Abstract
We describe the synthesis and characterization of metal-encoded polystyrene microspheres by multiple-stage dispersion polymerization with diameters on the order of 2 µm and a very narrow size distribution. Different lanthanides were loaded into these microspheres through the addition of a mixture of LnCl3 salts and excess acrylic acid or acetoacetylethyl methacrylate (AAEM) dissolved in ethanol to the reaction after about 10% conversion of styrene, i.e., well after the particle nucleation stage was complete. Individual microspheres contain ca. 106 – 108 chelated lanthanide ions, of either a single element or a mixture of elements. These microspheres were characterized one-by-one utilizing a novel mass cytometer with an inductively coupled plasma (ICP) ionization source and time-of-flight (TOF) mass spectrometry detection. Microspheres containing a range of different metals at different levels of concentration were synthesized to meet the requirements of binary encoding and enumeration encoding protocols. With four different metals at five levels of concentration, we could achieve a variability of 624, and the strategy we report should allow one to obtain much larger variability. To demonstrate the usefulness of element-encoded beads for highly multiplexed immunoassays, we carried out a proof-of-principle model bioassay involving conjugation of mouse IgG to the surface of La and Tm containing particles, and its detection by an anti-mouse IgG bearing a metal-chelating polymer with Pr.
1. INTRODUCTION
One of the most significant challenges in contemporary biotechnology is the simultaneous detection and quantitative determination of multiple biomarkers in a single assay. The goal of these highly multiplexed assays is to be able to extract large amounts of data from smaller samples with increasing efficiency.1–8 A variety of different formats has been proposed for these high-throughput approaches. These include multi-well microtiter plates, modified polymer surfaces (chips), and micrometer-sized polymer beads. Multiplexed bead-based arrays are an attractive option for supporting surface chemistries of immuno-9 and gene expression assays.10 In a manner similar to microtiter plates, various compositions, coatings or conjugation groups can be constructed or added to the microspheres to provide the requisite surface chemistry. These beads are then analyzed individually, often by flow cytometry. Cytometric fluorescent bead-based assays have demonstrated the increased sensitivity, specificity and dynamic range obtainable over standard enzyme immunoassays. 11–14
Traditional flow cytometry is based upon fluorescence or photoluminescence detection.4 Fluorescence refers to the photo-excited emission from typical organic dyes, whereas the more general term photoluminescence incorporates emission from quantum dots and the phosphorescence-like emission from lanthanide chelates. Cytometric assays require two types of markers. The bead itself carries one or more dyes in various levels of concentration that acts as a code for the type of biomolecule attached to its surface. This type of marker is often referred to as a classifier tag, which is the identification marker within the microspheres to indicate its type. In addition, one needs a reporter tag to indicate successful binding of analytes to the particle surface. The reporter tag (also a fluorescent dye or quantum dot) is attached either to the analyte itself, or more commonly, to a secondary reagent, such as an antibody, peptide or other type of biomolecule to provide a signal associated with a successful binding event. For example, the Luminex system15 employs classifier beads containing two dyes at ten levels of concentration, which theoretically allows 100 analytes to be identified by this bead set in one sample. The instrument is a flow cytometer equipped with two lasers, a 635-nm diode laser to excite the red and infrared dyes embedded in the beads, and a 523-nm Nd:YAG laser to excite the orange reporter, pycoerythrin (PE) attached to the reporter molecules.
Using such systems, many successful immuno- and gene expression assays have been reported. For example, Yang et al.16 could quantify gene expression at the level of RNA transcripts by demonstrating the multiplexing of 20 genes with a lower detection limit of 100 attomole. A recently published paper describes the use of a color-coded bead mixture for testing antibody specificity.17 A powerful high-throughput multiplex immunobead assay was used to test simultaneously 29 cytokines, chemokines, angiogenic as well as growth factors, and soluble receptors in the sera of patients diagnosed with high-risk melanoma.18
One of the limitations of photoluminescence-based assays is the limited number of different dyes and different emission intensities that can be read simultaneously. The analysis is complicated because different dyes often have to be excited at different wavelengths. There is also a finite bandwidth to the emission that limits the number of dyes that can be examined simultaneously. Some of these problems can be mitigated by using quantum dots with a very narrow size distribution, leading to narrower emission bands. Quantitative analysis of analyte concentration in a sample is difficult, and it is also a challenge to measure simultaneously intensity ratios that differ by more than a factor of ten.
A much larger amount of information can be coded if one employs different metal atoms or isotopes as labels. The type of metal and the amount present can be measured with high sensitivity by inductively coupled plasma mass spectrometry (ICP-MS).19 In ICP-MS, samples are burned in Ar plasma torch at ca. 7000 K, and in the process, metals are atomized and ionized with quantitative efficiency. Metal ions are detected with unit mass resolution and with a very wide dynamic range employing the quadrupole mass analyzer. Bioassays that are based on metal encoded beads and elemental reporter tags need a detection system capable of registering a fast transient signal individually. The fast data acquisition system should also be able to register at least hundreds transient events per second. These goals can be met with a novel instrument which injects cells or beads stochastically and analyzes them one-by-one. Here the beads are nebulized and injected into the plasma torch of the ICP ion source linked with time-of-flight (TOF) detection.20–22 We refer to this technique as mass cytometry (or FC-MS), and employ a prototype commercial instrument produced by DVS Sciences Inc. (Toronto Canada) designed to collect multiple mass spectra for single-cell or single-particle induced transient ion clouds, typically of 200–300 µs duration. The mass range of the instrument is limited to m/z = 100–220.
In this paper, we describe the synthesis and characterization of metal-encoded polystyrene microspheres designed for high throughput and highly multiplexed bioassays based upon analysis by mass cytometry. The metal-encoded polystyrene (PS) microspheres we describe represent the classifier reagents. Our challenge is to synthesize these microspheres with very narrow size distribution in a way that they contain large (e.g., 107) and uniform numbers of metal ions per bead. We focus on the lanthanide (Ln) series of elements because of the large number of stable commercially available isotopes and similar chemistry that facilitate their incorporation into the beads using a common synthesis methodology.23 Because the lanthanides have low natural abundance, we anticipate very low background signals in our assays. For the reporter tag, which signals a successful binding event, we employ metal chelating polymers described previously by our group,24 attached to an appropriate affinity reagent. The metal chelating polymer (MCP) carries ca. 30 copies of a single-isotope lanthanide such as terbium (169Tb) or praseodymium (141Pr). The reporter tag may carry several copies of the Ln-MCP. This combination opens the possibility that, depending on the number and concentration of metals per bead, assays could be developed utilizing bead sets containing a thousand or even millions of distinguishable codes.
There are a variety of strategies one can envision for obtaining functional polymer microspheres labeled with Ln ions. There are examples in the literature, where the motivation was unconnected to mass spectrometry detection. In experiments directed to Magnetic Resonance Imaging (MRI) enhancement, Paschkunova-Martic et al.25 doubly derivatized commercial carboxylated polystyrene (PS) microspheres (d = 100, 400, 900 nm) with tomato lectin (to target epithelial and endothelial glycocalyceal N-glycans) and with diethylenetriamine penta-acetic acid (DTPA) (as a ligand for Gd3+). The particles were then treated with a solution of a Gd3+-NTA (NTA = nitriloacetic acid) complex to generate contrast enhancement in MRI. Matsuya et al.26 used a solvent swelling method to promote uptake of a Eu(III) chelate by PS microspheres. This group was interested in bioassays based upon photoluminescence detection of Eu3+ emission, in which the Eu3+ was tightly bound to a chromophoric ligand that served as an antenna for enhanced photo-excitation. In our group, Vancaeyzeele et al.19 reported the synthesis of Ln-containing PS nanoparticles by miniemulsion polymerization. These were used for cell uptake by non-specific endocytosis and employed in bulk assays of cell adhesion by ICP-MS. These particles are too small for typical immunoassay protocols. The corresponding FC-MS transient signal intensities were too low for statistically robust encoding.
3. RESULTS AND DISCUSSION
Designing and synthesizing the classifier beads
There are three major requirements for polymer beads to be used in mass cytometric analyses. First, the beads must have a very narrow size distribution. We will characterize the size distribution in terms of the coefficient of variation of the particle diameter (CVd), or more related to encoding, the particle volume (CVV). Second, to be easily classifiable, the beads must have a very small bead-to-bead variation in the content of each lanthanide metal. We will characterize the Ln ion content in terms of its coefficient of variation CVLn. It is the magnitude of CVLn that will limit the ability of FC-MS to classify beads using different levels of concentration of various metals or isotopes in the beads. We expect that CVLn ≥ CVV. Finally, each microsphere must contain a sufficient number of ions for robust detection using counting statistics as a criterion. The transmission efficiency of FC-MS is on the order of 10−4, which means that on the order of only 1 in 104 ions generated in the plasma reaches the detector. This is balanced by a noise level of only 0.01 counts per second, meaning that a signal of several counts per bead recorded over a time frame less than 1 ms is significant. Thus the target for ion incorporation is between several times 104 to 108 Ln ions per particle.
There are two other important requirements. First, the particles should have mean diameters on the order of 1 to 5 µm. The size requirement is dictated in part by typical immunostaining protocols, where centrifugation and redispersion steps are used repeatedly. Smaller particles also may not contain sufficient numbers of metal ions for our target range of codes, and larger particles may not burn completely in the ICP ion source. In addition, these microspheres must have appropriate surface functionality for the covalent attachment of proteins such as antibodies or oligonucleotides, with minimal tendency for non-specific adsorption.
To meet these requirements, we chose two-stage dispersion polymerization of styrene in ethanol in the presence of polyvinylpyrrolidone (PVP).27 It is widely recognized that dispersion polymerization of styrene in ethanol leads to particles of the proper size range with an exceptionally narrow size distribution. It is relatively easy to obtain particles with CVd ≈ 3% (corresponding to CVV ≈ 10%), and with care, even narrower distributions can be obtained.28 One of the difficulties with this method is that the introduction of functional co-monomers (“problematic reagents”) can lead to loss of control over particle size and size distribution.29, 30 Several years ago, Song et al.31 showed that many of these problems could be avoided if the addition of the co-monomers was delayed until the particle nucleation step was complete (a few percent monomer conversion). He referred to this method as “two-stage” dispersion polymerization (which we denote as 2-DisP). Thus in our design, we initiate dispersion polymerization of styrene in ethanol, and after approximately 10% monomer conversion, add known amounts of LnCl3 in ethanol in the presence of an excess of a co-monomer that can serve as a ligand for Ln3+. We examine two different ligands: acetoacetylethyl methacrylate (AAEM), a β-ketoester, and acrylic acid (AA). AAEM was previously used as a chelating ligand for zirconium propoxide to incorporate it into hybrid nanoparticles through a combination of a sol–gel reaction and emulsifier free emulsion polymerization.32–35 The carboxylate group of AA is known to interact strongly with lanthanide ions.
The reagents used in these 2-DisP reactions are presented in Table 1, and the type and amounts of lanthanide salts employed are collected in Table 2. Experimental details are reported in Supporting Information. The characteristics of the particle samples synthesized are collected in Table 3.
Table 1.
Recipes for the dispersion polymerization of styrene with PVP55 and acrylic acid in ethanol
| Two-stage reactions | Three-stage reactions | ||||
|---|---|---|---|---|---|
| Materials (g added) | 1st stage | 2nd stage | 1st stage | 2nd stage | 3rd stage |
| Styrene | 6.25 | --a | 6.25 | -- | -- |
| PVP55 b | 1.0 | -- | 1.0 | -- | -- |
| TX305 c | 0.35 | -- | 0.35 | -- | -- |
| AMBN d | 0.25 | -- | 0.25 | -- | -- |
| Ethanol | 18.75 | 18.75 | 18.75 | 10.0 | 10.0 |
| Acrylic acide | -- | 0.125e | -- | 0.125 | 0.25 |
| LnCl3 | -- | f | -- | f | -- |
| EGDMA | -- | -- | -- | -- | 0.063 |
In the synthesis of samples AA050, AA068 and AA069, 6.25 g styrene were added during the second stage.
polyvinylpyrrolidone, M ≈ 55 kDa.
Triton X-305
azomethylbutyronitrile
In the synthesis of samples AA050 and AA068, AAEM was used in place of acrylic acid: 0.25 g (4 wt%/sty) AAEM for AA050 and 0.50 g (8 wt%/sty) AAEM for AA068.
Some combination of LaCl3.6H20, TbCl3.6H20, HoCl3.6H20, and TmCl3.6H20. for specific amounts, see Table 2.
TABLE 2.
Lanthanide contenta of the reaction mixture for synthesis of PS-PAA microspheres (2 wt % AA/styrene)
| Sample | Laa | Tma | Hoa | Tba | Eua | Ln totala |
|---|---|---|---|---|---|---|
| AA050 | -- | -- | -- | -- | 1.0 | 1.0 |
| AA068 | -- | -- | -- | -- | 1.0 | 1.0 |
| AA069 | -- | -- | -- | -- | 1.0 | 1.0 |
| AA087 | 0.10 | 0.10 | 0.10 | 0.10 | -- | 0.40 |
| AA088b | 0.10 | -- | 0.10 | 0.10 | -- | 0.30 |
| AA089c | -- | 0.10 | 0.10 | 0.10 | -- | 0.30 |
| AA093 | 0.2 | 0.2 | 0.2 | 0.2 | -- | 0.8 |
| AA099 | 0.05 | 0.05 | 0.05 | 0.05 | -- | 0.20 |
| AA100 | 0.02 | 0.02 | 0.02 | 0.02 | -- | 0.08 |
| AA105 | 0.10 | 0.10 | -- | -- | -- | 0.20 |
wt % relative to styrene of LnCl3.6H2O;
d = 1.8 ± 0.032 µm, CVD = 1.2 %
d = 1.8 ± 0.041 µm, CVD= 1.3 %
TABLE 3.
Particle size, size distribution, and the variation of Tm intensities for some PS-PAA microspheres synthesized in the presence of LnCl3.6H2O.
| Sample | Lna | Tmb | d ± σdc(µm) | CVd%c | V, σV(µm3)d | CVV%d | CVTm%e | % incorp.f | −COOHg |
|---|---|---|---|---|---|---|---|---|---|
| AA087 | 0.40 | 0.10 | 2.1 ± 0.051 | 1.8 | 4.9 ± 0.3 | 5.5 | 25 | 89 % | 2.1 × 108 |
| AA099 | 0.20 | 0.05 | 1.8 ± 0.026 | 1.1 | 3.1 ± 0.1 | 3.5 | 42 | 88 % | 1.9 × 108 |
| AA100 | 0.08 | 0.02 | 1.6 ± 0.068 | 2.9 | 2.3 ± 0.3 | 9.2 | 54 | 87 % | 2.0 × 108 |
| AA105h | 0.20 | 0.10 | 2.2 ± 0.032 | 1.1 | 5.7 ± 0.2 | 3.7 | 11 | 92 % | 4.3 × 108 |
wt %/styrene of LnCl3.6H2O;
wt % of TmCl3.6H2O based upon styrene used in the particle synthesis, see Table 2.
d = mean diameter; σd = one standard deviation; CVd = coefficient of variation of the diameter.
V = mean particle volume; σV = one standard deviation; CVV refers to the microsphere volume.
CV of 169Tm intensity measured by FC-MS.
% of Tm in the reaction that is incorporated into the microspheres.
Number of −COOH groups/microsphere by titration.
Synthesized by 3-stage dispersion polymerization.
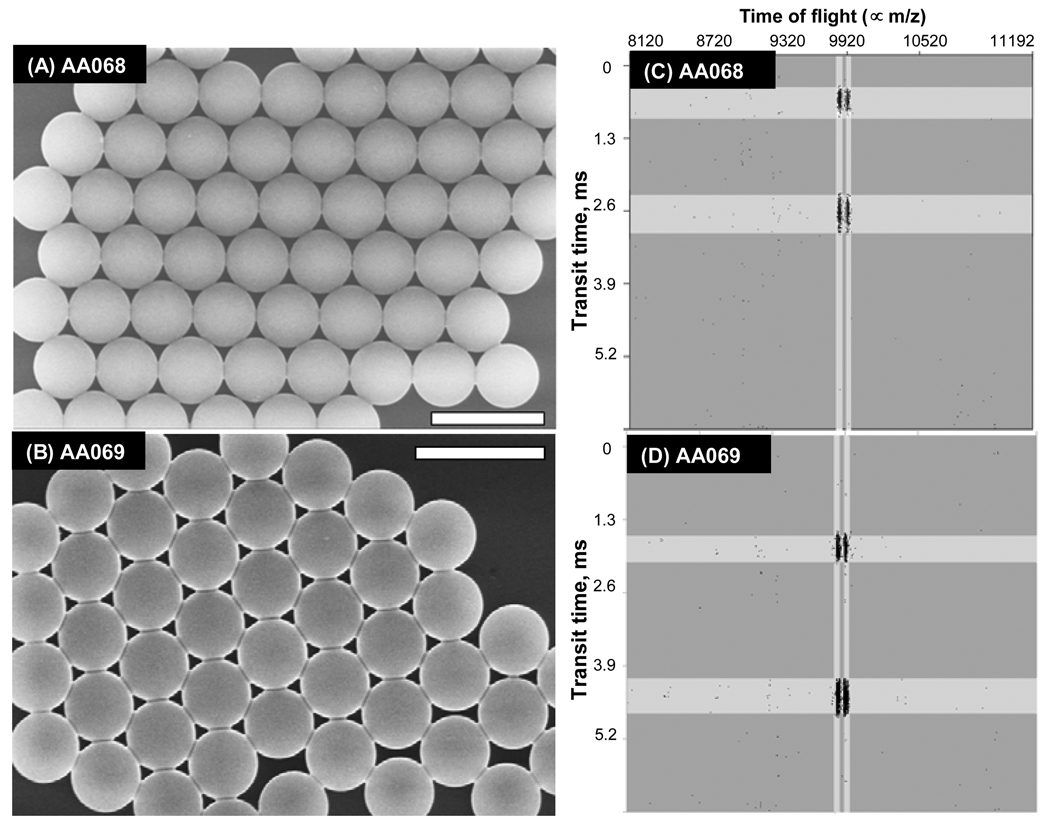
The reactions were clean, and yielded particles in high gravimetric yield, with no coagulum. Figure 1A shows a scanning electron microscope (SEM) image for sample AA068 in which 1.0 wt%/sty EuCl3 and 8.0 wt%/sty AAEM were added in the second stage. For comparison, Figure 1B presents an SEM image for sample AA069 in which 1.0 wt%/sty EuCl3 and 2.0 wt%/sty AA were added in the second stage. The two samples have nearly identical mean diameters (d = 2.7 µm) with a very narrow size distribution (CVd = 1.5 %). Similarly monodisperse microspheres were obtained when 1.0 wt%/sty EuCl3 and 4.0 wt%/sty AAEM were added in the second stage, as shown in Figure S1 of the Supporting Information). We conclude that the 2-Disp reaction works well to provide particles of the proper dimensions and appropriately narrow size distribution.
Figure 1.
SEM images and FC-MS screen captures for PS microsphere samples AA068 and AA069 synthesized in the presence of 1 % EuCl3 added in the second stage with (A and C) AAEM: 8 wt %/styrene (d = 2.7 µm, CVd = 1.5%), and (B and D) AA: 2 wt %/styrene (d = 2.7 µm, CVd = 1.4%). The white horizontal bars in the microphotographs represent 5 µm.
Determining lanthanide content by FC-MS
In this section, we describe how FC-MS is used to determine the bead-by-bead Ln ion content of the microspheres and averaged concentrations for distinguishably encoded populations. The microspheres were washed by several cycles of centrifugation and resuspension in water, and the resultant slurry (ca. 106 microspheres/mL) was nebulized into the mass cytometer sample introduction system, which in turn delivered microspheres individually but stochastically into the inductively coupled plasma torch. The high temperature of the plasma was found to be sufficient to atomize and then ionize the microspheres and the Ln ions embedded in them. The ion stream is then introduced into the time-of-flight mass analyzer. The transient signals corresponding to each microsphere ionization event are recorded by the detector and stored.
We first introduce the raw data to illustrate how the system works. To visualize the data stream, we use a presentation, sequentially stacked mass spectra, employed in LC-MS. Figure 1C shows about 5 ms of data following injection of a sample of AA068 (AAEM). Corresponding information for sample AA069 (AA) is shown in Figure 1D. The instrument captures sequential mass spectra taken at 13 µs intervals. Approximately 1000 microspheres per second were analyzed in this way. Full details are provided in ref 20. The y-axis reports the start time of acquisition of each mass spectrum. The x-axis corresponds to arrival time of ions and is related to the mass of the ions. Columns that correspond to Eu isotopes (151Eu and 153Eu) are highlighted by the vertical lines in the middle of each figure. An encoded bead is registered as a signal in 10 – 30 consecutive spectra over (0.1 – 0.4 ms), and is characterized by simultaneous transient signals for all encoding elements or isotopes. In Figure 1C and D the ion signals that correspond to individual microsphere ionization events are highlighted by horizontal bars. Each sample (AA068 and AA069) shows the signature of two individual particles. The qualitative information presented in Figures 1C and D is used for visualization of the transient signals, for their algorithmic bracketing and decoding. In addition, if free elements were present in the solution, caused, for example, by ion leakage from the beads, a continuous signal very different from the transient signal from individual beads would be observed.
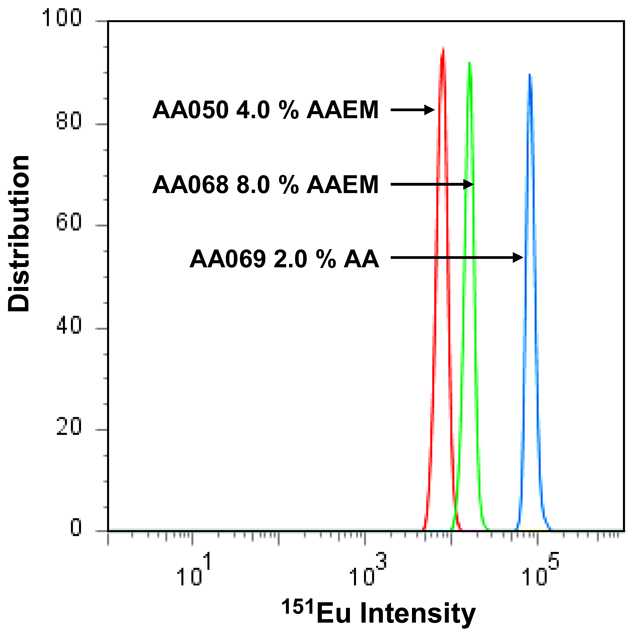
Quantitative information about the bead population can be drawn from a histogram representation of the frequency distribution of signal intensities for individual microspheres. In Figure 2, the population distribution is presented for the 151Eu ion signal collected for 3 min (ca. 4 × 105 microspheres) for three different samples, two containing AAEM and one containing AA. The x-axis of this plot is the “151Eu intensity” analog output of the TOF detector and is considered here as a relative number. The mean 151Eu intensities are 6900 for AA050, 15100 for AA068 and 90600 for AA069. These results indicate an increase of Eu ion content of the microspheres with increasing concentration of the AAEM co-monomer, and more importantly, a much higher Eu content for the AA-containing microspheres. Detailed calculations for the number of metal ions per microsphere are shown in the (Supporting Information Table S1). These data also allow the 151Eu content distribution (CVEu) of the microspheres to be determined.
Figure 2.
Distribution of FC-MS signal intensity for three different populations of PS microspheres prepared in presence of EuCl3 (1.0 wt%/styrene) plus (AA050) AAEM (4.0 wt%/styrene, 151Eu Intensity = 6900), (AA068) AAEM (8.0 % wt%/styrene, 151Eu Intensity = 15100) and (AA069) AA (2.0 wt%/styrene, 151Eu Intensity = 90600).
The higher Eu ion content for the AA-containing microspheres implies that, under our reaction conditions, carboxyl groups are more effective ligands than the acetoacetyl group for incorporating lanthanide ions into PS beads. As a consequence, for subsequent experiments, we focused on particles synthesized in the presence of acrylic acid.
3.2. Microsphere Encoding Protocols: Variability and Dimensionality
In this section, we consider encoding strategies for polymer beads based upon the idea that one can control lanthanide ion content per bead for a variety of lanthanide elements. There are several encoding strategies and for the purpose of this article, we discuss only the binary and enumeration protocols. Binary encoding represents the simplest system. The encoding elements are introduced in two concentrations (levels): 0 and 1. Thus binary encoding of beads with one or more of 10 encoding elements leads to a variability of 210 – 1 >103 distinguishable beads. The general formula is VR = 2N−1, where VR is the variability of a binary encoding system employing N elements. Enumeration encoding is a logical extension of the binary code. In this system, elements are introduced at several concentrations (K levels) with total variability VR = KN−1. This is the highest possible variability for any encoding strategy. For 10 elements and 5 levels of concentration, the variability is VR >>106.
We begin with an examination of binary encoding. Here the task is to synthesize a series of beads containing one or more lanthanide elements and to examine the ability of FC-MS to detect and resolve these elements. Our first set of examples is presented in the first 3 rows in Table 2. Each of the samples was synthesized with 2 wt%/sty in the second stage, along with different combinations of LnCl3 salts. These particles are similar in size (d ≈ 2 µm, CVd = 1.0 – 2.0 %) with a narrow size distribution.
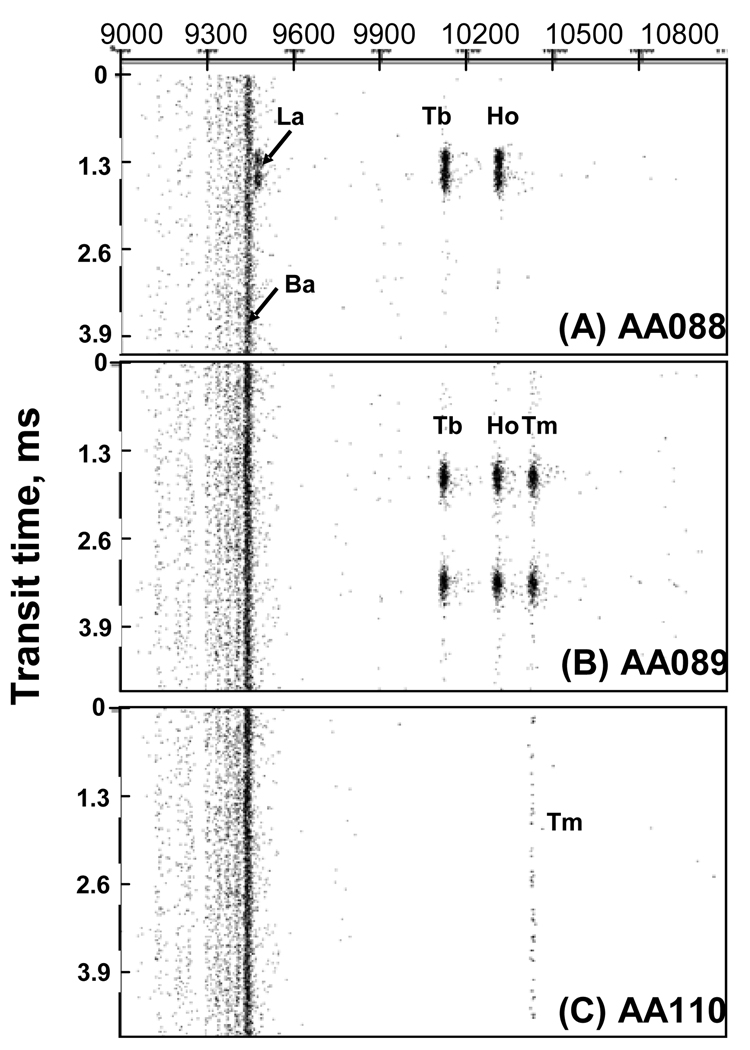
To demonstrate the ability of FC-MS to resolve the presence of multiple Ln ions in a single microsphere, we present data in Figures 3A and B, which represent screen captures of signals from samples AA088 (La, Tb, Ho) and AA089 (Tb, Ho, Tm). The dense vertical lines labeled “La,” “Tb” and “Ho” in Figure 3A (AA088) refer to signals from multiple mass spectra taken during the transit of a single microsphere through the plasma torch. The dark vertical line labeled “Ba” is due to trace amounts of barium in the buffer solution used for the introduction of this sample into the FC-MS instrument. Figure 3B shows signals from two successive microspheres of Sample AA089. One can see clear and distinct signals of the elements Tb, Ho, and Tm, but La is not detected. To test whether Ln ions associate or undergo non-specific binding to the PVP corona surrounding the particles, we synthesized a sample (AA110) in which TmCl3 but no acrylic acid was added in the second stage of the 2-DisP reaction. No signals were detected in the FC-MS analysis of this sample (Figure 3C) that could be associated with individual particles. One can see only a thin streak corresponding to weak traces 169Tm3+ ions present in the solution. These results demonstrate that particles suitable for binary encoding can be synthesized by 2-DisP and read by mass cytometry. A more complete development of a binary encoding algorithm for these particles is provided in the Supporting Information.
Figure 3.
FC-MS screen captures for the analysis of three types of PS microspheres, each containing similar amounts of different Ln metals. (A) sample AA088: AA 2.0 + LaCl3 0.1 + TbCl3 0.1 + HoCl3 0.1; (B) sample AA089: AA 2.0 + TbCl3 0.1 + HoCl3 0.1 + TmCl3 0.1 wt%/styrene. The numbers following each species refers to the wt%/styrene using in the particle synthesis. (C) Sample AA110: a control experiment in which TmCl3 0.1 wt%/styrene and no AA was added in the particle synthesis.
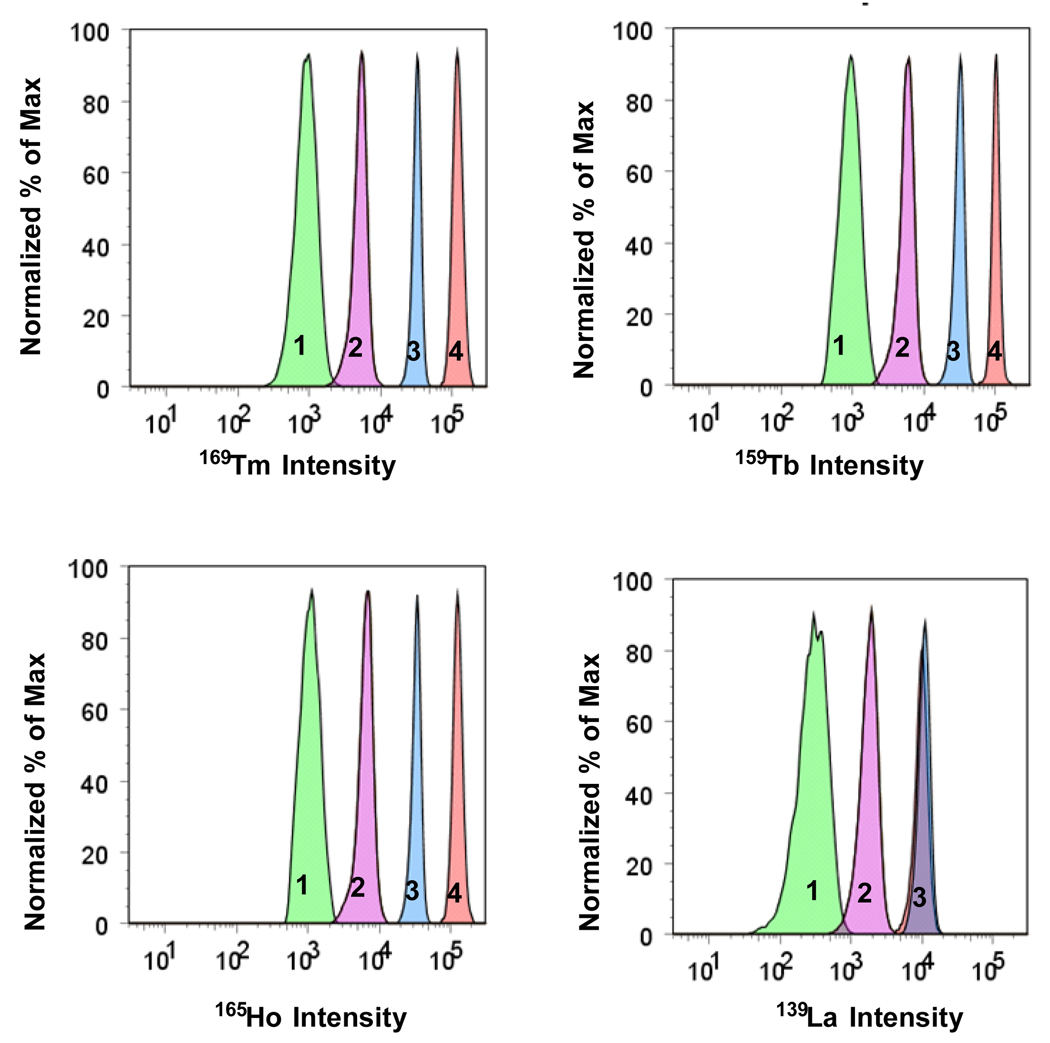
The next objective in building a multiplexed assay by FC-MS detection is to move to enumeration encoding which incorporates a variety of metals and a range of different levels of concentration. To meet this goal, we have to demonstrate the ability of FC-MS to detect and quantify different levels of metal ion concentration. To test this idea, we synthesized four sets of PS microsphere samples using in the synthesis five levels of Ln concentration (0, 0.02, 0.05, 0.10 and 0.20 wt%/sty) introduced in the second stage of the dispersion polymerization reaction. In these reactions, we used four elements (La, Tb, Ho, and Tm). Data for four sets of particles are presented in Figure 4. Here the y-axis represents the normalized distribution of each element. The x-axis reports on the signal intensity on a log scale associated with each element. The panels are grouped by element. The normalized peaks in each panel are labeled “1”, “2”, “3” and “4”, respectively, to indicate that they originate from PS beads prepared in the presence of 0.02, 0.05, 0.10 and 0.20 wt% LnCl3/sty. The label “0” refers to the absence of a Ln element, and, of course, there is no peak in the mass spectrum corresponding to concentration level “0”.
Figure 4.
Distribution of signal intensity for encoding elements for four different populations of microspheres. The microspheres were encoded with four elements (La, Ho, Tb, and Tm) and five levels of concentration (coded from “0” to “4”). For this system of encoding the variability is equal to 624. The green distribution is for “1”,”1”,”1”,”1” (sample AA100, #156) corresponding to La, Ho, Tb, and Tm, respectively. The violet distribution is for microspheres “2”,”2”,”2”,”2” (AA099, #312). The blue distribution is for microspheres “3”,”3”,”3”,”3” (AA087, #468). and the orange distribution is for microspheres “3”,”4”,”4”,”4” (AA093, #499). Note that two samples, AA087 and AA093, both have concentration level “3” of La. The encoding numbers (#156, #312, #468, #499) were assigned using the formalism presented in Supporting Information.
The data in Figure 4, collected for several thousand microspheres, demonstrate essentially baseline resolution in the ability to detect 169Tm, 159Tb, 165Ho, and 139La over a concentration range of three orders of magnitude. Thus the enumeration encoded microspheres can be distinguished successfully with minimal overlap. This example of enumeration encoded microspheres has a variability of 624 (i.e. 54 – 1) and with a larger range of elements or isotopes, opens the possibility to resolve an extremely large number of unique biomarkers. One can assign an encoding number to each type of bead, depending upon the element and its concentration in the particles. These numbers span the variability range from #001 to #624. Using the formalism presented in Supporting Information, and described in detail for binary encoding, the four samples presented in Figure 4 were assigned code numbers of #156, #312, #468, and #499,
3.3. Lanthanide Incorporation, Bead-to-Bead Variability and Surface Functionality
The widths of the peaks in Figure 4 provide information on the bead-to-bead variability of the Ln content of the beads. It appears that the greatest variation is in sample AA100 with the lowest concentration of Ln ions. More quantitative information about this variability is presented in Table 3, where we list values for the coefficient of variation of the Tm content (CVTm) for four PS bead samples. We see that these values are rather large for the first three entries in Table 3, and range from 25% to 54%, and are substantially larger than the variation in volume of the bead samples (CVV). The other elements present in these beads show similar variability.
Other information about these metal-encoded polymer beads is also presented in Table 3. We see that the variation in particle size is small. The first three entries, prepared by 2-DisP, have approximately 2 × 108 titratable carboxyl groups (see Figure S2 in the Supporting Information), which we assume are on the particle surface. These groups are intended for covalent attachment of biomolecules such as antibodies, and this level is comparable to that of commercial beads intended for bioassays. We use the data for Thulium as representative of the incorporation efficiency in the reaction. We see that ca. 88% of the Tm3+ ions introduced as TmCl3 in the reaction are incorporated into the polymer particles.
As we describe below, we had difficulty attaching representative biomolecules to the surface of particles prepared by 2-DisP. As a step to overcome this problem, and also in an attempt to obtain particles with a narrower range of Ln ions per particle, we examined the idea of adding a third stage to the dispersion polymerization reaction. The 3-DisP reaction was carried out as for 2-DisP, but at approximately 60% styrene conversion, additional acrylic acid and a small amount of ethylene glycol dimethacrylate as a cross-linking agent were added to the reaction as a warm solution in ethanol. The recipe is presented in Table 1 and the characteristics of sample AA105 prepared in this way are listed as the fourth entry in Table 3. A scanning electron microscope image of these particles is presented as Figure S5 in Supporting Information. Among the key results for this methodology is that the overall particle size is similar, with a very narrow distribution of sizes. Because of the addition of acrylic acid added in the third stage of the reaction, the number of titratable –COOH groups per microsphere increased by nearly a factor of two compared to microspheres prepared by 2-DisP. In addition, the Ln incorporation efficiency is somewhat improved (92% compared to 88%), but there is a substantial improvement in the variation in lanthanide content per particle. Here CVTm is now 11%, still larger than CVV = 3.7% for this sample. The FC-MS distribution curves for the La and Tm content of this sample are presented in Figure S6 in Supporting Information.
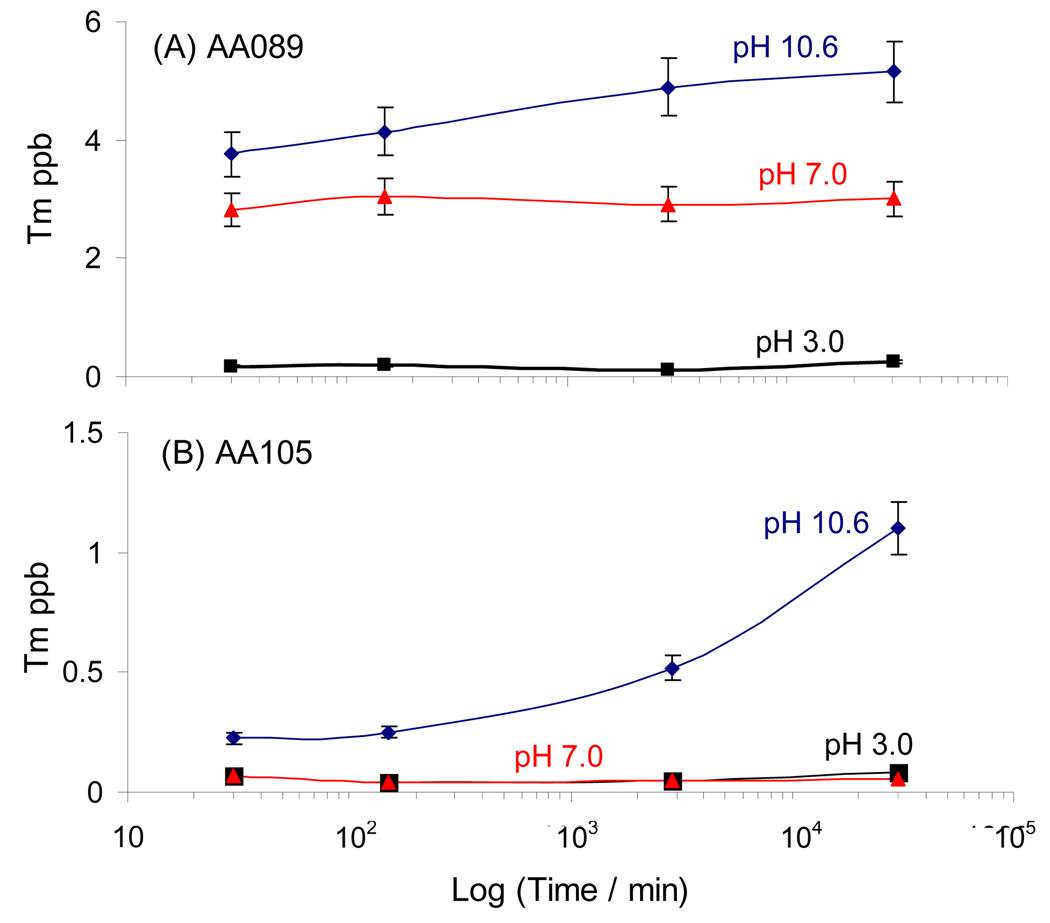
Before we turn our attention to the protein functionalization of the particles, we need to assess the stability of the particles toward leakage of the Ln ions into the aqueous medium, particularly under the experimental conditions associated with attaching biomolecules to the particles. We used traditional ICP-MS to follow the loss of 169Tm ion into the aqueous medium as a function of time. Experiments were carried out on two samples, AA089 prepared by 2-DisP, and sample AA105 prepared by 3-DisP. The results are presented in Figure 5 for particles suspended in three aqueous solutions buffered at pH 3.0, 7.0 and 10.6. At pH 3, there is essentially no detectable leakage of Tm3+ in either sample. At pH 7, the particles prepared by 2-DisP leak a small amount of Tm3+ when placed in buffer, but the amount present in the water does not increase over time. This loss of Tm3+ is significantly reduced for the particles prepared by 3-DisP. Leakage of Tm3+ ions is more significant at high pH, and again is substantially reduced for sample AA105. Note that the time scale in Figure 5 is logarithmic, and the times sampled are very long. After 3 weeks at pH 10.6, sample AA089 had lost only 0.1% of its Tm content, and sample AA105 had lost even less. Thus we conclude that leakage of embedded Ln ions into the aqueous medium is unlikely to be a source of problems in using or functionalizing these particles for bioassays.
Figure 5.
Tm ion release into the aqueous phase from colloidal suspensions of two Tm-containing PS microsphere samples in three different buffer solutions. (A) AA089, synthesized by 2-stage DisP and (B) AA105 synthesized by 3-stage DisP. Both microsphere samples contain 260 ppm Tm ion (w/w styrene). The pH 10.6, pH 7.0 and pH 3.0 buffer solutions are 200mM sodium carbonate/bicarbonate, 10 mM ammonium acetate, and 50 mM sodium acetate, respectively.
3.4. Covalent attachment of proteins to the surface of carboxylated microspheres
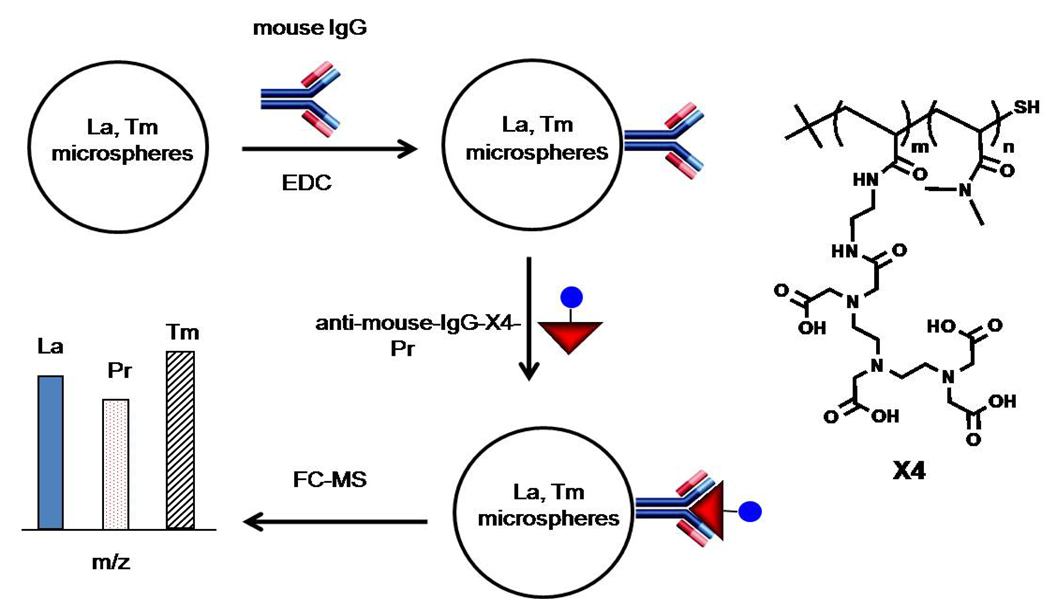
One application of metal encoded microspheres would be in sandwich-type immunoassays. As a first step in this direction, we carried out proof-of-principle experiments involving covalent attachment of proteins to the surface of these carboxylated microspheres. Our experimental design is depicted in Figure 6. In this example, the microspheres, as the classifier beads, are labeled with 139La and 169Tm. These microspheres are configured to capture antigens of interest with antibodies immobilized on the bead (here mouse IgG). The analyte (anti-mouse IgG) is labeled with a metal-chelating polymer (MCP) to act as a reporter tag. In the example, the MCP carries multiple copies of Pr3+ ions. Analysis of a successful binding event by FC-MS would detect the 139La and 169Tm characteristic of the PS microsphere as well as the 141Pr signal from the analyte. In our experiments, we use a MCP (X4, Figure 6) closely related to that described in ref. 24 with different ligands as pendant groups. Identical chemistry was used for attachment of the polymer to the antibody. This type of assay lends itself to multiplexing, in which particles bearing different antibodies are encoded with different metals and different concentrations of these metals. Digital signal processing identifies the unique bead set and quantifies the reporter metal, thus providing information on the amounts and types of analytes in the test sample.
Figure 6.
Schematic representation of antigen capture and detection using metal-encoded microspheres and FC-MS. Carboxylated La,Tm-encoded PS microspheres were conjugated to a mouse IgG using carbodiimide chemistry. Microspheres were then washed and incubated with anti-mouse antigen that is labeled with a Pr-containing polymer tag (anti-mouse-IgG-Pr) to identify the presence of captured antigen on the bead. Stringently washed microspheres were analyzed for concomitant signals of La, Tm, and Pr as an indication of a successful immunoreactions. At the right, we present the chemical structure of the polymer X4 before attachment to the antibody. X4 was reacted with a bismaleimide coupling agent and then covalently attached to the antibody via reaction with –SH groups produced by selective reduction of a disulfide bond in the hinge region of the antibody. Details are given in Ref. 24. Each polymer carries ca. 30 Pr3+ ions.
In experiments carried out on particle sample AA087, we were unable to conjugate significant amounts of mouse IgG to the surface using standard EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide)) coupling conditions.36, 37 Nor were we able to conjugate sufficient FITC-avidin to the beads (using EDC) to detect the fluorescein emission. These experiments point to the interference of the PVP corona surrounding the particles with access of the biomolecules to the -COOH groups at the surface of the particles. These disappointing results with 2-DisP particles were one motivation for us to design the three-stage dispersion polymerization protocol used to synthesize sample AA105. Here we were able to attach FITC-avidin to AA105 particles. Details are provided in Supporting Information. It is likely that the higher titratable –COOH content for these particles facilitates the protein conjugation reaction.
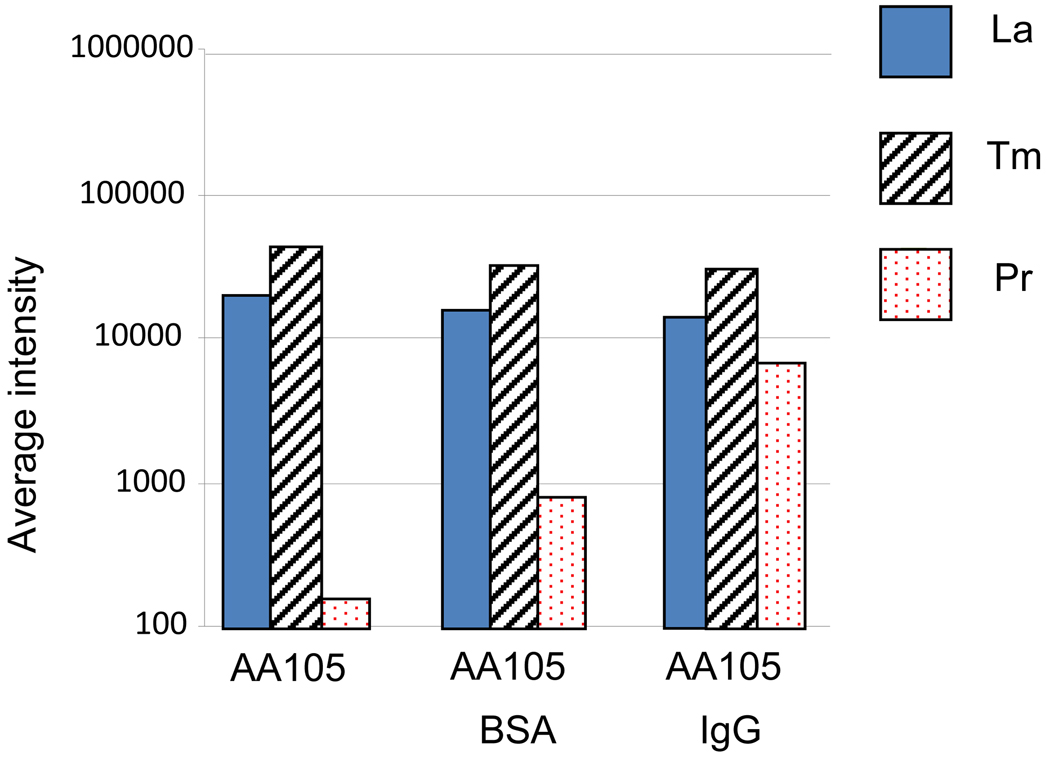
In the next set of experiments, a sample of EDC-activated AA105 beads (labeled with 139La and 169Tm) were treated with mouse IgG or with bovine serum albumin (BSA) in PBS buffer as control for non-specific reporter tag binding. These samples were mixed gently, allowed to react on a shaker for 2 h at 23 °C and then washed. After blocking the microspheres with 0.5% BSA/PBS buffer for 1 h, the samples were incubated with anti-mouse-IgG-X4-Pr (100 µg/mL). After several washes, the samples were analyzed by FC-MS. Between 30,000 and 60,000 beads were analyzed for each sample. The data for this experiment is presented in Figure 7. The left-hand column shows the result for a control experiment in which untreated AA105 beads were exposed to anti-mouse-IgG-X4-Pr (100 µg/mL) and tested for non-specific adsorption. This signal is very weak. The middle column reports on the interaction of anti-mouse-IgG-X4-Pr with the BSA-labeled beads. The right-hand column presents the data for the interaction of antimouse-IgG-X4-Pr with the particles conjugated to mouse IgG. The strong signal here indicates the success of the experiment. The binding of the reporter tag to the metal-encoded beads was measured effectively by FC-MS and yielded a signal two orders of magnitude greater than that due to non-specific adsorption of anti-mouse IgG to the particles, and one order of magnitude stronger than the non-specific interaction of the analyte with BSA conjugated to the particle surface.
Figure 7.
Log intensity values for 139La, 169Tm, and 141Pr from FC-MS measurements for unmodified AA105 particles, BSA-modified AA105 particles and AA105 particles to which anti-mouse-IgG-X4-Pr was covalently attached. These data come from the analysis of approximately 20,000 particles for each sample.
4. SUMMARY
We report the synthesis of a series of lanthanide-containing polystyrene (PS) microspheres with a very narrow size distribution, designed for highly multiplexed bioassays based upon a mass cytometer comprising a time-of-flight (TOF) inductively coupled plasma mass spectrometer. This instrument carries out bead-by-bead analysis at a rate of about 1000 beads per second. Upon entry into the plasma torch, individual beads are atomized in an Ar plasma, ionized, and then analyzed by TOF mass spectrometry. Approximately 10 to 30 mass spectra are taken during the transit time of a bead through the plasma. In bioassays based upon mass cytometry (FC-MS) analysis of bead arrays, the nature of the metals and their individual concentrations within each bead serve as a signature for the type of biomolecule attached to the bead surface.
The PS microsphere samples reported here were synthesized by dispersion polymerization of styrene in ethanol. They contain covalently grafted polyvinylpyrrolidone chains at the surface to suppress non-specific protein adsorption, and carboxylic acid groups for the attachment of biomolecules. They also contain up to 108 Ln ions per particle. Theses ions were introduced at different levels of concentration to meet the needs of an enumeration encoding formalism. For enumeration encoding with metal atoms or isotopes, the variability (VR) of the encoding depends upon the number of different clearly distinguishable levels of concentration (K) of N different elements or isotopes that can be introduced synthetically and detected by FC-MS. The magnitude of the variability is given by the expression VR = KN – 1. The data in Figure 4 show that four levels of finite concentration, varying over three orders of magnitude, for four different elements (La, Tb, Ho, Tm) can be distinguished and analyzed quantitatively. Since encoding encompasses zero concentration of each element, K = 5 and N = 4 for this sample set, and V = 624. FC-MS is capable of analyzing 20 or more lanthanide ions or discrete isotopes simultaneously. Thus extending this methodology to N = 10 and K = 5 should be straight forward (V > 106).
Values of K are limited by our ability to control the particle-to-particle variation of Ln ions. We obtained values of CVLn of about 20% to 25% for particles synthesized by two-stage dispersion polymerization, but closer to 10% for a sample (AA105) in which additional acrylic acid and a cross linking agent were added later in the polymerization reaction.
As a test for protein binding capabilities of these particles, we conjugated three different proteins to aliquots of sample AA105, which was encoded with 139La and 169Tm. FITC-avidin, BSA, and mouse IgG. A very simple model bioassay was constructed by choosing as an analyte anti-mouse IgG labeled with a 141Pr-containing metal chelating polymer. In FC-MS experiments, a strong signal for Pr accompanied the signals for La and Tm associated with mouse IgG conjugated particle. The Pr signal was two orders of magnitude stronger than that due to nonspecific adsorption of the metal chelating polymer onto the AA105 beads themselves and an order of magnitude stronger than that from non-specific adsorption to the BSA-functionalized beads.
We hope that this integrated, massively multiplexed reagent encoding/support system represents the start of research that will simplify and enhance the diagnostic, prognostic and therapeutic efficacy available to physicians and their patients. This will not only increase the effectiveness of healthcare but substantially reduce the human and financial costs of modern personalized treatment.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank NSERC Canada, the National Institutes of Health [NIH grant R01-GM076127], Genome Canada, and the Ontario Genomics Institute for their support of this research.
Footnotes
SUPPORTING INFORMATION
Experimental details, SEM images of particles of samples AA050 and AA105, titration curve for sample AA087, details of the calculation of the number of metal atoms per particle, and the binary encoding library for PS particles containing monoisotopic Ln ions, an integrated mass spectrum showing the resolving power of mass cytometry, the signal intensity distribution for 139La and 169Tm for sample AA105, the FC-MS signals for attempts to conjugate biomolecules to sample AA069, and fluorescence signals for the conjugation of FITC-avidin to AA105 microbeads. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Zheng W, He L. 2009;131:3432–3433. doi: 10.1021/ja809175q. [DOI] [PubMed] [Google Scholar]
- 2.Hardenbol P, et al. Genome Research. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldman ER, Clapp AR, Anderson GP, Uyeda HT, Mauro JM, Medintz IL, Mattoussi H. Analytical Chemistry. 2004;76:684–688. doi: 10.1021/ac035083r. [DOI] [PubMed] [Google Scholar]
- 4.Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. Journal of Microbiological Methods. 2003;53:245–252. doi: 10.1016/s0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- 5.Nolan JP, Sklar LA. Trends in Biotechnology. 2002;20:9–12. doi: 10.1016/s0167-7799(01)01844-3. [DOI] [PubMed] [Google Scholar]
- 6.Han M, Gao X, Su JZ, Nie S. Nature Biotechnology. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 7.Cook EB, Stahl JL, Lowe L, Chen R, Morgan E, Wilson J, Varro R, Chan A, Graziano FM, Barney NP. Journal of Immunological Methods. 2001;254:109–118. doi: 10.1016/s0022-1759(01)00407-0. [DOI] [PubMed] [Google Scholar]
- 8.Brenner S, et al. Nature Biotechnology. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 9.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein JR, Ferguson JA, Lee KH, Walt DR. Journal of the American Chemical Society. 2003;125:13753–13759. doi: 10.1021/ja0365577. [DOI] [PubMed] [Google Scholar]
- 11.Harma H, Soukka T, Lovgren T. Clinical Chemistry. 2001;47:561–568. [PubMed] [Google Scholar]
- 12.Morgan E, Varro R, Sepulveda H, Ember JA, Apgar J, Wilson J, Lowe L, Chen R, Shivraj L, Agadir A, Campos R, Ernst D, Gaur A. Clinical Immunology. 2004;110:252–266. doi: 10.1016/j.clim.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Na K, Kim S, Park K, Kim K, Dae GW, Ick CK, Chung HM, Park KH. Journal of the American Chemical Society. 2007;129:5788–5789. doi: 10.1021/ja067707r. [DOI] [PubMed] [Google Scholar]
- 14.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 15.Luminex Corp. 2009 http://www.luminexcorp.com/products.
- 16.Yang L, Tran DK, Wang X. Genome Research. 2001;11:1888–1898. doi: 10.1101/gr.190901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwenk JM, Lindberg J, Sundberg M, Uhlén M, Nilsson P. Molecular and Cellular Proteomics. 2007;6:125–132. doi: 10.1074/mcp.T600035-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Yurkovetsky ZR, Kirkwood JM, Edington HD, Marrangoni AM, Velikokhatnaya L, Winans MT, Gorelik E, Lokshin AE. Clinical Cancer Research. 2007;13:2422–2428. doi: 10.1158/1078-0432.CCR-06-1805. [DOI] [PubMed] [Google Scholar]
- 19.Vancaeyzeele C, Ornatsky O, Baranov V, Shen L, Abdelrahman A, Winnik MA. Journal of the American Chemical Society. 2007;129:13653–13660. doi: 10.1021/ja073970w. [DOI] [PubMed] [Google Scholar]
- 20.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Analytical Chemistry. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 21.Ornatsky OI, Lou X, Nitz M, Sch نfer S, Sheldrick WS, Baranov VI, Bandura DR, Tanner SD. Analytical Chemistry. 2008;80:2539–2547. doi: 10.1021/ac702128m. [DOI] [PubMed] [Google Scholar]
- 22.Tanner SD, Bandura DR, Ornatsky O, Baranov VI, Nitz M, Winnik MA. Pure and Applied Chemistry. 2008;80:2627–2641. [Google Scholar]
- 23.Kagan HB. Chemical Reviews. 2002;102:1805–1806. [Google Scholar]
- 24.Lou X, Zhang G, Herrera I, Kinach R, Ornatsky O, Baranov V, Nitz M, Winnik MA. Angewandte Chemie - International Edition. 2007;46:6111–6114. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paschkunova-Martic I, Kremser C, Mistlberger K, Shcherbakova N, Dietrich H, Talasz H, Zou Y, Hugl B, Galanski M, So ج ˆlder E, Pfaller K, Ho ج ˆliner I, Buchberger W, Keppler B, Debbage P. Histochemistry and Cell Biology. 2005;123:283–301. doi: 10.1007/s00418-005-0780-7. [DOI] [PubMed] [Google Scholar]
- 26.Matsuya T, Tashiro S, Hoshino N, Shibata N, Nagasaki Y, Kataoka K. Analytical Chemistry. 2003;75:6124–6132. doi: 10.1021/ac034346e. [DOI] [PubMed] [Google Scholar]
- 27.Ober CK, Lok KP, Hair ML. Journal of polymer science. Polymer letters edition. 1985;23:103–108. [Google Scholar]
- 28.Tseng CM, Lu YY, El-Aasser MS, Vanderhoff JW. Journal of Polymer Science, Part A: Polymer Chemistry. 1986;24:2995–3007. [Google Scholar]
- 29.Kim JW, Kim BS, Suh KD. Colloid and Polymer Science. 2000;278:591–594. [Google Scholar]
- 30.Yasuda M, Seki H, Yokoyama H, Ogino H, Ishimi K, Ishikawa H. Macromolecules. 2001;34:3261–3270. [Google Scholar]
- 31.Song JS, Tronc F, Winnik MA. Journal of the American Chemical Society. 2004;126:6562–6563. doi: 10.1021/ja048862d. [DOI] [PubMed] [Google Scholar]
- 32.In M, Gerardin C, Lambard J, Sanchez C. Journal of Sol-gel Science and Technology. 1995;5:101–114. [Google Scholar]
- 33.Pich A, Zhang F, Shen L, Berger S, Ornatsky O, Baranov V, Winnik MA. Small. 2008;4:2171–2175. doi: 10.1002/smll.200801159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez C, In M. Journal of Non-Crystalline Solids. 1992;147–148:1–12. [Google Scholar]
- 35.Wang J, Shi TJ, Jiang XC. Nanoscale Research Letters. 2009;4:240–246. doi: 10.1007/s11671-008-9232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jennings TL, Rahman KS, Fournier-Bidoz S, Chan WCW. Analytical Chemistry. 2008;80:2849–2856. doi: 10.1021/ac7026035. [DOI] [PubMed] [Google Scholar]
- 37.Raez J, Biais DR, Zhang Y, Alvarez-Puebla RA, Bravo-Vasquez JP, Pezacki JP, Fenniri H. Langmuir. 2007;23:6482–6485. doi: 10.1021/la700701x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.