Abstract
The hypersensitive response (HR) is a form of programmed cell death (PCD) commonly associated with the immune response in plants. HR cell death is often characterized by DNA fragmentation, loss of plasma membrane integrity, protein degradation and typical morphological changes such as plasma membrane shrinkage and nuclear condensation. Initiation of HR cell death requires de novo protein synthesis, suggesting that HR cell death induction involves a transcriptional network regulated by a key factor. We recently identified the OsNAC4 gene, which encodes a plant-specific transcription factor that exhibited rapid but transient transcriptional activation during the early stages of HR cell death. Overexpression of OsNAC4 in rice plants induced cell death accompanied by all characteristics of HR cell death: DNA fragmentation, loss of plasma membrane integrity, and protein degradation. In OsNAC4 RNAi knock-down lines exposed to an avirulent bacterial strain, the cellular response was characterized by a marked decrease in HR cell death compared to wild-type rice cells. Gene expression profiling, which compared rice cells and OsNAC4 knock-down transformants using a rice cDNA microarray, demonstrated that OsNAC4 controls the transcription of at least 139 genes including OsHSP90, involved in loss of plasma membrane integrity, and IREN, which encodes novel plant nuclease involved in cleavage of nuclear DNA. Here we report that although OsNAC4 overexpression caused rapid protein degradation during HR cell death, neither IREN nor OsHSP90 were involved. Thus, three important processes that accompany HR cell death are regulated by independent signaling pathways that are collectively induced by OsNAC4.
Key words: programmed cell death, plant immune responses, hypersensitive cell death, cultured rice cells, transcriptional network
Plants respond to infection using a two-branched innate immune system that shares features with animal innate immunity.1 The first branch, referred to as the pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), involves the recognition of PAMPs (conserved microbial factors) and subsequent induction of basal defenses.2,3 The second branch involves molecular recognition of virulence factors, either directly or through their effects on host targets. A class of nucleotide binding leucine-rich repeat (NB-LRR) resistance R proteins has been implicated in this layer of resistance, which is referred to as effector-triggered immunity (ETI).3 This type of immunity is also known as pathogen race-host plant cultivar-specific disease resistance. ETI seems to be an accelerated and amplified PTI response and is often associated with a type of cell death that is referred to as hypersensitive response (HR).3
HR cell death is often accompanied by 180-bp nucleosomal DNA laddering, cell morphological changes such as plasma membrane shrinkage and nuclear condensation, loss of plasma membrane integrity and protein degradation.4,5 HR cell death results from overexpression of certain transgenes in plants, and its initiation requires de novo protein synthesis, suggesting that HR cell death is induced by a transcriptional network regulated by a key factor.6,7
We previously identified several genes that are upregulated during HR cell death using PCR subtraction analysis and microarray analysis.7,8 Of these, OsNAC4 exhibits rapid and transient transcriptional activation during the early stages of HR cell death. The OsNAC4 gene encodes a plant-specific transcription activation region (TAR) and an NAC domain, which was originally identified in a consensus sequence found in petunia NAM and Arabidopsis ATAF1.9–11 The presence of these domains suggests that OsNAC4 is a plant-specific transcription factor functioning as a component of the cell death pathway conditioning HR. Overexpression of OsNAC4 leads to cell death accompanied by the loss of plasma membrane integrity, nuclear DNA fragmentation, protein degradation and the morphological changes specific to HR cell death. In cell lines in which OsNAC4 levels have been suppressed by RNAi, HR cell death is markedly decreased in response to avirulent bacterial strain.12 Gene expression profiling, comparing rice cells and OsNAC4 knock-down transformants using a rice 22 k oligo microarray, demonstrated that OsNAC4 controls the transcription of at least 139 genes, including OsHSP90 and IREN, which encodes a Ca2+-dependent nuclease. During the induction of HR cell death, OsHSP90 is involved in loss of plasma membrane integrity, while IREN causes cleavage of nuclear DNA, suggesting that these two events occurring during HR cell death are regulated by independent pathways that are both induced by OsNAC4.12
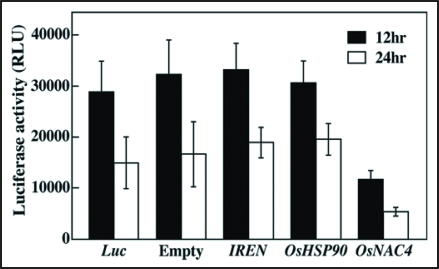
To examine the roles of OsHSP90 and IREN in protein degradation during cell death, we developed a transient assay system in rice protoplasts. Two plasmids were co-introduced into rice protoplasts, one of which encoded the test gene (OsNAC4, OsHSP90 or IREN) and the other contained the Luc gene that encodes firefly luciferase. Both genes were under the control of the constitutive ubiquitin promoter derived from maize.13 If overexpression of the test gene induces protein degradation, then little accumulation of luciferase activity would be observes in the transformed rice due to degradation of the luciferase proteins. If overexpression did not induce protein degradation, the transformed cells would be expected to exhibit high levels of luciferase activity.
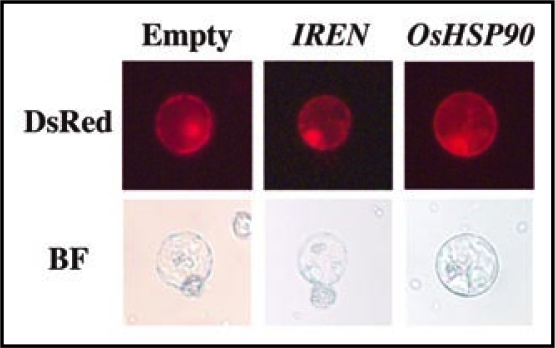
When the Luc vector was co-introduced with an empty vector into rice protoplasts, a high level of accumulated luciferase activity was observed 12 and 24 h after transformation. The accumulation of luciferase activity in OsNAC4 co-introduced cells was dramatically reduced in comparison to that of the empty vector-transformed cells (Fig. 1). Interestingly, co-transformation of IREN or OsHSP90 with the Luc vector caused no reduction of luciferase activity. Similar phenomena were also observed when a vector containing the DsRed gene, which encodes red fluorescence protein downstream of the cauliflower mosaic virus 35S promoter, was used as a monitor for protein degradation. When the IREN or OsHSP90 and DsRed genes were co-introduced into the rice protoplasts using polyethylene glycol (PEG), transformed rice protoplasts exhibited the same level of DsRed fluorescence as was observed in empty vector-introduced cells (Fig. 2). These results indicate that although OsNAC4 overexpression causes rapid protein degradation accompanied by HR cell death, IREN and OsHSP90, which are transcriptionally regulated by OsNAC4, are not involved in protein degradation during OsNAC4-mediated HR cell death.
Figure 1.
Cell death induction by OsNAC4, IREN and OsHSP90. Luc vector co-introduced with empty vector, IREN, OsHSP90 or OsNAC4 into rice protoplasts. After 12 h (solid bars) and 24 h (open bars), luciferase activity was measured by Luciferase assay system kit according to the manufacturer's instructions. Each data point represents the mean and standard deviation of five replicate samples.
Figure 2.
Contribution of OsHSP90 and IREN to the protein degradation accompanied by OsNAC4-mediated HR Cell Death. DsRed vector cointroduced with empty vector, IREN and OsHSP90 into rice protoplasts. After 12 h transformation, DsRed fluorescence was observed at an excitation wavelength of 540 nm and an emission wavelength of 600 nm. BF, bright field image.
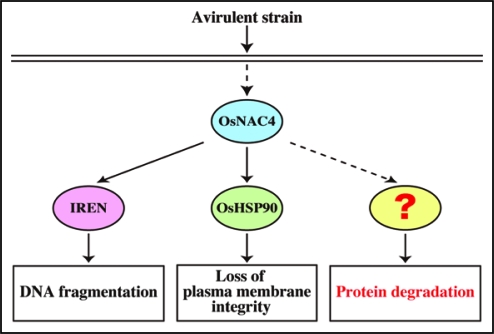
Our results suggest an attractive and simplified model for HR cell death induction. Following pathogen recognition signaling during immune response, OsNAC4 induces the expression of at least 139 genes including OsHSP90 and IREN. The OsHSP90 protein is located in the endoplasmic reticulum (ER) and appears to cause a loss of plasma membrane integrity either directly or indirectly, while IREN is translocated into the nucleus where it cleaves nuclear DNA via its endonuclease activity. Although OsHSP90 and IREN independently regulate two events in HR cell death (loss of plasma membrane integrity and DNA fragmentation), protein degradation, the other important event in HR cell death, is caused by another factor (Fig. 3). The protein degradation factor is assumed to be transcriptionally controlled by the OsNAC4 transcription factor because overexpression of OsNAC4 in plant cells caused rapid protein degradation. Indeed 139 genes that are upregulated by OsNAC4 contain several genes encoding a proteasome such as the F-box protein and ubiquitin. An identification of the protein degradation factor will help further our understanding of the induction mechanism OsNAC4-mediated HR cell death.
Figure 3.
Induction model of DNA fragmentation, loss of plasma membrane integrity, and protein degradation during OsNAC4-mediated HR cell death.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/9087
References
- 1.Nürnberger T, Brunner F, Kemmerling B, Piater L. Links. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 2.Zipfel C, Felix G. Plants and animals: a different taste for microbes? Curr Opin Plant Biol. 2005;8:353–360. doi: 10.1016/j.pbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 4.Che FS, Iwano M, Tanaka N, Takayama S, Minami E, Shibuya N, et al. Biochemical and morphological features of rice cell death induced by Pseudomonas avenae. Plant Cell Physiol. 1999;40:1036–1045. [Google Scholar]
- 5.Tanaka N, Nakajima Y, Kaneda T, Takayama S, Che FS, Isogai A. DNA laddering during hypersensitive cell death in cultured rice cells induced by an incompatible strain of Pseudomonas avenae. Plant Biotech. 2001;18:295–299. [Google Scholar]
- 6.Levine A, Pennell RI, Alvarez ME, Palmer R, Lamb C. Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr Biol. 1996;6:427–437. doi: 10.1016/s0960-9822(02)00510-9. [DOI] [PubMed] [Google Scholar]
- 7.Kaneda T, Fujiwara S, Takai R, Takayama S, Isogai A, Che FS. Identification of genes involved in induction of plant hypersensitive cell death. Plant Biotech. 2007;24:191–200. [Google Scholar]
- 8.Fujiwara S, Tanaka N, Kaneda T, Takayama S, Isogai A, Che FS. Rice cDNA microarraybased gene expression profiling of the response to flagellin perception in cultured rice cells. Mol Plant Microbe Interact. 2004;17:986–998. doi: 10.1094/MPMI.2004.17.9.986. [DOI] [PubMed] [Google Scholar]
- 9.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 10.Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 11.Collinge M, Boller T. Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol. 2001;46:521–529. doi: 10.1023/a:1010639225091. [DOI] [PubMed] [Google Scholar]
- 12.Kaneda K, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, et al. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 2009;28:926–936. doi: 10.1038/emboj.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornejo MJ, Luth D, Blankenship KM, Anderson OD, Blechl AE. Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol Biol. 1993;23:567–581. doi: 10.1007/BF00019304. [DOI] [PubMed] [Google Scholar]