Abstract
The small phenolic molecule salicylic acid (SA) plays a key role in plant defense. Significant progress has been made recently in understanding SA-mediated defense signaling networks. Functional analysis of a large number of genes involved in SA biosynthesis and regulation of SA accumulation and signal transduction has revealed distinct but interconnecting pathways that orchestrate the control of plant defense. Further studies utilizing combinatorial approaches in genetics, molecular biology, biochemistry and genomics will uncover finer details of SA-mediated defense networks as well as further insights into the crosstalk of SA with other defense signaling pathways. The complexity of defense networks illustrates the capacity of plants to integrate multiple developmental and environmental signals into a tight control of the costly defense responses.
Key words: salicylic acid, disease resistance, signal transduction, Arabidopsis, Pseudomonas syringae
Plants have evolved sophisticated defense mechanisms to ward off attacks from pathogens. In addition to pre-formed physical/chemical barriers, plants can actively monitor the presence of pathogens and subsequently activate defense signaling networks, which in turn restrict the further growth and spread of pathogens.
The small phenolic compound salicylic acid (SA) plays a central role in plant defense signaling. It is required for recognition of pathogen-derived components and subsequent establishment of local resistance in the infected region as well as systemic resistance at the whole plant level.1–3 SA accumulation is inducible upon infections of various pathogens, treatment with elicitors from pathogens, and stress conditions.3–5 Exogenous application of SA and its synthetic analogs to plants is sufficient to invoke disease resistance.6–9 Disruption of SA accumulation and/or signaling by mutations or by a transgenic SA hydrolase encoded by the bacterial gene nahG greatly compromises defense against pathogens.10 In addition, the phytohormones jasmonic acid (JA) and ethylene (ET) regulate SA-mediated defense as well as many aspects of plant development. Emerging evidence also implicates additional phytohormones in plant defense, two of which, auxin and abscisic acid, were recently shown to impact the SA pathway.11,12
The past two decades have witnessed exciting progress made towards a comprehensive understanding of defense networks in the model plant Arabidopsis, especially those regulated by SA. The discovery of an expanding array of genes involved in SA-mediated defense suggests the complexity of defense networks. Surprisingly, information on functional relationships among many defense genes is sparse. Connecting the dots (genes) on the defense map to form pathways, which are further interconnected into complex defense networks, still remains a challenging task. This review focuses on our current understanding of the interactions among genes that regulate three key sub-circuits of the SA pathway: SA biosynthesis, SA accumulation and SA signal transduction. Discussions of the crosstalk between components involved in the SA pathway and those in other defense pathways can be found in some excellent reviews.13–17
SID2-Dependent and SID2-Independent SA Biosynthesis
The Arabidopsis SA INDUCTION-DEFICIENT 2 (SID2) gene encodes isochorismate synthase, which presumably converts chorismate to isochorismate. In addition to reduced SA accumulation, sid2 mutants show enhanced disease susceptibility (eds) to various pathogens, which can be rescued by SA treatment.18,19 These observations provide the first genetic evidence that a chorismate-derived pathway is critical for SA biosynthesis. This pathway is likely evolutionarily conserved since a prokaryotic SID2 homologue is functional in producing SA when expressed in plants.20,21
A number of studies indicate that the amino acid phenylalanine, itself a derivative of chorismate, also contributes to SA production in a SID2-independent pathway.22–24 Recently, using the Arabidopsis mutant accelerated cell death 6-1 (acd6-1) that constitutively expresses high SA levels, Lu et al. observed that the sid2-1 mutation, when introduced into the acd6-1 background, suppressed the majority of but not all SA accumulation. This result highlights the activation of SID2-independent SA production in acd6-1.25 It is possible that one or more SID2-independent pathway(s) produce small amount of SA while SID2 mediates the bulk synthesis of SA (Fig. 1).
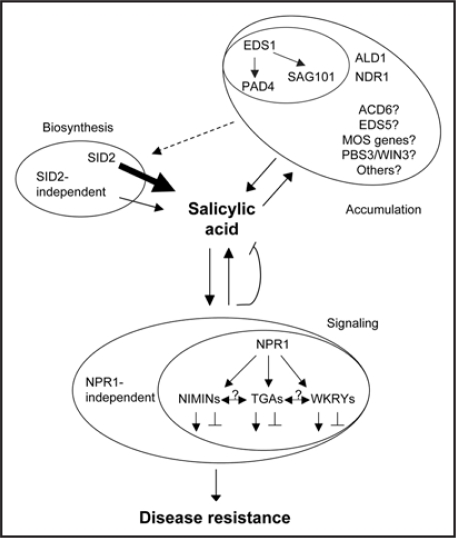
Figure 1.
A model for SA-mediated defense networks. The networks are grouped into three intricately interconnected sectors, SA biosynthesis, SA accumulation and SA signaling. For SA biosynthesis, SID2 contributes to the majority of SA production while SID2-independent pathway(s) plays a minor role, as denoted by the thickness of the arrows. For SA accumulation, there are multiple independent regulatory pathways. PAD4 and SAG101 physically interact with EDS1, likely acting downstream of EDS1 in two separate pathways. NDR1 is known to act independently of EDS1, likewise, ALD1 and PAD4 function in different pathways. Expression of many SA regulators in this group is inducible with SA treatment, suggestion that these regulators and SA form signal amplification loops. For SA signaling, there are both NPR1-dependent and -independent pathways. The NPR1 node includes NIMIN proteins and transcriptions factors, such as TGAs and WRKYs. Components in the NPR1 node can both positively and negatively regulate plant defense. Question mark indicates that the functional relationship of a SA regulator with other regulators is unclear. Dotted arrow indicates the possibility that components regulating SA accumulation may directly or indirectly affect the biosynthetic pathways. Note not all SA regulators are shown because of space limitation. LMM genes are not included because it is unclear if they play a direct role in regulating SA-mediated defense.
Multiple Pathways Feed into the Regulation of SA Accumulation
Protein sequence analysis indicates that the majority of SA regulators lack distinct enzymatic motifs, suggesting that, unlike SID2, these proteins are not directly involved in SA biosynthesis. Just how these regulators affect SA accumulation still remains to be determined, but mutant analysis has revealed two types of regulatory mechanisms. Loss of function mutations (LOFs) in the positive regulators lead to reduced SA accumulation, coinciding with an eds phenotype, whereas in the negative regulators, LOFs often give rise to increased SA accumulation associated with an enhanced disease resistance (edr) phenotype.
The positive SA regulator EDS1 represents an important node in the defense networks26 (Fig. 1). EDS1 is a putative lipase that confers basal defense against viral, bacterial and fungal pathogens in addition to being a necessary component required by a subset of plant resistance (R) genes.27–31 EDS1 physically interacts with two putative lipases, PHYTOALEXIN DEFICIENT 4 (PAD4) and SENESCENCE-ASSOCIATED GENE 101 (SAG101), both of which share homology with EDS1.32,33 Several lines of evidence indicate that PAD4 and SAG101 act in separate pathways to transduce EDS1 signaling: (1) expression of PAD4 and SAG101 at the RNA and/or protein levels is EDS1-dependent; (2) the three proteins are not found in the same complex; and (3) double mutant pad4-1sag101 plants exhibit an additive eds phenotype upon pathogen infection, compared with the single mutants.33,34 Like eds1 mutants, pad4-1 is severely impaired in SA accumulation.35 It is possible that the EDS1/PAD4 complex plays a major role in SA-mediated defense, but the role of the EDS1/SAG101 complex in SA-mediated processes awaits further investigation.
Another positive SA regulator, NONRACE-SPECIFIC DISEASE RESISTANCE 1 (NDR1), appears to act independently of EDS1 since the two proteins are differentially required for the function of distinct classes of R proteins.30,36,37 Beyond EDS1 and NDR1, the list of positive SA regulators is rapidly growing; for instance, ACD6, AGD2-like DEFENSE RESPONSE PROTEIN 1 (ALD1), EDS5/SID1, AVRPPHB SUSCEPTIBLE 3 (PBS3)/HOPW1-1-INTERACTING 3 (WIN3), and the MODIFIER OF SNC1 (MOS) proteins are also required for SA accumulation.38–48 The existence of so many SA regulators poses some interesting questions. For instance, are there additional SA regulators that physically interact with EDS1? Do other SA regulators exist in protein complexes that do not involve EDS1? Do any of the other SA regulators function together in the same pathway? And how many different pathways regulate SA accumulation?
To address some of these questions, microarray analysis has been used to explore the hierarchical relationship of SA regulators. The fact that eds1 and pad4 mutants share a similar global gene expression pattern corroborates the idea that these regulators act in the same branch of SA-mediated defense.49 In addition, a mini-microarray that contains 337 defense genes50 was used to analyze expression profiles of several SA mutants, and the results placed the corresponding proteins in the following pathway, ordered upstream to downstream: EDS1/PAD4, NDR1, PBS3, NONEXPRESSOR of PR GENES 1 (NPR1; an ankyrin-repeat protein that transduces SA signaling, as described below), and EDS5/SID2.51 Note that the biosynthetic protein SID2 lies downstream of the other regulators, emphasizing the inter-connectedness of the SA pathway sub-circuits. It is worth mentioning that, due to the small array size, the above ordered pathway should be further tested with alternative methods, such as traditional epitasis analysis. However, a potential problem with such an approach is that knockouts of individual SA positive regulators already exhibit an eds phenotype, and it can be difficult to detect an additive eds phenotype with pathogen infection when two or more mutants are combined in a plant. Therefore, more sensitive assays should be developed to assess the functional relationship between SA positive regulators.
In addition to affecting SA accumulation and disease resistance, mutations in many negative SA regulators are often associated with developmental defects, such as cell death and/or dwarfism. Plants that carry such mutations are called lesion mimic mutants (LMMs).52,53 To gain insights into the function of the corresponding genes, the LMMs are often crossed to mutants defective in genes that positively regulate SA, ET or JA signaling. Analysis of these double or triple mutants indicates that SA plays an important role in regulating phenotypes conferred by many LMMs. In addition, such genetic analyses have also revealed interesting interactions between SA regulators. In some cases, multiple positive SA regulators differentially affect phenotypes caused by a particular LMM. For instance, mutations in EDS1 or PAD4, but not NDR1, completely suppress cell death and disease resistance conferred by lsd1,54 supporting the idea that EDS1 and PAD4 operate in a different pathway from NDR1.30 In other cases, a certain positive SA regulator is differentially required for different LMMs. For instance, the eds1 mutant completely suppresses cell death and disease resistance phenotypes conferred by edr1 and lsd1 but only partially affect vad1 phenotypes,54–56 suggesting that VAD1 function differently from EDR1 and LSD1. Although an understanding of the precise functions of the LMM genes remains elusive, many studies clearly indicate that LMMs can be utilized to genetically dissect the interactions among defense genes.
Indeed, one of the most closely scrutinized LMMs, acd6-1, has proven to be an excellent tool for this sort of genetic analysis. Although wild type ACD6 acts as a positive SA regulator, a gain-of-function mutation renders acd6-1 with hallmarks of the LMMs.43,57 Intriguingly, the extreme dwarfism of acd6-1 is inversely correlated with SA-mediated defense.25 Therefore, the size of acd6-1 can be used in genetic analyses as a convenient phenotypic readout to assess the functional relationships among SA regulators. For instance, Song et al. reported that in the acd6-1 background, two positive SA regulators, ALD1 and PAD4, when mutated, additively affected size and defense, suggesting that these regulators act in separate pathways to affect SA-mediated defense44 (Fig. 1). In addition, the triple mutant acd6-1eds1-2pad4-1 shares similar morphology with the two parental double mutants (Salimian and Lu, unpublished data), yet again reinforcing the idea that EDS1 and PAD4 act in the same pathway. However, one must use caution interpreting the analysis of acd6-1 triple mutants. At least one of the two mutants introduced into the acd6-1 background should be null, since otherwise an observed additive effect could result from quantitative difference in the mutant alleles, rather than the actions of two genes from different pathways. Such caveats notwithstanding, acd6-1 is a powerful tool that should enable systematic dissection of the genetic complexity of SA-mediated defense networks. This analysis can also be extended to study the crosstalk between SA and other defense signaling pathways.
NPR1-Dependent and NPR1-Independent Pathways Transduce SA Signaling
NPR1 represents a key node in SA signal transduction (Fig. 1). npr1 mutants are compromised in disease resistance and insensitive to inducers activating SA signaling.58–61 Conversely, overexpression of Arabidopsis NPR1 or its homologues from tomato, wheat and apple confers broad resistance against diverse bacterial and fungal pathogens.62–65 SA-dependent, NPR1-independent pathways also exist, one of which is mediated by the transcription factor AtWhirly1.66 In addition, genetic analyses of the LMMs have also revealed the existence of additional NPR1-independent pathways, though the molecular identities of the corresponding players are largely unknown.56,57,67–71
NPR1 responds to SA-induced redox changes by undergoing a protein conformation change, which subsequently regulates its cytoplasm-nucleus translocation and activity in gene expression control.72,73 Protein-protein interaction assays revealed that NPR1 interacts with seven out of the ten members of the TGA transcription factor family, in addition to three structurally related NIMIN proteins.74–78 In addition, global gene expression profiling analysis showed that members of the WRKY transcription factor family act downstream of NPR1.79 While most NPR1 interactors or downstream components play a positive role in regulating plant defense, negative regulators, such as TGA2, NIMIN1 and WRKY58, are also involved in relaying NPR1-mediated signaling79–82 (Fig. 1). This complexity is consistent with the multiple roles of NPR1 reported previously, which suggest it not only positively regulates SA signal transduction, but also regulates SA accumulation in both negative and positive ways.25 Different NPR1 functions are probably dependent on input signals from pathogens and plant genetic background, allowing plants the flexibility to integrate intrinsic factors with extrinsic ones in fine-tuning disease resistance.
SA-Mediated Defense Networks are Interconnected
Although it is convenient to think of SA-mediated defense networks in terms of three separate sub-circuits (biosynthesis, accumulation and signaling), emerging evidence indicates that these sub-circuits are intricately interconnected. While multiple genes regulate SA accumulation, expression of some of these genes, ACD6, ALD1, PAD4, EDS1, EDS5 and PBS3/WIN3, are inducible by SA, suggesting a mechanism of signal amplification involving both upstream and downstream components in the SA pathway.29,35,43–47 Consistent with this idea, microarray analysis by Wang et al.51 placed NPR1 both downstream and upstream of several SA regulators. In addition, there also appears to be a negative feedback loop involving NPR1 and SA that keeps the signaling networks in check (Fig. 1). Although the precise molecular functions of many SA regulators are still mysterious, it is conceivable that the non-enzyme SA regulators could directly affect the activities of SA biosynthetic enzymes; alternatively, their effects might be indirect, via the control of precursor availability for SA biosynthesis, or SA stability, sequestration, transport or conjugation. In addition, SA signal transducers might affect expression of both enzyme and non-enzyme SA regulators, besides other defense genes. It should be interesting to tease out which SA regulators act in a SID2- or NPR1-dependent manner, and which ones act in a SID2- or NPR1-independent manner. Further investigation of the SA networks should also reveal additional positive and negative feedback loops, some possibly interlocked with one another, in regulating plant defense.
Conclusions
The framework by which we understand SA-mediated defense networks has become increasingly more complex with the ever-expanding discovery of additional SA pathway components, not to mention interactions among these components. This complexity is augmented by the fact that SA also cross-talks with multiple other defense signals. The complexity of defense networks reveals the capacity of plants to integrate genetic information and development with environmental factors into a tight control of energetically costly defense responses. Studies involving combinatorial approaches, including molecular biology, genetics, biochemistry and global gene expression profiling, will continue to shed light on multifaceted defense signaling networks and provide new perspectives that will aid the combat against pathogens and improve disease resistance traits in crops.
Acknowledgements
I would like to thank Drs. Stephen Miller and Mauricio Bustos at UMBC for critical reading of the manuscript and Dr. Barbara Kunkel at Washington University for helpful discussions on the genetic analysis of acd6-1 triple mutants. I apologize to colleagues whose work was not included. This work was supported by startup funds from UMBC and a grant from the National Science Foundation (NSF RIG-0818651) to H.L.
Abbreviations
- EDS
enhanced disease susceptibility
- ET
ethylene
- JA
jasmonic acid
- LMM
lesion mimic mutant
- SA
salicylic acid
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/9173
References
- 1.Hammond-Kosack KE, Jones JD. Resistance gene-dependent plant defense responses. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008;53:763–775. doi: 10.1111/j.1365-313X.2007.03369.x. [DOI] [PubMed] [Google Scholar]
- 4.Vernooij B, Uknes S, Ward E, Ryals J. Salicylic acid as a signal molecule in plant-pathogen interactions. Curr Opin Cell Biol. 1994;6:275–279. doi: 10.1016/0955-0674(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 5.Sharma YK, Davis KR. The effects of ozone on antioxidant responses in plants. Free Radic Biol Med. 1997;23:480–488. doi: 10.1016/s0891-5849(97)00108-1. [DOI] [PubMed] [Google Scholar]
- 6.White RF. Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology. 1979;99:410–412. doi: 10.1016/0042-6822(79)90019-9. [DOI] [PubMed] [Google Scholar]
- 7.Metraux JP, Ahl-Goy P, Staub T, Speich J, Steinemann A, Ryals J, et al. Induced resistance in cucumber in response to 2,6-dichloroisonicotinic acid and pathogens. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. [Google Scholar]
- 8.Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, et al. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- 9.Gorlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, et al. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 1996;8:629–643. doi: 10.1105/tpc.8.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 11.Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol. 2007;17:1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 12.de Torres Zabala M, Bennett MH, Truman WH, Grant MR. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03875.x. [DOI] [PubMed] [Google Scholar]
- 13.Koornneef A, Pieterse CM. Cross talk in defense signaling. Plant Physiol. 2008;146:839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feys BJ, Parker JE. Interplay of signaling pathways in plant disease resistance. Trends Genet. 2000;16:449–455. doi: 10.1016/s0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 16.Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 18.Nawrath C, Metraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 20.Mauch F, Mauch-Mani B, Gaille C, Kull B, Haas D, Reimmann C. Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J. 2001;25:67–77. doi: 10.1046/j.1365-313x.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- 21.Verberne MC, Verpoorte R, Bol JF, Mercado-Blanco J, Linthorst HJ. Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat Biotechnol. 2000;18:779–783. doi: 10.1038/77347. [DOI] [PubMed] [Google Scholar]
- 22.Yalpani N, Leon J, Lawton MA, Raskin I. Pathway of Salicylic Acid Biosynthesis in Healthy and Virus-Inoculated Tobacco. Plant Physiol. 1993;103:315–321. doi: 10.1104/pp.103.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribnicky DM, Shulaev VV, Raskin II. Intermediates of salicylic acid biosynthesis in tobacco. Plant Physiol. 1998;118:565–572. doi: 10.1104/pp.118.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong J, Pierrel MA, Atanassova R, Werck-Reichhart D, Fritig B, Saindrenan P. Free and conjugated benzoic acid in tobacco plants and cell cultures. Induced accumulation upon elicitation of defense responses and role as salicylic acid precursors. Plant Physiol. 2001;125:318–328. doi: 10.1104/pp.125.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu H, Salimian S, Gamelin E, Wang G, Fedorowski J, LaCourse W, et al. Genetic analysis of acd6-1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J. 2009;58:401–412. doi: 10.1111/j.1365-313X.2009.03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiermer M, Feys BJ, Parker JE. Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol. 2005;8:383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 1996;8:2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandra-Shekara AC, Navarre D, Kachroo A, Kang HG, Klessig D, Kachroo P. Signaling requirements and role of salicylic acid in HRT- and rrt-mediated resistance to turnip crinkle virus in Arabidopsis. Plant J. 2004;40:647–659. doi: 10.1111/j.1365-313X.2004.02241.x. [DOI] [PubMed] [Google Scholar]
- 29.Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao S, Calis O, Patrick E, Zhang G, Charoenwattana P, Muskett P, et al. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 2005;42:95–110. doi: 10.1111/j.1365-313X.2005.02356.x. [DOI] [PubMed] [Google Scholar]
- 32.Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, et al. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell. 2005;17:2601–2613. doi: 10.1105/tpc.105.033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 35.Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science. 1997;278:1963–1965. doi: 10.1126/science.278.5345.1963. [DOI] [PubMed] [Google Scholar]
- 37.Shapiro AD, Zhang C. The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 2001;127:1089–1101. [PMC free article] [PubMed] [Google Scholar]
- 38.Goritschnig S, Zhang Y, Li X. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 2007;49:540–551. doi: 10.1111/j.1365-313X.2006.02978.x. [DOI] [PubMed] [Google Scholar]
- 39.Palma K, Zhao Q, Cheng YT, Bi D, Monaghan J, Cheng W, et al. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 2007;21:1484–1493. doi: 10.1101/gad.1559607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Cheng YT, Bi D, Palma K, Li X. MOS2, a protein containing G-patch and KOW motifs, is essential for innate immunity in Arabidopsis thaliana. Curr Biol. 2005;15:1936–1942. doi: 10.1016/j.cub.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 42.Palma K, Zhang Y, Li X. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol. 2005;15:1129–1135. doi: 10.1016/j.cub.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Lu H, Rate DN, Song JT, Greenberg JT. ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell. 2003;15:2408–2420. doi: 10.1105/tpc.015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song JT, Lu H, McDowell JM, Greenberg JT. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 2004;40:200–212. doi: 10.1111/j.1365-313X.2004.02200.x. [DOI] [PubMed] [Google Scholar]
- 45.Nawrath C, Heck S, Parinthawong N, Metraux JP. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell. 2002;14:275–286. doi: 10.1105/tpc.010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee MW, Lu H, Jung HW, Greenberg JT. A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. Submitted 2007. [DOI] [PubMed]
- 47.Jagadeeswaran G, Raina S, Acharya BR, Maqbool SB, Mosher SL, Appel HM, et al. Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 2007;51:234–246. doi: 10.1111/j.1365-313X.2007.03130.x. [DOI] [PubMed] [Google Scholar]
- 48.Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L, Wildermuth MC, et al. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 2007;144:1144–1156. doi: 10.1104/pp.107.097691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato M, Mitra RM, Coller J, Wang D, Spivey NW, Dewdney J, et al. A high-performance, small-scale microarray for expression profiling of many samples in Arabidopsis-pathogen studies. Plant J. 2007;49:565–577. doi: 10.1111/j.1365-313X.2006.02972.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang L, Mitra RM, Hasselmann KD, Sato M, Lenarz-Wyatt L, Cohen JD, et al. The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: roles of major regulators and the phytotoxin coronatine. Mol Plant Microbe Interact. 2008;21:1408–1420. doi: 10.1094/MPMI-21-11-1408. [DOI] [PubMed] [Google Scholar]
- 52.Lorrain S, Vailleau F, Balague C, Roby D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8:263–271. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- 53.Yoshioka K, Moeder W. Lesion mimic mutants. Plant Signal Behav. 2008;3:1–4. doi: 10.4161/psb.3.10.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rusterucci C, Aviv DH, Holt BF, 3rd, Dangl JL, Parker JE. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell. 2001;13:2211–2224. doi: 10.1105/tpc.010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang D, Christiansen KM, Innes RW. Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiol. 2005;138:1018–1026. doi: 10.1104/pp.105.060400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorrain S, Lin B, Auriac MC, Kroj T, Saindrenan P, Nicole M, et al. Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell. 2004;16:2217–2232. doi: 10.1105/tpc.104.022038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses and cell growth. Plant Cell. 1999;11:1695–1708. doi: 10.1105/tpc.11.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 59.Shah J, Tsui F, Klessig DF. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant-Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- 60.Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner HY, et al. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IkappaB. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 62.Fitzgerald HA, Chern MS, Navarre R, Ronald PC. Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol Plant Microbe Interact. 2004;17:140–151. doi: 10.1094/MPMI.2004.17.2.140. [DOI] [PubMed] [Google Scholar]
- 63.Lin WC, Lu CF, Wu JW, Cheng ML, Lin YM, Yang NS, et al. Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 2004;13:567–581. doi: 10.1007/s11248-004-2375-9. [DOI] [PubMed] [Google Scholar]
- 64.Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah J. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Microbe Interact. 2006;19:123–129. doi: 10.1094/MPMI-19-0123. [DOI] [PubMed] [Google Scholar]
- 65.Malnoy M, Jin Q, Borejsza-Wysocka EE, He SY, Aldwinckle HS. Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus x domestica. Mol Plant Microbe Interact. 2007;20:1568–1580. doi: 10.1094/MPMI-20-12-1568. [DOI] [PubMed] [Google Scholar]
- 66.Desveaux D, Subramaniam R, Despres C, Mess JN, Levesque C, Fobert PR, et al. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell. 2004;6:229–240. doi: 10.1016/s1534-5807(04)00028-0. [DOI] [PubMed] [Google Scholar]
- 67.Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. Roles of salicylic acid, jasmonic acid and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell. 2000;12:2175–2190. doi: 10.1105/tpc.12.11.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jirage D, Zhou N, Cooper B, Clarke JD, Dong X, Glazebrook J. Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J. 2001;26:395–407. doi: 10.1046/j.1365-313x.2001.2641040.x. [DOI] [PubMed] [Google Scholar]
- 70.Tang D, Ade J, Frye CA, Innes RW. Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 2005;44:245–257. doi: 10.1111/j.1365-313X.2005.02523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 72.Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 73.Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Dong X. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science. 2008 doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weigel RR, Bauscher C, Pfitzner AJ, Pfitzner UM. NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol Biol. 2001;46:143–160. doi: 10.1023/a:1010652620115. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, et al. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact. 2000;13:191–202. doi: 10.1094/MPMI.2000.13.2.191. [DOI] [PubMed] [Google Scholar]
- 77.Niggeweg R, Thurow C, Weigel R, Pfitzner U, Gatz C. Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol Biol. 2000;42:775–788. doi: 10.1023/a:1006319113205. [DOI] [PubMed] [Google Scholar]
- 78.Despres C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12:279–290. [PMC free article] [PubMed] [Google Scholar]
- 79.Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2:123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weigel RR, Pfitzner UM, Gatz C. Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell. 2005;17:1279–1291. doi: 10.1105/tpc.104.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kesarwani M, Yoo J, Dong X. Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 2007;144:336–346. doi: 10.1104/pp.106.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fitzgerald HA, Canlas PE, Chern MS, Ronald PC. Alteration of TGA factor activity in rice results in enhanced tolerance to Xanthomonas oryzae pv. oryzae. Plant J. 2005;43:335–347. doi: 10.1111/j.1365-313X.2005.02457.x. [DOI] [PubMed] [Google Scholar]