Abstract
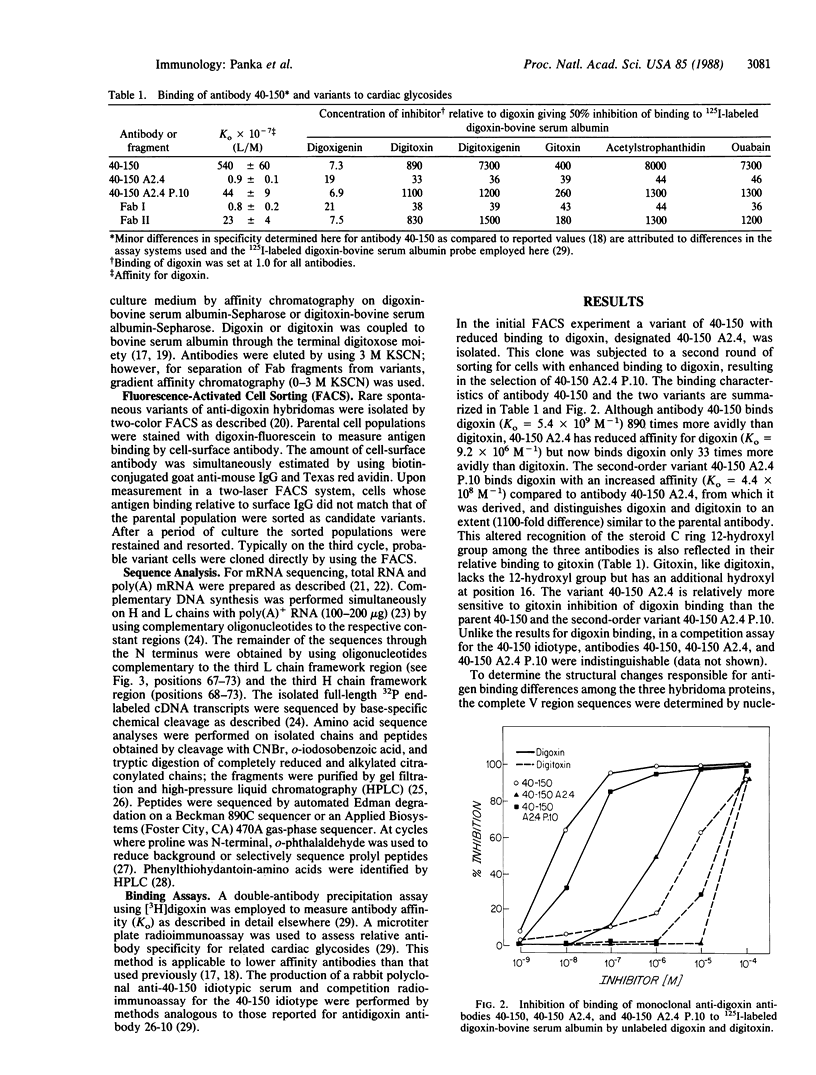
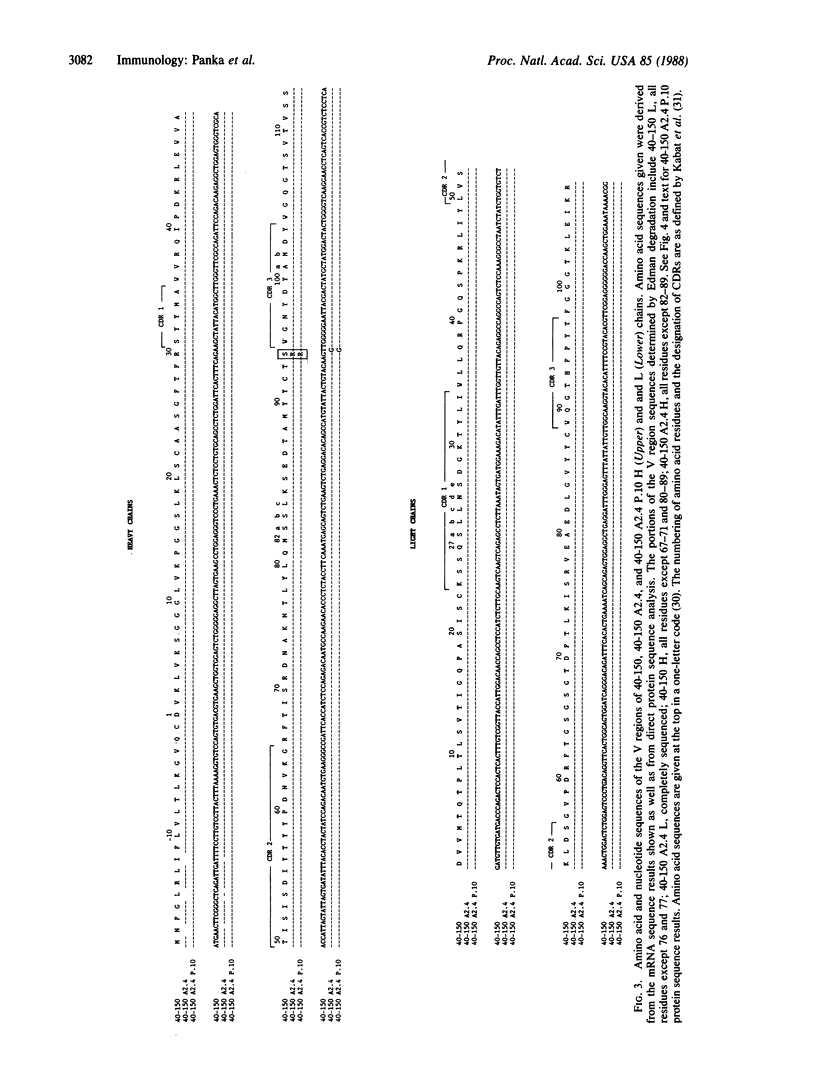
Rare spontaneous variants of the anti-digoxin antibody-producing hybridoma 40-150 (Ko = 5.4 x 10(9) M-1) were selected for altered antigen binding by two-color fluorescence-activated cell sorting. The parent antibody binds digoxin 890-fold greater than digitoxin. The variant 40-150 A2.4 has reduced affinity for digoxin (Ko = 9.2 x 10(6) M-1) and binds digoxin 33-fold greater than digitoxin. A second-order variant, derived from 40-150 A2.4 (designated 40-150 A2.4 P.10), demonstrated partial regain of digoxin binding (Ko = 4.4 x 10(8) M-1). The altered binding of the variant 40-150 A2.4 was accounted for by a point mutation resulting in substitution of arginine for serine at position 94 in the heavy chain variable region. Antibody 40-150 A2.4 P.10 also contains this arginine but owes its enhanced antigen binding to deletion of two amino acids from the heavy chain amino terminus. This unusual sequence alteration in an immunoglobulin framework region confers increased affinity for antigen.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achstetter T., Ehmann C., Wolf D. H. Proteolysis in eucaryotic cells: aminopeptidases and dipeptidyl aminopeptidases of yeast revisited. Arch Biochem Biophys. 1983 Oct 1;226(1):292–305. doi: 10.1016/0003-9861(83)90296-5. [DOI] [PubMed] [Google Scholar]
- Amit A. G., Mariuzza R. A., Phillips S. E., Poljak R. J. Three-dimensional structure of an antigen-antibody complex at 2.8 A resolution. Science. 1986 Aug 15;233(4765):747–753. doi: 10.1126/science.2426778. [DOI] [PubMed] [Google Scholar]
- Brauer A. W., Oman C. L., Margolies M. N. Use of o-phthalaldehyde to reduce background during automated Edman degradation. Anal Biochem. 1984 Feb;137(1):134–142. doi: 10.1016/0003-2697(84)90359-2. [DOI] [PubMed] [Google Scholar]
- Brüggemann M., Radbruch A., Rajewsky K. Immunoglobulin V region variants in hybridoma cells. I. Isolation of a variant with altered idiotypic and antigen binding specificity. EMBO J. 1982;1(5):629–634. doi: 10.1002/j.1460-2075.1982.tb01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cook W. D., Rudikoff S., Giusti A. M., Scharff M. D. Somatic mutation in a cultured mouse myeloma cell affects antigen binding. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1240–1244. doi: 10.1073/pnas.79.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dildrop R., Brüggemann M., Radbruch A., Rajewsky K., Beyreuther K. Immunoglobulin V region variants in hybridoma cells. II. Recombination between V genes. EMBO J. 1982;1(5):635–640. doi: 10.1002/j.1460-2075.1982.tb01220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J., Röhm K. H. Subcellular localization and levels of aminopeptidases and dipeptidase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1978 Nov 10;527(1):31–41. doi: 10.1016/0005-2744(78)90253-x. [DOI] [PubMed] [Google Scholar]
- Honjo T., Habu S. Origin of immune diversity: genetic variation and selection. Annu Rev Biochem. 1985;54:803–830. doi: 10.1146/annurev.bi.54.070185.004103. [DOI] [PubMed] [Google Scholar]
- Hudson N. W., Mudgett-Hunter M., Panka D. J., Margolies M. N. Immunoglobulin chain recombination among antidigoxin antibodies by hybridoma-hybridoma fusion. J Immunol. 1987 Oct 15;139(8):2715–2723. [PubMed] [Google Scholar]
- Hunter M. M., Margolies M. N., Ju A., Haber E. High-affinity monoclonal antibodies to the cardiac glycoside, digoxin. J Immunol. 1982 Sep;129(3):1165–1172. [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975 Aug 7;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Marquart M., Deisenhofer J., Huber R., Palm W. Crystallographic refinement and atomic models of the intact immunoglobulin molecule Kol and its antigen-binding fragment at 3.0 A and 1.0 A resolution. J Mol Biol. 1980 Aug 25;141(4):369–391. doi: 10.1016/0022-2836(80)90252-1. [DOI] [PubMed] [Google Scholar]
- Mudgett-Hunter M., Anderson W., Haber E., Margolies M. N. Binding and structural diversity among high-affinity monoclonal anti-digoxin antibodies. Mol Immunol. 1985 Apr;22(4):477–488. doi: 10.1016/0161-5890(85)90132-4. [DOI] [PubMed] [Google Scholar]
- Padlan E. A., Davies D. R. Variability of three-dimensional structure in immunoglobulins. Proc Natl Acad Sci U S A. 1975 Mar;72(3):819–823. doi: 10.1073/pnas.72.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panka D. J., Margolies M. N. Complete variable region sequences of five homologous high affinity anti-digoxin antibodies. J Immunol. 1987 Oct 1;139(7):2385–2391. [PubMed] [Google Scholar]
- Porter R. R. Structural studies of immunoglobulins. Science. 1973 May 18;180(4087):713–716. doi: 10.1126/science.180.4087.713. [DOI] [PubMed] [Google Scholar]
- Radbruch A., Zaiss S., Kappen C., Brüggemann M., Beyreuther K., Rajewsky K. Drastic change in idiotypic but not antigen-binding specificity of an antibody by a single amino-acid substitution. Nature. 1985 Jun 6;315(6019):506–508. doi: 10.1038/315506a0. [DOI] [PubMed] [Google Scholar]
- Rudikoff S., Giusti A. M., Cook W. D., Scharff M. D. Single amino acid substitution altering antigen-binding specificity. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1979–1983. doi: 10.1073/pnas.79.6.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satow Y., Cohen G. H., Padlan E. A., Davies D. R. Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A. J Mol Biol. 1986 Aug 20;190(4):593–604. doi: 10.1016/0022-2836(86)90245-7. [DOI] [PubMed] [Google Scholar]
- Saul F. A., Amzel L. M., Poljak R. J. Preliminary refinement and structural analysis of the Fab fragment from human immunoglobulin new at 2.0 A resolution. J Biol Chem. 1978 Jan 25;253(2):585–597. [PubMed] [Google Scholar]
- Shlomchik M. J., Nemazee D. A., Sato V. L., Van Snick J., Carson D. A., Weigert M. G. Variable region sequences of murine IgM anti-IgG monoclonal autoantibodies (rheumatoid factors). A structural explanation for the high frequency of IgM anti-IgG B cells. J Exp Med. 1986 Aug 1;164(2):407–427. doi: 10.1084/jem.164.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. A., Margolies M. N. Complete amino acid sequence of the heavy-chain variable region from an A/J mouse antigen-nonbinding monoclonal antibody bearing the predominant arsonate idiotype. Biochemistry. 1984 Sep 25;23(20):4726–4732. doi: 10.1021/bi00315a031. [DOI] [PubMed] [Google Scholar]
- Smith J. A., Margolies M. N. Complete amino acid sequences of the heavy and light chain variable regions from two A/J mouse antigen nonbinding monoclonal antibodies bearing the predominant p-azophenyl arsonate idiotype. Biochemistry. 1987 Jan 27;26(2):604–612. doi: 10.1021/bi00376a036. [DOI] [PubMed] [Google Scholar]
- Smith T. W., Butler V. P., Jr, Haber E. Characterization of antibodies of high affinity and specificity for the digitalis glycoside digoxin. Biochemistry. 1970 Jan 20;9(2):331–337. doi: 10.1021/bi00804a020. [DOI] [PubMed] [Google Scholar]
- Suh S. W., Bhat T. N., Navia M. A., Cohen G. H., Rao D. N., Rudikoff S., Davies D. R. The galactan-binding immunoglobulin Fab J539: an X-ray diffraction study at 2.6-A resolution. Proteins. 1986 Sep;1(1):74–80. doi: 10.1002/prot.340010112. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Wingard M., Matsueda G., Wolfe R. S. Myxobacter AL-1 protease II: specific peptide bond cleavage on the amino side of lysine. J Bacteriol. 1972 Nov;112(2):940–949. doi: 10.1128/jb.112.2.940-949.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T., Kabat E. A. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med. 1970 Aug 1;132(2):211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]


