Abstract
The mouse has long been an important model for the study of human genetic disease. Through the application of genetic engineering and mutagenesis techniques, the number of unique mutant mouse models and the amount of phenotypic data describing them are growing exponentially. Describing phenotypes of mutant mice in a computationally useful manner that will facilitate data mining is a major challenge for bioinformatics. Here we describe a tool, the Mammalian Phenotype Ontology (MP), for classifying and organizing phenotypic information related to the mouse and other mammalian species. The MP Ontology has been applied to mouse phenotype descriptions in the Mouse Genome Informatics Database (MGI, http://www.informatics.jax.org/), the Rat Genome Database (RGD, http://rgd.mcw.edu), the Online Mendelian Inheritance in Animals (OMIA, http://omia.angis.org.au/) and elsewhere. Use of this ontology allows comparisons of data from diverse sources, can facilitate comparisons across mammalian species, assists in identifying appropriate experimental disease models, and aids in the discovery of candidate disease genes and molecular signaling pathways.
Keywords: Ontology, Phenotype, Mammal, Annotation, Model System
Phenotype refers to the observable morphological, physiological and behavioral characteristics of an individual in the context of the environment where such characteristics are studied. Many phenotypic characters can change throughout the lifespan of an individual - they can appear or disappear, or increase or decrease in severity. Changing environmental inputs can influence phenotypic characteristics. Phenotypic variation is an expression of genotype, or the sum of an individual’s entire genetic makeup and environmental history (the sum of environmental inputs over the life of the individual). Thousands of human diseases associated with phenotypic and genetic variations have been described, with more underlying genetic causes of disease being discovered continually. In addition, some inherited syndromes characterized clinically may be associated with combinations of different genetic variants in an individual.
The mouse is a significant model organism in the study of human genetic disorders. Mice and humans are physiologically and genetically very similar. Mice are experimentally tractable: they are small and easy to maintain mammals; mice breed quickly and have a short lifespan; and, unlike humans, all life stages, including embryogenesis, are accessible to study. The environment in which mice live can be controlled to carefully introduce exogenous factors, such as diet, drugs, social interactions, or stimuli that may influence phenotype. Importantly, in addition to many spontaneous and induced mouse models available, the genome of mice can be directly and specifically manipulated to introduce exogenous genetic material, reporters, and specific genetic mutations found in human disease.
Information on mouse strains and mutants, and their associated genetic alleles or QTL (quantitative trait loci) has been collected for many years. Tens of thousands of mouse mutations already exist and their phenotypes have been systematically curated in the Mouse Genome Informatics (http://www.informatics.jax.org) database. Hundreds of new mutations continue to be reported each month in the scientific literature. Most mouse phenotype data appear in publications as a combination of textual description, figures, and tables with no standardization, other than adherence to the publishing format. In addition to these published reports, a wealth of data have been or are being generated internationally from large-scale ENU mutagenesis centers [c.f., 10, 14, 17, 22] and the International Mouse Knockout Consortium projects, KOMP (US Knockout Mouse Project), NORCOMM (North American Conditional Mouse Mutagenesis), EUCOMM (European Conditional Mouse Mutagenesis), and TIGM (Texas A&M Institute for Genomic Medicine) [1, 2, 9, 18], much of which will only be available in electronic form. Given the richness and complexity of written language, that by its nature is inherently inconsistent in usage and content, and the avalanche of new data being generated by large-scale projects, there needs to be a standardized, robust way of representing phenotype information in a manner that allows for computational methodologies to fully exploit and realize the potential of these data.
Ontologies are being developed in many domains of biology to facilitate information extraction from literature, aid in scientific discovery and experimental analysis, and to support database interoperability (e.g., Gene Ontology (GO) [6]; Mouse Adult Anatomy (MA) [15]; and Cell Line Ontology (CL) [5]). Ontologies describe the concepts that exist in an area of knowledge and the relationships that exist between these concepts [4]. We have built an ontology describing mammalian phenotype to classify and organize these descriptions for mouse and other mammalian species and for facilitating comparisons across species.
MAMMALIAN PHENOTYPE ONTOLOGY
The description of mouse phenotypes in databases should be captured such that we can compare, combine and analyze biologically relevant information from divergent sources. For example, published reports vary significantly in the level of detail and breadth of data, while high-throughput phenotyping centers thoroughly test a limited number of specific parameters. Phenotype descriptions should be standardized to allow complex queries using other integrated data types such as sequence, genomic location and/or biological function of gene products. Such standard phenotype descriptions must have accurate and precise lexical syntax, yet be applicable to multiple organisms to allow cross-species comparisons. To address this problem, we developed the Mammalian Phenotype Ontology (MP) to describe abnormal mammalian phenotypes. [27].
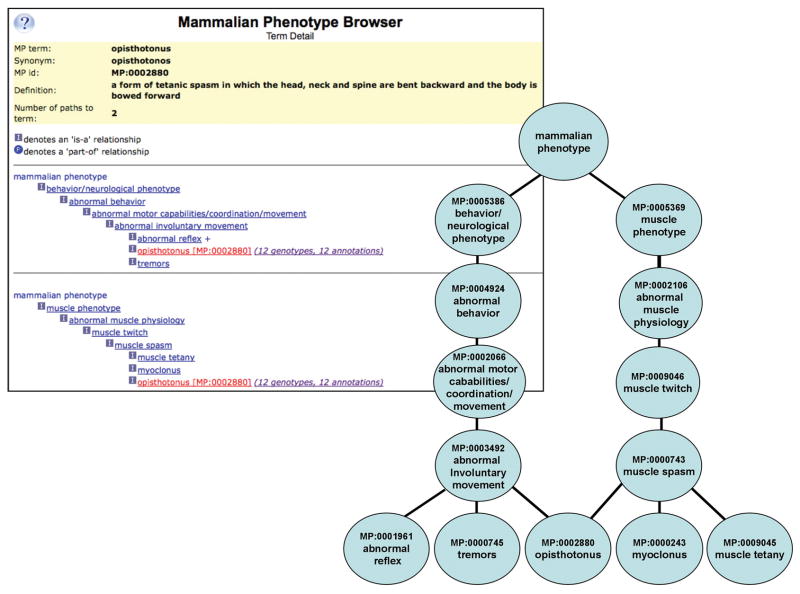
The Mammalian Phenotype Ontology (MP) is a standardized structured vocabulary. MP terms are organized into a directed acyclic graph (DAG) with terms placed hierarchically from the very general at the top of the graph to more specific terms as one moves down the structure. The highest level terms describe physiological systems, survival, and behavior. The physiological systems branch into morphological and physiological phenotype terms at the next node level. A browser is available to view the MP at (http://www.informatics.jax.org/searches/MP_form.shtml) and a sample page is shown in Figure 1. Each vocabulary term includes a unique accession ID, term name, synonyms and definition. The relationship to child nodes and parent nodes is also represented. Any term with more than one parent is shown in multiple sequential graphs.
Figure 1.
Mammalian Phenotype Ontology, showing details for the term “opisthotonus” [MP:0002880]. Left, MGI Browser display shows the term name, common synonyms, the MP ID and the term definition. Each term is followed by a link indicating the number of genotypes and annotations to the term or any children of the term in MGI. The term path is displayed as a hierarchical tree. Note that since there are two paths to the term “opisthotonus”, each path appears as a unique hierarchical tree. Right, a graphical representation for the term “opisthotonus”. The two paths from the term to the top of the ontology can be seen.
The organization of the MP allows annotation of data to the level of resolution available, whether general or highly specific. For example, a published phenotype referred to as “moderate behavioral impairment” may be annotated to the high level term “abnormal behavior” [MP:0004924] while a more detailed description of “opisthotonus” [MP:0002880] could be used if more specific observations or studies had been done. Each term shown in the browser is followed by a hypertext link listing the number of genotype annotations in MGI to each term or to a child of that term (Figure 1, “opisthotonus” has 12 annotations to 12 different genotypes, as of this writing). This hypertext link leads to a list of mouse genotypes annotated in MGI and the annotated term and reference(s) cited for that annotation. This feature allows a narrow or broad search that can return genotypes annotated to detailed phenotypic descriptions or to genotypes that have less well-characterized data.
In MGI, mouse phenotype data are annotated to genotype objects. Each genotype consists of one or more mutant allele pairs combined with associated genetic background strain information. Each MP term annotated to a genotype is supported by one or more references and may also include additional unstructured notes that describe specific comparative details such as incidence, experimental conditions, e.g., diet or stressor, age of onset or quantitative values. Figure 2 shows an example of MP annotations to genotypes involving the Engtm1Mle mutant allele (the first targeted mutation of the endoglin gene made in the Michelle Letarte laboratory). Users can view information about the Engtm1Mle mutant allele from two perspectives on its allele detail page: 1) The phenotype summary section presents a matrix view showing phenotypes (MP Ontology terms) on one axis and genotypes involving Engtm1Mle on the other axis. Using the expansion toggles to view more or fewer specific MP terms, it is easy to compare across genotypes to observe similarities or differences. 2) The phenotype data by genotype section lists each genotype that involves Engtm1Mle that has been phenotypically characterized. Links in the genotype column lead to the greatest detail available for descriptions of that genotype, including MP annotations and additional annotated detail. If an author states that a particular mouse genotype is a model of human disease or syndrome, an association with an Online Mendelian Inheritance in Man (OMIM) human disease term is made, and links to both OMIM and MGI’s Human Disease and Mouse Model Detail pages are provided. In addition, published images highlighting phenotype data are frequently associated with both allele and genotype records. Each of these phenotype sections is organized by the biological system affected and is based on the structure of the MP Ontology. As of 12/20/08, MGI contained 139,751 MP annotations to 27,778 genotypes, representing 20,987 independent alleles, and 8,286 genes and 4,023 QTL (Table 1).
Figure 2.
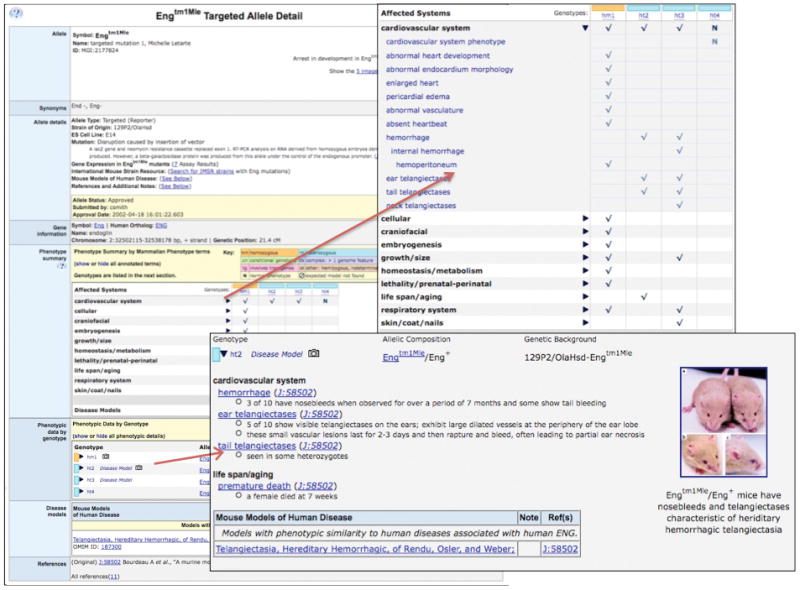
Example of a MGI phenotype web page featuring MP term annotations. The Engtm1Mle (endoglin gene, targeted mutation 1, Michelle Letarte) allele record shows phenotype data organized in two ways. A tabular Phenotype summary lists systems affected by the mutation organized by genotype. Specific systems can be expanded to reveal annotations to more granular terms. The Phenotypic data by genotype section lists the genotypes reported for this mutation, and expansion of this section reveals detailed annotations organized by systems and includes further descriptive annotations and reference data. Note that three cohorts of mice heterozygous for Engtm1Mle on different genetic backgrounds have been phenotypically analyzed. Distinct phenotypes were observed for this mutation on each of these unique genetic backgrounds.
Table 1.
Phenotypes, Alleles & Disease Models at MGI
Contents of phenotype data in MGI as of 12/20/2008. Additions to the MGI database are made daily.
| Records | Phenotype, Allele & Disease Models at MGI |
|---|---|
| 20,987 | Mutant alleles |
| 8,286 | Genes with mutant alleles |
| 27,778 | Genotypes with phenotype annotations |
| 889 | Human diseases with genotypic mouse models |
| 12,493 | Targeted alleles |
| 817 | Cre/Flp expressing Transgenes & Knock-Ins |
| 6,491 | Mammalian Phenotype (MP) ontology terms |
| 139,751 | MP annotations total |
| 4,023 | QTL |
In addition to mouse data, the Mammalian Phenotype Ontology has been used to classify phenotype data from rat and other mammalian species (RGD, OMIA). Species-specific terms are included in the MP. For example, species such as rodents, felines and others exhibit vibrissae while primates do not. In such cases, annotations can be made to vibrissae-related terms when appropriate to a species and ignored when the terms do not apply. The MP also contains related species-specific terms such as the primate-specific term abnormal menstrual cycle [MP:0003375 and the term abnormal estrous cycle [MP:0001927] that is applicable to many other non-primate mammals. By using the parent term abnormal ovulation cycle [MP:0009344] data annotated to this parent term and both of its children could be retrieved from multiple sources.
The MP Ontology, as well as other commonly used biomedical ontologies (e.g., GO, MA, and CL), is maintained using the OBO-Edit software developed by Lewis and Richter [11]. Daily edits include the addition of new term and synonyms, new relationships based on comparisons to orthogonal ontologies and revisions to existing terms. We adhere to Open Biomedical Ontologies (OBO) Foundry principles (http://www.obofoundry.org/about.shtml) in the construction and revision of the ontology, such as orthogonality, public availability and collaborative development [26]. In addition to the MP browser mentioned above, the MP ontology is available in OBO format from the MGI ftp site (ftp://ftp.informatics.jax.org/pub/reports/index.html#pheno) and in OBO and OWL (Web Ontology Language) format from the OBO Foundry site (http://www.obofoundry.org/cgi-bin/detail.cgi?id=mammalian_phenotype).
We continue to refine and expand the MP. Current users of the ontology, including curators from MGI, RGD and the Neuromice Consortium have contributed terms. In addition to curator and community suggestions, we are actively soliciting participation from collaborators and researchers in various fields of specialty to review existing portions of the ontology for accuracy in terminology, completeness and the correct hierarchical structure. We have recently completed a review of the hearing and ear morphology section of the ontology with assistance from Dr. Kenneth Johnson at The Jackson Laboratory and Dr. Karen Steel at the Wellcome Trust Sanger Institute. With this review, 253 new hearing and ear morphology and physiology terms were added, and the associated structural hierarchy for all 387 hearing and ear related terms was reorganized. Review of the cardiovascular and immunology sections of the MP Ontology is underway. A term tracker for all community MP Ontology requests has been established at Sourceforge (https://sourceforge.net/tracker/?atid=1109502&group_id=76834&func=browse). Additional information may be obtained by emailing pheno@informatics.jax.org.
ANNOTATING PHENOTYPE USING A PRECOORDINATED VERSUS A POSTCOORDINATED APPROACH
An alternate method for encoding phenotypic data is the EQ (entity + quality) approach [12,13]. Under this schema, phenotypic data are represented as qualifications (“Q” or attribute) of descriptive nouns or phrases (“E” or entity). For each noun, represented from distinct biomedical ontology such as anatomy ontologies (MA for mouse and others), the cell line ontology (CL) and the GO ontology, there is a set of qualities or attributes represented in the Phenotype and Trait Ontology (PATO, http://www.obofoundry.org/cgi-bin/detail.cgi?id=quality) [12]. These instances are linked together to describe a single phenotypic character, termed the postcoordinated approach.
The advantage of the EQ approach is that existing ontologies are used and standardized in coordination with other vocabularies that provide supporting data for phenotypic information. It is a useful method in high-throughput screens when phenotypic data are collected for many individuals, where phenotyping protocols are tightly controlled, and phenotyping is generally done at a very broad (vs. deep) level. However, data described in publications generally uses precise clinical terminology that describes summary results for cohorts of animals that have been phenotyped at a very deep and precise level. Further, clinical terms are not always easily represented by EQ statements and may require multiple statements to describe them, the sum of which may not accurately reflect the complexity and precision of clinical term’s meaning. Table 2 shows one straightforward example and one complex example of MP terms and how they could be represented in EQ format. The term cataracts [MP:0001304], defined as “complete or partial opacity of the lens,” can be related to the anatomy term lens [MA:0000275, or FMA:58241] and the qualifier opaque [PATO: 0000963]. However, the term jaundice [MP:0000611], defined as “clinical manifestation of hyperbilirubinemia, with deposition of bile pigments in the skin, resulting in yellowish staining of the skin and mucous membranes” needs several EQ statements to describe the phenotype that together do not accurately reflect the biological meaning that “jaundice” imparts to a clinical researcher.
Table 2.
Mammalian Phenotype (MP) Ontology terms and their logical definitions
| MP term | MP Definition | Entity | Quality |
|---|---|---|---|
| cataract MP:0001304 |
complete or partial opacity of the lens | lens MA:0000275 FMA:58241 |
opaque PATO:0000963 |
| jaundice MP:0000611 |
clinical manifestation of hyperbilirubinemia, with deposition of bile pigments in the skin, resulting in yellowish staining of the skin and mucous membranes | skin MA:0000151 FMA:7163 |
yellow PATO:0000324 |
| skin mucous gland MA:0000148 mucous gland FMA:62888 |
yellow PATO:0000324 |
||
| pigment accumulation in tissues GO:0043480 |
yellow PATO:0000324 |
||
| pigment accumulation in tissues GO:0043480 |
mislocalized PATO:0000628 |
The Mammalian Phenotype Ontology uses precoordinated terms that include both the entity and quality concepts. A primary consideration is the use of terms by clinicians and scientists to describe phenotypes and use of terms commonly seen in scientific literature. However, there are two other practical reasons in using the precoordinated approach for mouse phenotype data, both focused on efficiency in the curation process, which is the most costly part of maintaining a database [16]. First, the precoordinated approach provides specific defined terms for curators to use, and thus increases curation consistency. And, second, the EQ approach requires look-up of terms from multiple ontologies for the correct “entity” (and sometimes requesting new terms to be added by ontology providers), thus slowing the curation process.
The EQ term mappings of the Mammalian Phenotype Ontology to other foundational ontology terms (i.e. the logical definitions) may be stored, allowing ontological reasoning and interoperability with other ontological resources such as the GO, FMA, MA, PATO, CL and others. Phenotypic comparisons to data annotated with other precoordinated phenotype ontologies, such as the recently described Human Phenotype Ontology (HPO) [25] can be made if such logical definitions are provided, or by storing term mappings to the HPO directly. Creating logical definitions will also allow comparisons to phenotypic data annotated using the postcoordinated approach. We are assisting in collaboration with others in the preparation of a logical definitions file for the MP (http://www.obofoundry.org/cgi-bin/detail.cgi?id=mammalian_phenotype_xp, unpublished data).
At MGI, we have used the precoordinated MP Ontology to curate specific mutant alleles in genotypes from literature and from user contributions. The MP Ontology is also applicable to high-throughput data from mutagenesis and knockout centers. We have applied this ontology to data from large-scale phenotyping sets generated by Lexicon Genetics and Deltagen (http://www.nih.gov/science/models/mouse/deltagenlexicon/factsheet.html) and provided to the National Institutes of Health. In addition to providing the raw data files (http://www.informatics.jax.org/external/ko/), we have annotated this information with the MP Ontology to make these data integrated and searchable with all other mouse phenotype data in MGI.
USE OF PHENOTYPE ONTOLOGIES FOR DATA RETRIEVAL AND ANALYSIS
Structured phenotype information in MGI is integrated with many other diverse types of data. Phenotype information can be easily retrieved and complex queries can be constructed in a variety of ways.
A summary of mouse phenotype data for any gene at MGI may be viewed on a gene detail page. A gene detail page for Pax6 (paired box gene 6) is shown in Figure 3. In addition to summary information about sequence, genetic location, expression, domain structure, Gene Ontology classifications and references, the phenotype data section contains a hyperlink to a listing of the thirty-two phenotypic alleles of Pax6. A brief description of the phenotypic effects of the mutations in Pax6 is presented. Phenotype images are available for mutations in this gene and the hyperlink leads to a summary page for all images.
Figure 3.
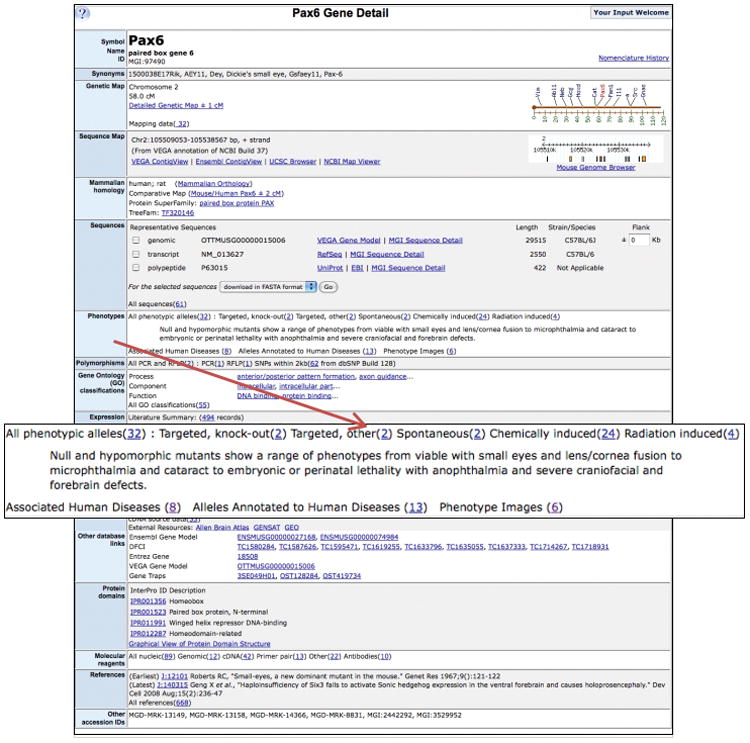
Example of a MGI gene detail page highlighting summary mouse phenotype information. Gene Detail pages contain summary information about the gene with hyperlinks provided to more detailed information. Summary phenotype data includes the number of alleles listed by type of allele, a brief description of the generalized phenotypic effects of many mutations in the gene and links to mouse model of human disease information and to phenotype image data.
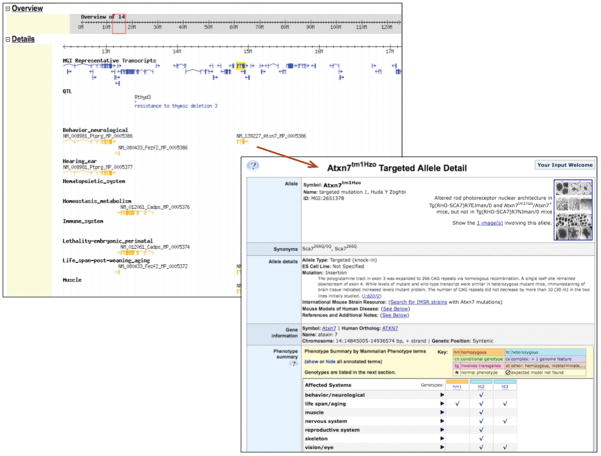
High-level phenotype data can be viewed on the mouse Genome Browser (Figure 4). Specific genome tracks can be viewed representing top-level phenotype terms in the MP Ontology. Annotations to any term in that class will be represented on a separate track in the genome browser. For each icon represented on the track, a link is present that will return a phenotype detail page representing that allele or QTL. Large regions of the genome can be scanned for phenotypes of interest to predict candidate genes for QTL or spontaneous or induced mutations.
Figure 4.
Example of phenotype annotations displayed on the Mouse Genome Browser. Left, a region of chromosome 14 including the Atxn7 gene is shown. Tracks representing high-level phenotypes of genes in this region are shown along with tracks indicating known alleles of the genes. The gene model graphics in the phenotype or allele tracks are hyperlinked to the allele record that describes that mutation, shown on the right.
The MP Ontology and the phenotype annotations in MGI have served the scientific community as an analysis tool and information resource. For example, Martinez-Morales, et al. (2007) used MGI’s mouse phenotype annotations describing neural crest cells and neural crest derivatives to select genes that control differentiation of these structures. Phylogenetic comparison of these genes showed that 9% are unique to vertebrates, much higher than for other tissues. Over half of these unique genes encode soluble ligands that control specification of neural crest cells into derivative lineages [21]. Ramanathan et al. (2008) have used comparative expression profiling from inbred strains with high and low lactation capacity to identify signaling pathways involved in enhanced lactation performance. Many differentially expressed genes identified had phenotypic alleles annotated to Mammalian Phenotype Ontology terms related to maternal performance or mammary-gland development [23]. Reed et al. (2008) analyzed mouse body weight annotations in MGI and showed that body weight is frequently affected in mice carrying knockout alleles. Based on a sample set of 1,977 viable mouse knockout strains, 34% of these were associated with annotations to body weight changes and suggests that thousands of genes may influence this complex trait [24].
Mammalian Phenotype ontology annotations in MGI have also been used for candidate and comparative gene analysis in mouse and human. Bao L, et al. (2007) identified a set of mouse genes with alleles that are known to affect behavioral and neurological phenotypes. Analysis of expression of these genes in recombinant inbred strains identified expression quantitative trait loci (eQTL) that affected expression of members of the gene set. Mapping of these eQTL led to the identification of novel genetic loci and causal genes for a number of behavioral and neurological phenotypes [3]. Chen, et al. describe a method using comparisons to mouse Mammalian Phenotype annotations to prioritize candidates for novel human disease gene identification [8].
Conclusion
The Mammalian Phenotype Ontology is designed to apply phenotype descriptions in a standardized way in mouse and other mammalian species. It is in use at Mouse Genome Informatics (MGI; http://www.informatics.jax.org/) [7], the Rat Genome Database (RGD; http://rgd.mcw.edu/), [28] at Online Mendelian Inheritance in Animals (OMIA; http://omia.angis.org.au/) [19], by several mouse mutagenesis centers [c.f., 14], and by the European Mouse Disease Clinic (EUMODIC; http://www.eumodic.org/) [20] for its Resource of Standardized Phenotype Screens).
MGI offers a number of tools to access and query mouse phenotype data annotated to the MP ontology:
Gene detail pages offer links to all mutations/models for a particular gene (Figure 3).
The MP Ontology Browser provides the ability to find, genome-wide, all annotated alleles that have a particular phenotype (Figure 1).
The Phenotypes, Alleles & Disease Models Query Form allows simultaneous querying of multiple parameters of phenotypes, attributes, and diseases.
The Human Disease Vocabulary can be used to find mouse models of a particular disease.
The Mouse GBrowse (genome browser) visually displays genome tracks for alleles and phenotypes (Figure 4).
The MGI Batch Query allows selection of gene sets and display or download of these with accompanying phenotype and disease annotations.
Direct data access can also be obtained by computational users through data downloads from the MGI FTP site, Web services, or via personal SQL access.
All of these tools and associated tutorials are accessible from the Phenotypes, Alleles & Disease Models section of the MGI database (http://prodwww.informatics.jax.org/mgihome/homepages/phenotypes.shtml).
The MP Ontology can assist in identifying novel models of human disease. By querying standardized and integrated data about phenotypes, expression, biochemical function, sequence and/or genomic location, potential new targets can be identified. These data provide new insights into genetic systems and signaling pathways and will improve our ability to predict and build better animal models for investigating processes involved in human health and disease.
Acknowledgments
We thank Drs. Michael Sasner, Molly Bogue and Susan Bello for helpful comments on the manuscript. This work was supported by grant HG000330 from the National Human Genome Research Institute.
References
- 1.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KC, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, von Melchner H, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B. The knockout mouse project. Nat Genet. 2004 Sep;36(9):921–4. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, Chambon P, Cox R, Davidson D, Davies K, Duboule D, Forejt J, Granucci F, Hastie N, de Angelis MH, Jackson I, Kioussis D, Kollias G, Lathrop M, Lendahl U, Malumbres M, von Melchner H, Müller W, Partanen J, Ricciardi-Castagnoli P, Rigby P, Rosen B, Rosenthal N, Skarnes B, Stewart AF, Thornton J, Tocchini-Valentini G, Wagner E, Wahli W, Wurst W. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004 Sep;36(9):925–7. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao L, Peirce JL, Zhou M, Li H, Goldowitz D, Williams RW, Lu L, Cui Y. An integrative genomics strategy for systematic characterization of genetic loci modulating phenotypes. Hum Mol Genet. 2007 Jun 1;16(11):1381–90. doi: 10.1093/hmg/ddm089. [DOI] [PubMed] [Google Scholar]
- 4.Bard JB, Rhee SY. Ontologies in biology: design, applications and future challenges. Nat Rev Genet. 2004 Mar;5(3):213–22. doi: 10.1038/nrg1295. [DOI] [PubMed] [Google Scholar]
- 5.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6(2):R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blake JA, Harris MA. The Gene Ontology (GO) project: structured vocabularies for molecular biology and their application to genome and expression analysis. Curr Protoc Bioinformatics. 2008 Sep;Chapter 7(Unit 72) doi: 10.1002/0471250953.bi0702s00. [DOI] [PubMed] [Google Scholar]
- 7.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA Mouse Genome Database Group. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008 Jan;36(Database issue):D724–8. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007 Oct 16;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins FS, Finnell RH, Rossant J, Wurst W. A new partner for the international knockout mouse consortium. Cell. 2007 Apr 20;129(2):235. doi: 10.1016/j.cell.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Cordes SP. N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express. Microbiol Mol Biol Rev. 2005 Sep;69(3):426–39. doi: 10.1128/MMBR.69.3.426-439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day-Richter J, Harris MA, Haendel M, Lewis S Gene Ontology OBO-Edit Working Group. OBO-Edit--an ontology editor for biologists. Bioinformatics. 2007 Aug 15;23(16):2198–200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- 12.Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2005;6(1):R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gkoutos GV, Green EC, Mallon AM, Blake A, Greenaway S, Hancock JM, Davidson D. Ontologies for the description of mouse phenotypes. Comp Funct Genomics. 2004;5(6–7):545–51. doi: 10.1002/cfg.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldowitz D, Frankel WN, Takahashi JS, Holtz-Vitaterna M, Bult C, Kibbe WA, Snoddy J, Li Y, Pretel S, Yates J, Swanson DJ. Large-scale mutagenesis of the mouse to understand the genetic bases of nervous system structure and function. Brain Res Mol Brain Res. 2004 Dec 20;132(2):105–15. doi: 10.1016/j.molbrainres.2004.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M. The Adult Mouse Anatomical Dictionary: a tool for annotating and integrating data. Genome Biol. 2005;6(3):R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, St Pierre S, Twigger S, White O, Yon Rhee S. Big data: The future of biocuration. Nature. 2008 Sep 4;455(7209):47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hrabé de Angelis M, Balling R. Large scale ENU screens in the mouse: genetics meets genomics. Mutat Res. 1998 May 25;400(1–2):25–32. doi: 10.1016/s0027-5107(98)00061-x. [DOI] [PubMed] [Google Scholar]
- 18.International Mouse Knockout Consortium. Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007 Jan 12;128(1):9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Lenffer J, Nicholas FW, Castle K, Rao A, Gregory S, Poidinger M, Mailman MD, Ranganathan S. OMIA (Online Mendelian Inheritance in Animals): an enhanced platform and integration into the Entrez search interface at NCBI. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D599–601. doi: 10.1093/nar/gkj152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallon AM, Blake A, Hancock JM. EuroPhenome and EMPReSS: online mouse phenotyping resource. Nucleic Acids Res. 2008 Jan;36(Database issue):D715–8. doi: 10.1093/nar/gkm728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Morales JR, Henrich T, Ramialison M, Wittbrodt J. New genes in the evolution of the neural crest differentiation program. Genome Biol. 2007;8(3):R36. doi: 10.1186/gb-2007-8-3-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadeau JH, Balling R, Barsh G, Beier D, Brown SD, Bucan M, Camper S, Carlson G, Copeland N, Eppig J, Fletcher C, Frankel WN, Ganten D, Goldowitz D, Goodnow C, Guenet JL, Hicks G, Hrabe de Angelis M, Jackson I, Jacob HJ, Jenkins N, Johnson D, Justice M, Kay S, Kingsley D, Lehrach H, Magnuson T, Meisler M, Poustka A, Rinchik EM, Rossant J, Russell LB, Schimenti J, Shiroishi T, Skarnes WC, Soriano P, Stanford W, Takahashi JS, Wurst W, Zimmer A International Mouse Mutagenesis Consortium. Sequence interpretation. Functional annotation of mouse genome sequences. Science. 2001 Feb 16;291(5507):1251–5. doi: 10.1126/science.1058244. [DOI] [PubMed] [Google Scholar]
- 23.Ramanathan P, Martin IC, Gardiner-Garden M, Thomson PC, Taylor RM, Ormandy CJ, Moran C, Williamson P. Transcriptome analysis identifies pathways associated with enhanced maternal performance in QSi5 mice. BMC Genomics. 2008 Apr 29;9:197. doi: 10.1186/1471-2164-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed DR, Lawler MP, Tordoff MG. Reduced body weight is a common effect of gene knockout in mice. BMC Genet. 2008 Jan 8;9:4. doi: 10.1186/1471-2156-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: A Tool for Annotating and Analyzing Human Hereditary Disease. Am J Hum Genet. 2008 Nov;83(5):610–5. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S OBI Consortium. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007 Nov;25(11):1251–5. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6(1):R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ RGD Team. The Rat Genome Database, update 2007--easing the path from disease to data and back again. Nucleic Acids Res. 2007 Jan;35:D658–62. doi: 10.1093/nar/gkl988. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]