Abstract
Plants adapt to and survive in some of the harshest environments. Their success can be ascribed to an ability to maintain an optimal subcellular redox environment. Peroxisomes, ubiquitous ROS producing and scavenging organelles in eukaryotes play an important role in cellular homeostasis. Recently the formation of thin membrane extensions called peroxules has provided further evidence for peroxisomal role in rapidly sensing and responding to alterations in subcellular ROS. Within a cell the transient extension and retraction of peroxules is asynchronous but takes place within seconds. Peroxules follow tracks defined by tubules of the endoplasmic reticulum and their formation does not appear to involve an elaborate transcriptional-translational machinery. Rather the rapidity of peroxisomal responses suggests ROS instigated membrane modifications aimed at local ROS scavenging or leading to peroxisome elongation prior to their fission for increasing peroxisome numbers within a cell. A model on post-biogenesis peroxisomal life-cycle taking cognizance of rapid peroxisomal responses is presented.
Key words: peroxisome, peroxules, live-imaging, H2O2, endomembrane, ROS
Rapid Peroxisomal Responses to Selective ROS
Recently we have drawn attention to a rapid subcellular response of plants to changes in their redox environment.1 Within seconds of exposure to H2O2 and -OH radicals peroxisomes produce membrane extensions. The morphological similarity between matrixules emanating from miotchondria and stromules extending from plastids has resulted in naming these peroxisomal extensions peroxules.2 Peroxule formation is transient as they characteristically extend and retract, frequently connect with chloroplasts and mitochondria. In general, peroxules may be construed as part of a ROS responsive machinery aimed at relieving subcellular stress created by toxic ROS. However, a further increase in subcellular ROS in cells displaying peroxules leads to the complete elongation of individual peroxisomes into 4–6 µm long tubules with diameters of 0.4 ± 0.2 µm. These elongated tubules subsequently break up into smaller peroxisomes.1 Thus peroxule formation and elongation of peroxisomes in response to different intensities or duration of the same ROS also points to differential subcellular responses to ROS. Clearly a threshold sensing mechanism exists for determining whether the amount of subcellular ROS within a cell can be taken care of locally or if more global measures involving the entire cell or even a group of cells are required. Higher stress levels usually result in an increase in peroxisome numbers.
The rapid activity of the peroxisomal ROS sensing and response machinery creates new questions on the mechanisms that might be involved in the phenomenon. Presently two major notions are used to explain the peroxisomal life cycle in eukaryotes. The first model considers peroxisomes to be of endosymbiont origin and capable of growth and division.3 The second rejects their endosymbiogenic origins and advocates de novo peroxisomal biogenesis from the ER.4,5 Evidence that suggests condition dependent de novo biogenesis as well as growth and division has also been presented in yeast.6 Recently Mullen and Trelease have merged both major concepts to provide a reasonable ‘ER semi-autonomous peroxisome maturation and replication’ model.7 The merged model and its minor modifications8,9 suggest that pre-peroxisomal vesicles born on the ER break off from it into the cytoplasm and undergo homotypic fusion to form early peroxisomes. These nascent peroxisomes import matrix and membrane proteins directly from the cytosol and thus grow in size. Upon receiving an internal cue mature peroxisomes undergo elongation and divide.10 The elongation and division steps are believed to be mediated by membrane proteins belonging to the peroxin11 family, fis1 and dynamin related proteins 3A and 3B (DRP3A/DRP3B).11–13
The consensus model thus considers both de novo biogenesis of peroxisomes and explains their purported semi-autonomy. However, a critical perusal of literature spanning more than 5 decades does not provide any succinct reasons for maintaining the strong belief in peroxisomal independence. On the contrary numerous original reports and reviews point to the possibility of there being connectivity between peroxisomes and the ER.14–16 Notably the generalized models rely largely on yeast and mammalian cells. As Trelease and Lingard point out,17 in plants there is no evidence for de novo biogenesis of peroxisomes from the ER or for the scission and subsequent fusion of pre-peroxisomal vesicles. Similarly, while elongated peroxisomes are observed routinely in diverse organisms the rapidity of peroxisome elongation had not been appreciated so far. In most cases the elongation of peroxisomes has been perceived as ‘growth’ with the ER being loosely pointed to as a source of the required membrane phospholipids and peroxisomal membrane proteins.7,17,18 However, the observation of rapidly extending and retracting peroxules in sub-second intervals defies the notion of acquiring membranes from the ER, assimilating them to form peroxules and then just as quickly discarding them to achieve tubule retraction. Rapid subcellular responses to ROS such as peroxule formation and the tubulation of single peroxisomes requires an alternative explanation.
ROS Mediated Modification of Membranes can Explain Peroxule Formation as well as Rapid Elongation of Peroxisomes
ROS in general, and H2O2 and the more toxic hydroxyl radical in particular alter membrane properties considerably.19 H2O2 is relatively more stable in vivo as compared to other ROS molecules with its half-life being in the order of milliseconds as opposed to micro-seconds for superoxide radicals.20 Membrane protein modifications brought about by H2O2 include an effect on disulfide bonding of proteins in a concentration dependent effect.21,22 H2O2 can contribute to the carbonylation of proteins, oxidation of methionine residues and thiol groups of cysteines. The oxidation of the sulfahydryl group (-SH) of one cysteine molecule can lead to a sulfenic (-SOH), sulfinic (-SO2H) or sulfonic (-SO3H) derivative. This can change the enzyme activity of a protein or the binding capacity of a transcription factor.21,23 Thus the rapid changes in peroxisome morphology observed during peroxule formation or peroxisome elongation might reflect the direct effects of H2O2 stress on membrane architecture. Indeed dilation of tubules making up the endoplasmic reticulum has been reported as a transient effect of increase in subcellular H2O224 and a high degree of membrane protein conservation exists between peroxisomes and the ER.25 Moreover, an indication of peroxisome-ER connectivity is seen in our observations of peroxules tracking over ER tubules in a complete reflection of ER dynamics.1 An interpretation resulting from our observations that takes the rapidity of peroxisomal response to ROS into account is summarized here in a model (Fig. 1). A key feature of our model based on peroxisomal behaviour in plant cells is that it discards the view of independent peroxisomes. On the contrary it advocates peroxisome-ER connectivity.1 As suggested by our observations peroxisomes can be generated or absorbed, can grow by protein import, can send out peroxules along ER defined paths or elongate rapidly and break-up by membrane fission without there being a need for them to be free of the ER.
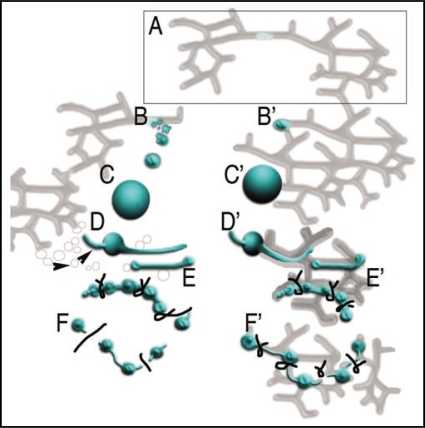
Figure 1.
Model depicting contemporary views4–9 on the post-biogenesis phase of peroxisomal life cycle and its modification based on our observations.1 Peroxisome biogenesis (box A) has been maintained in the ER in accordance with compelling evidence from different organisms.4–8 Pathway (B–F) depicts a contemporaneous consensus viewpoint based on.5–13 The (B′–F′) portion of the model presents a modified viewpoint that takes rapid peroxisomal and ER responses to ROS.1,24 Notably peroxisomes, post-biogenesis, are considered independent in contemporary views whereas our observations refute their independence and advocate post-biogenesis peroxisome ER connectivity. Pre-peroxisomal vesicles (B) undergo fusion to create a proper peroxisome. The view is based largely on the yeast model8,9 and no evidence for pre-peroxisomal vesicles, their scission or subsequent fusion to produce a peroxisome exists in plants.17 Nevertheless in both viewpoints (C and C′) peroxisomes are able to grow in size by importing membrane and matrix proteins from the cytosol. Contemporary models do not tackle the peroxule formation but peroxisomal elongation is known to precede their fission into smaller peroxisomes (D and D′). Whereas (D) depicts elongation to involve ER-derived vesicle-mediated growth7 (arrowheads point to ER derived vesicles) our observations1 supported by independent studies24,25 suggest that peroxule formation and peroxisome elongation (D′) are both transient phenomenon. As depicted in (D′–E′) peroxisome and ER dilation occur due to ROS instigated membrane modifications. The fission of elongated peroxisomes is believed to be mediated by DRP3 proteins.8,9 Accordingly (E) depicts pinching of an elongated peroxisome followed by scission (F) of independent peroxisomes into the cytoplasm. In our viewpoint, there is no evidence to suggest DRP3 role in scission though their tubulation and pinching activity has been demonstrated. Thus (F′) depicts DRP3 activity that serves to pinch membranes and limit continuity between peroxisomal domains and the ER lumen.1 The new model advocating peroxisome-ER connectivity explains peroxisomal functionality as well as its rapid responsiveness to ROS. Peroxisome fission in our model (F′) occurs in tandem with the fission of ER tubules.
Acknowledgements
Funding on Early Intracellular Response Profiling in Plants (EIRPP) by Natural Sciences and Engineering Research Council (NSERC), Canada, The Canadian Foundation for Innovation (CFI) and Ontario Ministry of Research and Innovation is gratefully acknowledged.
Addendum to: Sinclair AM, Trobacher CP, Mathur N, Greenwood JS, Mathur J. Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J. 2009 doi: 10.1111/j.1365-313X.2009.03863.x. In press.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/9232
References
- 1.Sinclair AM, Trobacher CP, Mathur N, Greenwood JS, Mathur J. Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J. 2009;58 doi: 10.1111/j.1365-313X.2009.03863.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Scott I, Sparkes IA, Logan DC. The missing link: inter-organellar connections in mitochondria and peroxisomes? Trends Plant Sci. 2007;12:380–381. doi: 10.1016/j.tplants.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Lazarow PB, Fujiki Y. Biogenesis of Peroxisomes. Ann Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 4.Geuze HJ, Murk JL, Stroobants AK, Griffith JM, Kleijmeer MJ, Koster AJ, et al. Involvement of the endoplasmic reticulum in peroxisome formation. Mol Biol Cell. 2003;14:2900–2907. doi: 10.1091/mbc.E02-11-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullen RT, Trelease RN. The ER-peroxisome connection in plants: Development of the “ER semi-autonomous peroxisome maturation and replication” model for plant peroxisome biogenesis. Biochim Biophys Acta. 2006;1763:1655–1668. doi: 10.1016/j.bbamcr.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Titorenko VI, Mullen RT. Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J Cell Biol. 2006;174:11–17. doi: 10.1083/jcb.200604036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagarasanu A, Fagarasanu M, Rachubinski RA. Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- 10.Guo T, Gregg C, Boukh-Viner T, Kyryakov P, Goldberg A, Bourque S, et al. A signal from inside the peroxisome initiates its division by promoting the remodeling of the peroxisomal membrane. J Cell Biol. 2007;177:289–303. doi: 10.1083/jcb.200609072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingard MJ, Trelease RN. Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J Cell Sci. 2006;119:1961–1972. doi: 10.1242/jcs.02904. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Hu J. Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 2009;57:146–159. doi: 10.1111/j.1365-313X.2008.03677.x. [DOI] [PubMed] [Google Scholar]
- 13.Lingard MJ, Gidda SK, Bingham S, Rothstein SJ, Mullen RT, Trelease RN. Arabidopsis PEROXIN11c-e, FISSION1b and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle—associated replication of peroxisomes. Plant Cell. 2008;20:1567–1585. doi: 10.1105/tpc.107.057679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabak HF, Murk JL, Braakman I, Geuze HJ. Peroxisomes start their life in the endoplasmic reticulum. Traffic. 2003;4:512–518. doi: 10.1034/j.1600-0854.2003.00110.x. [DOI] [PubMed] [Google Scholar]
- 15.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 16.Bernhard JM, Bowser SS. Peroxisome proliferation in Foraminifera inhabiting the Chemocline: an adaptation to reactive oxygen species exposure? J Eukar Microbiol. 2008;55:135–144. doi: 10.1111/j.1550-7408.2008.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trelease RN, Lingard MJ. Participation of the plant ER in peroxisomal biogenesis. In: Robinson DG, editor. Plant Cell Monogr 4. The plant endoplasmic reticulum. Springer Verlag: Berlin Heidelberg; 2006. pp. 205–232. [DOI] [Google Scholar]
- 18.Schrader M, Fahimi HD. The peroxisome: still a mysterious organelle. Histochem Cell Bio. 2008;129:421–440. doi: 10.1007/s00418-008-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magder S. Reactive oxygen species: toxic molecules or spark of life? Crit Care. 2006;10:208. doi: 10.1186/cc3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reth M. Hydrogen peroxide as second messenger in lymphocyte activation. Nat Immunol. 2002;3:1129–1134. doi: 10.1038/ni1202-1129. [DOI] [PubMed] [Google Scholar]
- 21.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. 3rd ed. NY: Oxford Univ Press; 1999. [Google Scholar]
- 22.Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 23.Apel K, Hirt H. Reactive oxygen species metabolism, oxidative stress and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 24.Margittai E, Löw P, Szarka A, Csala M, Benedetti A, Bánhegyi G. Intraluminal hydrogen peroxide induces a permeability change of the endoplasmic reticulum membrane. FEBS Letts. 2008;582:4131–4136. doi: 10.1016/j.febslet.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Schlüter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A. The evolutionary origin of peroxisomes: An ER-peroxisome connection. Mol Biol Evol. 2006;23:838–845. doi: 10.1093/molbev/msj103. [DOI] [PubMed] [Google Scholar]