: 10.1105/tpc.105.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Wei Q, Wu W, Cheng Y, Hu G, Hu F, et al. Activation of the heterotrimeric G protein α-subunit, GPA1, suppresses ftsh-mediated inhibition of chloroplast development in Arabidopsis. Plant J. 2009;58:1041–1053. doi: 10.1111/j.1365-313X.2009.03843.x. [DOI] [PubMed] [Google Scholar]