Abstract
Cholangiocarcinoma (CCA) and hepatocellular carcinoma (HCC) occur with relatively high incidence in Thailand. The secretome, proteins secreted from cancer cells, are potentially useful as biomarkers of the diseases. Proteomic analysis was performed on the secreted proteins of cholangiocarcinoma (HuCCA-1) and hepatocellular carcinoma (HCC-S102, HepG2, SK-Hep-1, and Alexander) cell lines. The secretomes of the five cancer cell lines were analyzed by SDS-PAGE combined with LC/MS/MS. Sixty-eight proteins were found to be expressed only in HuCCA-1. Examples include neutrophil gelatinase-associated lipocalin (lipocalin 2), laminin 5 beta 3, cathepsin D precursor, desmoplakin, annexin IV variant, and annexin A5. Immunoblotting was used to confirm the presence of lipocalin 2 in conditioned media and cell lysate of 5 cell lines. The results showed that lipocalin 2 was a secreted protein which is expressed only in the conditioned media of the cholangiocarcinoma cell line. Study of lipocalin 2 expression in different types of cancer and normal tissues from cholangiocarcinoma patients showed that lipocalin 2 was expressed only in the cancer tissues. We suggest that lipocalin 2 may be a potential biomarker for cholangiocarcinoma.
1. Introduction
In Thailand, cholangiocarcinoma (CCA), a malignant tumor derived from bile duct epithelium, occurs with a high incidence in tropical countries where it is associated with liver fluke (Opisthorchis viverrini) infestation and nitrosamine ingestion [1]. CCA shows high mortality and presents challenges in diagnosis; so prognosis for CCA patients is rather poor. There is still need for better tumor markers for early diagnosis. The most widely used circulating marker for CCA is carbohydrate antigen (CA) 19-9 [2]. However, (CA) 19-9 is also elevated in pancreatic cancer, gastric cancer, and primary biliary cirrhosis and has been shown it gives false positive results [3]. Carcinoembryonic antigen (CEA) is the other common tumor marker used for detecting CCA. CEA is not specific, being mainly used for colorectal cancers, and can be elevated in other types of cancer, such as gastrointestinal or gynecologic malignancies [4].
Differences in expression profiles between normal liver and CCA tissues were studied, because CCA is contained in liver tissue and is suggested to arise from the same stem cells as HCC [5]. We have previously compared CCA and HCC cell lines using proteomic techniques in order to investigate potential CCA markers for early diagnosis [6]. Comparison of 2D-PAGE patterns for a cholangiocarcinoma cell line (HuCCA-1) and two hepatocellular carcinoma cell lines (HepG2 and HCC-S102) showed that cytokeratin 7 (CK7), cytokeratin 19 (CK19), an unknown proteins (U2/2), and galectin-3 were found in CCA but not in HCC. An extension of this study investigated membrane proteins and cytosolic proteins [7], which showed that ten membrane proteins were found in HuCCA-1 but not in HCC-S102, including mitogen-activated protein kinase kinase kinase 2, calgizzarin, integrin alpha-6 precursor, ezrin, and hippocalcin-like protein 1. The subproteomic approach used here may be useful for developing potential biomarkers for early detection of CCA. However, proteomic studies using cell lines have limitations, since it is not known whether the differentially expressed proteins will be present in accessible biological fluids such as plasma, serum, or urine or not.
Cells and tissues secrete proteins into the extracellular environment, the secretome, which may reflect a large variety of pathological conditions and may be a useful source of biomarkers. The secreted proteins are known to regulate many biological processes. These proteins are not only components of the extracellular matrix and biological fluids but are also involved in blood coagulation, immune defense, signal transduction, and carcinogenesis [8]. Certain proteins secreted from cancer cells that enter the circulatory system can be utilized as targets for monitoring or screening for the presence of cancer cells. The cancer secretome, such as that from hepatocellular carcinoma [9], lung cancer [10], breast cancer [11], and oral squamous cell carcinoma [12], has been studied by many research groups to determine the release of the total proteins by cancer cells. This technique provides useful tools for the discovery of novel biomarkers, by using a cell culture model system in which the cells were grown in serum-free media for proteomic analysis [13, 14]. Secretome analysis of nasopharyngeal carcinoma (NPC) cell lines has been studied by SDS-PAGE and MALDI-TOF MS and revealed several potential NPC protein markers [15, 16].
Since the secretomes of cholangiocarcinoma have not previously been reported, we have used conditioned media to compare the secretomes of cholangiocarcinoma cell line (HuCCA-1) with those of hepatocellular carcinoma cell lines (HCC-S102, HepG2, SK-Hep-1, and Alexander). Expression of proteins was studied by SDS-PAGE and LC/MS/MS, and overexpression of proteins in HuCCA-1 was confirmed by using 1-DE and 2-DE immunodetection. Finally, the potential biomarker was validated in various types of cholangiocarcinoma tissues.
2. Materials and Methods
2.1. Sample Collection
Tissue samples were collected from Department of Surgery, and Liver Fluke and Cholangiocarcinoma Research Center, Faculty of Medicine, Khon Kaen University. Ethical clearance for the tissues was approved by the Ethics Committee for Human Research of Khon Kaen University (HE471214). After resection, specimens were immediately taken to a pathologist, who sampled both the tumor itself and adjacent normal-appearing bile duct. Both samples were then snap-frozen in liquid nitrogen within 10 minutes of removal from the patients and stored at −80°C until analysis. The details of types, ages, genders, grades, and histopathology are shown in Table 1.
Table 1.
Medical diagnosis of cholangiocarcinoma patients with tumor types and histopathology.
| Sample ID | Gender | Age | Tumor type | Staging | Histopathology |
|---|---|---|---|---|---|
| 1 | F | 62 | Peripheral, Single, Seg 7 | 4B | Well differentiated adenocarcinoma, CCA, invade intrahepatic vein |
| 2 | F | 67 | Combined, Single, Seg 7, Hilar, long | 4A | Well differentiated tubular type, CCA |
| 3 | M | 64 | Peripheral, Single, Seg 8 | 4B | Well differentiated tubular type, CCA |
| 4 | M | 43 | Peripheral, Single, Seg 6 | 4B | Moderately diff adenocarcinoma invade intrahepatic vessel |
| 5 | M | 65 | Peripheral, Multiple, Seg 1-2 | 4A | Moderately diff, CCA |
| 6 | M | 46 | Peripheral, Single, Seg 6 | 4A | Moderately diff adenocarcinoma invade intrahepatic vein |
| 7 | M | 39 | Peripheral, Single, Seg 6 | 4B | Moderately diff, CCA |
| 8 | F | 56 | Peripheral, Single, Seg 6 | 4B | Poorly diff CCA, invade intrahepatic vein |
| 9 | M | 67 | Combined, Single, Seg 8, Hilar | 4A | Poorly diff adenocarcinoma |
| 10 | M | 53 | Central, Hilar, long | 3 | Papillary mucinous carcinoma |
| 11 | F | 35 | Peripheral, Single | 4B | Papillary mucinous cyst adenocarcinoma |
| 12 | M | 49 | Peripheral, Multiple, Seg 2-4 | 4A | Papillary mucinous carcinoma |
2.2. Cell Cultures
Cholangiocarcinoma cell line, HuCCA-1 derived from bile duct tumor mass of Thai patient, was kindly provided by Professor Sirisinha [17] and grown in Ham's F12 culture medium (Hyclone laboratories, Logan, UT, USA), containing 15 mM HEPES and supplemented with 10% fetal bovine serum (FBS, Hyclone Laboratories), 100 U/mL penicillin, and 100 μg/mL streptomycin and 125 ng/ml amphotericin B. Hepatocellular carcinoma cells, HCC-S102, established from Thai patient [18] was kindly provided by Dr. Sumalee Tungpradakul and grown in RPMI 1640 (Gibco, Grand Island, NY, USA) containing 25 mM HEPES, supplemented with penicillin (100 U/mL), streptomycin (100 μg/mL) amphotericin B (125 ng/mL) (Gibco), and 10% FBS. Another human hepatocellular carcinoma cell line, the Alexander cell line, a human HCC-derived cell line secreting HBsAg, originally obtained from American Type Culture Collection (Rockville, MD, USA) was kindly provided by Dr. Mammen Mammen and Dr. Ananda Nisalak of the Department of Virology, the Armed Forces Research Institute of Medical Sciences (AFRIMS), Bangkok, Thailand. The hepatoblastoma cell line, HepG2 and hepatocellular cell line, SK-Hep-1 were purchased from American Type Culture Collection. The Alexander HepG2, and SK-Hep-1 cell lines were grown in DMEM (Gibco) with 10% FBS, 100 U/mL penicillin, 100 μg/mL Streptomycin, and 125 ng/mL amphotericin B. All cells were maintained at 37°C in a humidified atmosphere, 95% air, 5% CO2 at 37°C.
2.3. Cell Viability Assay
After incubating the cells in complete media or serum-free media for 24 hours, the numbers of viable cells and dead cells were counted by using the trypan blue dye exclusion method. The percentage of cell viability was expressed as the ratio of total viable cells to the sum of total viable and dead cells.
2.4. Harvesting Conditioned Media
To obtain culture supernatants, approximately 3 × 107 cancer cells were grown to 80% confluence and then the cells were washed with serum-free medium 2 times before incubation in serum-free medium for 24 hours. After incubation, the conditioned medium was harvested and centrifuged 800xg at 4°C for 10 minutes to remove suspended cells. The supernatant was dialyzed against distilled water using a dialysis membrane with molecular weight cutoff 3500 Da (Cellu Sep, Texas, USA) for 48 hours and then concentrated by SpeedVac. The concentrations of total proteins were determined by the Bradford protein assay reagent (Bio-Rad Laboratories, CA) [19].
2.5. SDS-PAGE of Conditioned Media from Cell Lines
Twenty micrograms of conditioned media from five cell lines were resolved on 8%–14% gradient SDS-PAGE (100 × 105 × 0.75 mm). Electrophoresis was performed in a Hoefer system (Hoefer, Inc., San Francisco, CA, USA) at 12 mA, room temperature for 2 hours. The protein was stained with Coomassie brilliant blue R-250 (SERVA Electrophoresis GmbH, Heidelberg, Germany).
2.6. Western Blotting and Immunodetection
Protein extracts, separated by 12% SDS-PAGE and transferred onto nitrocellulose membrane (Hybond ECL, GE Healthcare, Buckinghamshire, UK), were probed with monoclonal antibodies against NGAL (Santa Cruz Biotechnology, CA, USA). Proteins of interest were detected using the enhanced chemiluminescence (ECL) plus detection system, with high-performance film (Hyperfilm ECL; GE Healthcare).
2.7. Two-Dimensional Gel Electrophoresis (2-DE)
2D PAGE was performed using the immobiline/polyacrylamide system. Three hundred micrograms of conditioned media from HuCCA-1 were applied by overnight in-gel rehydration of 70 mm, using nonlinear pH 3–10 IPG gel strips (GE Healthcare). The first dimension (IEF) was performed to give a total of 8000 Vh on an IPGphor (GE Healthcare). Focused IPG strips were equilibrated for 10 minutes in a solution (6 M urea, 30% glycerol, 1% SDS 50 mM Tris-HCl buffer (pH 6.8), and 1% DTT), and then for an additional 10 minutes in the same solution except that DTT was replaced by 2.5% iodoacetamide. The IPG strips were then applied to the second dimension 12% T SDS-PAGE (100 × 105 × 1.5 mm). After electrophoresis, proteins were visualized by Coomassie blue R-250 staining.
2.8. Sample Preparation for SDS-PAGE Immunodetection
Tissues were homogenized in phosphate buffered saline containing 0.2 mM phenylmethanesulfonylfluoride, 2 μg/mL pepstatin A, 1 μg/mL bestatin, and centrifuged for 20 minutes at 15 000 rpm. Protein content of the supernatant was determined using the Bradford method [19]. Samples (10 μg protein) were mixed with sample buffer, boiled, and applied to 15% T SDS polyacrylamide gels (100 × 80 × 0.75 mm). Electrophoresis was performed in a Hoefer system (Hoefer, Inc., San Francisco, CA, USA) at 10 mA, room temperature for 1.5 hours, followed by electroblotting of proteins from the gel onto PVDF membranes (Immobilon-P; Millipore, Billerica, MA, USA) at 100 V for 30 minutes at 4°C. After blocking in 5% nonfat dry milk, membranes were probed with 1 : 200 diluted anti-NGAL monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), repeatedly washed in 20 mM Tris buffered saline, pH 7.6, containing 0.1% Tween 20, and then incubated in 1 : 5000 rabbit antimouse immunoglobulin G (IgG; Dako Cytomation, Glostrup, Denmark) for 1 hour. After washing, the reaction was developed using the ECL plus detection system, with high-performance film (Hyper-film ECL; GE Healthcare).
2.9. Tryptic In-Gel Digestion
Protein spots were excised and transferred to 0.5 mL microfuge tubes. Fifty μL of 0.1 M NH4HCO3 in 50% acetonitrile was added. The gel was incubated 3 times for 20 minutes at 30°C. The solvent were discarded and gel particles were dried completely by Speed Vac. Reduction and alkylation were performed by swelling the gel pieces in 50 μL buffer solution (0.1 M NH4HCO3, 10 mM DTT and 1 mM EDTA) and incubating at 60°C for 45 minutes. After cooling, the excess liquid was removed and quickly replaced by the same volume of freshly prepared 100 mM iodoacetamide in 0.1 M NH4HCO3 solution. The reaction was incubated at room temperature in the dark for 30 minutes. The iodoacetamide solution was removed and the gel pieces were washed with 50% acetonitrile in water, 3 times for 10 minutes each time, and the gel pieces were completely dried. Aliquots of trypsin (Promega Corporation, WI, USA) (1 μg trypsin /10 μL 1% acetic acid) were prepared and stored at −20°C. Fifty μL of digestion buffer (0.05 M Tris HCl, 10% acetonitrile, 1 mM CaCl2, pH 8.5) and 1 μL of trypsin were added to the gel pieces. After incubating the reaction mixture at 37°C overnight, the digestion buffer was removed and saved. The gel pieces were then extracted by adding 60 μL of 2% freshly prepared trifluoroacetic acid and incubating for 30 minutes at 60°C. The extract and the saved digestion buffer were finally pooled and dried.
2.10. Protein Identification by LC/MS/MS
LC/MS/MS analyses were carried out using a capillary LC system (Waters) coupled to a Q-TOF mass spectrometer (Micromass, Manchester, UK) equipped with a Zspray ion source working in the nanoelectrospray mode. Glu-fibrinopeptide was used to calibrate the instrument in MS/MS mode. The tryptic peptides were concentrated and desalted on a 75 μm ID × 150 mm C18 PepMap column (LC Packings, Amsterdam, The Netherlands). Eluents A and B were 0.1% formic acid in 97% water, 3% acetonitrile, and 0.1% formic acid in 97% acetonitrile, respectively. Six μL of sample was injected into the nanoLC system, and separation was performed using the following gradient: 0 minute 7% B, 35 minutes 50% B, 45 minutes 80% B, 49 minutes 80% B, 50 minutes 7% B, and 60 minutes 7% B. The database search was performed with ProteinLynx screening SWISS-PROT and NCBI. For some proteins that were difficult to find, the Mascot search tool available on the Matrix Science site screening NCBInr was used. The search parameters were set as follows: peptide mass tolerance = 1 Da; MS/MS ion mass tolerance = 1 Da; enzyme set as trypsin and allowance was set up to two missed cleavages; peptide charges were limited to 2+ and 3+. The proteins were identified with P-values ≤.05 and Mascot scores >35 were considered as promising hits.
3. Results
3.1. Comparison of Secretomes from Cholangiocarcinoma and Hepatocellular Carcinoma Cells
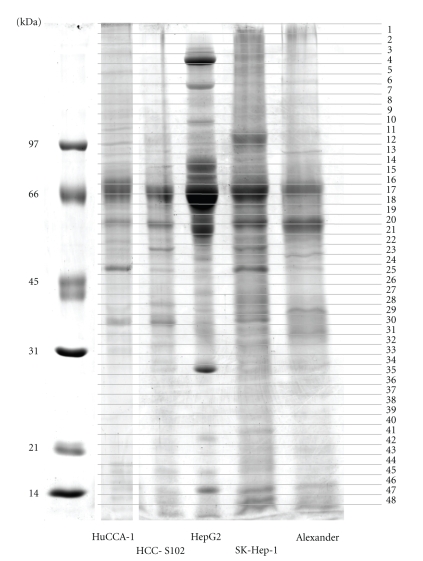
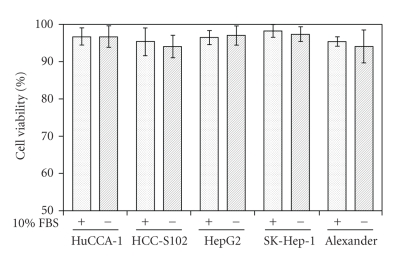
The secreted proteins from conditioned media of HuCCA-1, HCC-S102, HepG2, SK-Hep-1, and Alexander cells were resolved by SDS-PAGE and visualized by Coomassie blue as shown in Figure 1. The secreted protein patterns were enriched in the conditioned media and showed patterns that differed significantly in each cell line. The effect of 24-hour serum starvation on the viability of the five cancer cells cultured was tested and the results showed that this treatment had little effect on cell viability (Figure 2).
Figure 1.
SDS-PAGE analysis of secretomes obtained from five cell lines. HuCCA-1, HCC-S102, HepG2, SK-Hep-1, and Alexander are shown in order from left to right, with MW standards in the leftmost lane. Each gel was cut into 0.5 mm slices, numbered as shown from the top of the gel to the dye front. Proteins in each slice were reduced, alkylated, digested with trypsin, and subjected to LC/MS/MS analysis.
Figure 2.
Viability of cancer cells cultured in complete media or serum-free media for 24 hours. Cell viability was determined by trypan blue exclusion. Data are expressed as mean ± SD of from three independent experiments.
Because of the high sensitivity of mass spectrometry in identifying mixtures of proteins, the stained gels were marked and cut into 0.5 mm slices from the gel top to dye front as shown in Figure 1. Forty-eight total gel bands were subjected to in-gel tryptic digestion and the proteins were identified by LC/MS/MS. There are 83 proteins secreted from the HuCCA-1 cell line as shown in Table 2. The secreted proteins from 4 hepatocellular carcinoma cell lines (HCC-S102, HepG2, SK-Hep-1, and Alexander) were collected in the form of a database, together with those of other cell lines including lung, breast, cervical, and oral cell lines, used in our group (manuscript in preparation). The secreted proteins from 4 hepatocellular carcinoma cell lines are shown in Table 1 in supplementary Material available online at doi:10.1155/2010/437143. The molecular weights shown in Table 2 and Supplementary Table 1 are theoretical values from databases. Some experimental molecular weights differed from theoretical values because the bands were from SDS-PAGE, in which the proteins were denatured and found as isozymes. From Table 2 and Supplementary Table 1, some proteins containing one matched peptide were considered because the peptide score was greater than 35, higher than the Mascot score of 20, which Kristiansen et al. considered to be significant [20]. We also manually selected only the peptide match that had a length of at least 8 amino acids with sequence tag of at least three amino acids to be a good y-ion series. Relevant information on the functions of proteins was taken from Swiss-Prot and NCBI. Then proteins were classified by their functions into various categories, namely, chaperone/stress response, cell cycle, cytoskeleton/mobility, DNA replication/gene regulation, extracellular matrix, immunological response, ion channels, metabolism, protection and detoxification, protein synthesis and degradation, signal transduction, transport/binding proteins, tumor-associated proteins, and unannotated/function inferred.
Table 2.
Identified secreted proteins from CCA cell line (HuCCA-1).
| Bands | Accession numbers | Names | Proteins were not expressed in HCC cell lines | MW/pI | Score | Peptide matches | %Coverage | Matched sequences | Functions |
|---|---|---|---|---|---|---|---|---|---|
| 1 | TSP1_HUMAN | Thrombospondin-1 | 129.30/4.71 | 77 | 7 | 7 | K.GPDPSSPAFR.I | Cytoskeleton/mobility | |
| K.FQDLVDAVR.A | |||||||||
| K.GGVNDNFQGVLQNVR.F | |||||||||
| R.FVFGTTPEDILR.N | |||||||||
| R.FTGSQPFGQGVEHATANK.Q | |||||||||
| K.GTSQNDPNWVVR.H | |||||||||
| R.AQGYSGLSVK.V | |||||||||
| 2 | K2C6B_HUMAN | Keratin, type II cytoskeletal 6B | √ | 59.96/8.05 | 122 | 4 | 8 | R.SGFSSISVSR.S | Cytoskeleton/mobility |
| K.FASFIDKVR.F | |||||||||
| K.YEELQVTAGR.H | |||||||||
| R.ATGGGLSSVGGGSS | |||||||||
| TIK.Y | |||||||||
| 3 | MYH10_HUMAN | Myosin-10 | 228.80/5.44 | 35 | 2 | 0 | K.ESLTK.L | Cytoskeleton/mobility | |
| K.NDNSSR.F | |||||||||
| 4 | LAMC1_HUMAN | Laminin subunit gamma-1 | 177.49/4.82 | 42 | 1 | 4 | K.TEQQTADQLLAR.A | Cytoskeleton/mobility | |
| 5 | MYH10_HUMAN | Myosin-10 | 228.80/5.44 | 37 | 3 | 0 | K.NDNSSR.F | Cytoskeleton/mobility | |
| K.ESLTK.L | |||||||||
| R.GCLARK.A | |||||||||
| gi 4761370 | Immunoglobulin lambda light chain variable region | √ | 10.05/9.17 | 36 | 2 | 22 | R.AEDEAYYYCSSYAD | Immunological response | |
| TKCFGVR.R | |||||||||
| K.AFGVR.R | |||||||||
| ZNF28_HUMAN | Zinc finger protein 28 | √ | 98.64/9.44 | 38 | 2 | 5 | K.LEAVGTGIEPKAMS | DNA replication/gene regulation | |
| QGLVTFGDVAVDFSQ | |||||||||
| EEWEWLNPIQR.N | |||||||||
| K.NFQKSSVVIK.Q | |||||||||
| 6 | STK10_HUMAN | Serine/threonine-protein kinase 10 | √ | 112.07/6.52 | 36 | 1 | 1 | R.ELVAEAKAEVMEEEDGR.D | Metabolism |
| PTPRH_HUMAN | Receptor-type tyrosine-protein phosphatase H | √ | 122.28/5.21 | 35 | 1 | 0 | R.TETRNTTDTR.V | Metabolism | |
| Q9HC04_HUMAN | Cytochrome P450 monooxygenase | √ | 49.50/5.98 | 38 | 1 | 2 | K.TLDPFETMLK.S | Metabolism | |
| MYST4_HUMAN | Histone acetyltransferase MYST4 | √ | 199.70/5.05 | 39 | 2 | 2 | K.KPEELLSCADCGSSGH | DNA replication/gene regulation | |
| PSCLKFCPELTTNVK.A | |||||||||
| R.ANSRQSPAK.V | |||||||||
| 7 | PTPRF_HUMAN | Receptor-type tyrosine-protein phosphatase F | 213.83/5.96 | 37 | 2 | 1 | R.YSAPANLYVR.V | Transport/binding proteins | |
| R.AAGTEGPFQEVDGV | |||||||||
| ATTR.Y | |||||||||
| gi 119621568 | HCG1811249, isoform CRA_a | √ | 149.08/7.76 | 59 | 5 | 4 | R.ELTDLNQEFETLQEK.A | Unknown | |
| K.EINSLQSDFTK.Y | |||||||||
| R.EAELQVDQILTK.S | |||||||||
| R.LLIDDQLLR.N | |||||||||
| R.LSLSPEVLDLTSNSLK.R | |||||||||
| ITA2_HUMAN | Integrin alpha-2 | √ | 129.22/5.10 | 38 | 3 | 2 | R.FGIAVLGYLNR.N | Transport/binding proteins | |
| K.IPLLYDAEIHLTR.S | |||||||||
| K.IGQTSSSVSFK.S | |||||||||
| 8 | LAMB3_HUMAN | Laminin-5 beta 3 | √ | 129.33/7.30 | 36 | 2 | 1 | K.IQEVGEITNLR.V | Transport/binding proteins |
| R.LPNVDLVLSQTK.Q | |||||||||
| 9 | ITA6_HUMAN | Integrin alpha-6 | √ | 118.82/6.36 | 36 | 1 | 1 | K.DEITFVSGAPR.A | Transport/binding proteins |
| 10 | RAP2B_HUMAN | Ras-related protein Rap-2b | √ | 20.49/4.52 | 40 | 1 | 14 | K.EIEVDSSPSVLEILDTAGTEQFASMR.D | Signal transduction |
| RAP2C_HUMAN | Ras-related protein Rap-2c | √ | 20.73/4.67 | 40 | 1 | 14 | Signal transduction | ||
| RAP2A_HUMAN | Ras-related protein Rap-2a | √ | 20.60/4.52 | 40 | 1 | 14 | Signal transduction | ||
| 11 | EF2_HUMAN | Elongation factor 2 | 95.28/6.39 | 36 | 3 | 3 | R.VFSGLVSTGLK.V | Protein synthesis and degradation | |
| M.VNFTVDQIR.A | |||||||||
| K.YEWDVAEAR.K | |||||||||
| ACTN4_HUMAN | Alpha-actinin-4 | 102.20/5.27 | 87 | 7 | 9 | R.DLLLDPAWEK.Q | Transport/binding proteins | ||
| K.LASDLLEWIR.R | |||||||||
| K.ASIHEAWTDGK.E | |||||||||
| R.EAILAIHK.E | |||||||||
| R.QFASQANVVGPWIQ | |||||||||
| TK.M | |||||||||
| K.GISQEQMQEFR.A | |||||||||
| R.ETTDTDTADQVIASFK.V | |||||||||
| 12 | DSG4_HUMAN | Desmoglein-4 | √ | 113.75/4.42 | 43 | 1 | 2 | R.EGIKGSSLLNYVLGTY | Transport/binding proteins |
| TAIDLDTGNPATDVR.Y | |||||||||
| 13 | gi 153792590 | Heat shock 90 kDa protein 1 | √ | 98.10/5.07 | 97 | 6 | 10 | R.ELISNSSDALDK.I | Chaperone/stress response |
| K.EDQTEYLEER.R | |||||||||
| K.YIDQEELNK.T | |||||||||
| K.TKPIWTR.N | |||||||||
| K.HIYYITGETK.D | |||||||||
| K.DQVANSAFVER.L | |||||||||
| 14 | EZRI_HUMAN | Ezrin | 69.37/5.94 | 42 | 4 | 6 | K.APDFVFYAPR.L | Cytoskeleton/mobility | |
| K.IALLEEAR.R | |||||||||
| R.QLLTLSSELSQAR.D | |||||||||
| K.IGFPWSEIR.N | |||||||||
| gi 386758 | GRP78 precursor | √ | 72.29/5.07 | 61 | 4 | 7 | R.ITPSYVAFTPEGER.L | Chaperone/stress response | |
| K.TFAPEEISAMVLTK.M | |||||||||
| R.IINEPTAAAIAYGLDK.R | |||||||||
| R.LTPEEIER.M | |||||||||
| 15 | TRFL_HUMAN | Lactotransferrin | 78.13/8.01 | 35 | 1 | 2 | R.YYGYTGAFR.C | Transport/binding proteins | |
| MOES_HUMAN | Moesin | 67.78/6.08 | 43 | 2 | 3 | R.EDAVLEYLK.I | Cytoskeleton/mobility | ||
| K.APDFVFYAPR.L | |||||||||
| 16 | Q53GZ6_HUMAN | Heat shock 70 kDa protein 8 isoform 1 variant | √ | 70.85/5.37 | 96 | 5 | 8 | R.TTPSYVAFTDTER.L | Chaperone/stress response |
| K.SFYPEEVSSMVLTK.M | |||||||||
| K.DAGTIAGLNVLR.I | |||||||||
| R.FEELNADLFR.G | |||||||||
| K.LLQDFFNGK.E | |||||||||
| 17 | ALBU_HUMAN | Human serum albumin | 69.32/5.86 | 61 | 3 | 3 | K.VPQVSTPTLVEVSR.N | Transport/binding proteins | |
| R.LHCASLQK.F | |||||||||
| K.KVPQVSTPTLVEVSR.N | |||||||||
| 18 | Q16503_HUMAN | Thyrotropin receptor-I | √ | 28.44/6.95 | 25 | 1 | 2 | K.ELPLLK.F | Signal transduction |
| 19 | KPYM_HUMAN | Pyruvate kinase isozymes M1/M2 | 57.88/7.95 | 119 | 7 | 13 | K.GSGTAEVELKK.G | Metabolism | |
| K.ITLDNAYMEK.C | |||||||||
| K.IYVDDGLISLQVK.Q | |||||||||
| K.GVNLPGAAVDLPA | |||||||||
| VSEK.D | |||||||||
| K.DIQDLK.F | |||||||||
| R.GDLGIEIPAEK.V | |||||||||
| K.VFLAQK.M | |||||||||
| 20 | PDIA3_HUMAN | Protein disulfide-isomerase A3 | √ | 56.75/5.92 | 36 | 1 | 2 | R.ELSDFISYLQR.E | Metabolism |
| PDIA1_HUMAN | Protein disulfide-isomerase | 56.64/6.10 | 40 | 2 | 6 | K.TFSHELSDFGLESTA | Protein synthesis and degradation | ||
| GEIPVVAIR.T | |||||||||
| R.FLQDYFDGNLK.R | |||||||||
| 21 | HERC6_HUMAN | Probable E3 ubiquitin-protein ligase HERC6 | 46.39/7.19 | 39 | 1 | 2 | K.LTLDEK.K | Metabolism | |
| RBM27_HUMAN | RNA-binding protein 27 | √ | 44.78/9.54 | 41 | 1 | 3 | K.TSSAVSTPSKVK.T | DNA replication/gene regulation | |
| K2C7_HUMAN | Keratin, type II cytoskeletal 7 | 51.39/5.42 | 45 | 2 | 5 | R.EVTINQSLLAPLR.L | Cytoskeleton/mobility | ||
| R.LPDIFEAQIAGLR.G | |||||||||
| CATD_HUMAN | Cathepsin D | √ | 44.52/6.09 | 77 | 6 | 16 | R.TMSEVGGSVEDLIAK.G | Protein synthesis and degradation | |
| K.QPGITFIAAK.F | |||||||||
| K.FDGILGMAYPR.I | |||||||||
| K.LVDQNIFSFYLSR.D | |||||||||
| K.VSTLPAITLK.L | |||||||||
| K.LSPEDYTLK.V | |||||||||
| 22 | CATD_HUMAN | Cathepsin D | √ | 44.52/6.09 | 88 | 4 | 9 | K.LVDQNIFSFYLSR.D | Protein synthesis and degradation |
| K.QPGITFIAAK.F | |||||||||
| R.YYTVFDR.D | |||||||||
| R.VGFAEAAR.L | |||||||||
| EF1A1_HUMAN | Elongation factor 1-alpha 1 | 47.43/9.00 | 49 | 2 | 5 | R.EHALLAYTLGVK.Q | Protein synthesis and degradation | ||
| K.IGGIGTVPVGR.V | |||||||||
| TBB5_HUMAN | Tubulin beta chain | 49.73/4.75 | 43 | 2 | 4 | R.ISVYYNEATGGK.Y | Cytoskeleton/mobility | ||
| R.FPGQLNADLR.K | |||||||||
| 23 | ENOA_HUMAN | Alpha-enolase | 47.14/7.17 | 100 | 6 | 15 | R.AAVPSGASTGIYEA | Metabolism | |
| LELR.D | |||||||||
| K.KLNVTEQEK.I | |||||||||
| K.LMIEMDGTENK.S | |||||||||
| K.GVPLYR.H | |||||||||
| K.LAMQEFMILPVGAA | |||||||||
| NFR.E | |||||||||
| K.YDLDFK.S | |||||||||
| gi 119590453 | EDAR-associated death domain, isoform CRA_a | √ | 42.32/5.64 | 42 | 2 | 5 | K.KLNVTEQEK.I | Unknown | |
| K.LMIEMDGTENK.S | |||||||||
| 24 | ACTB_HUMAN | Actin, cytoplasmic 1 | 41.71/5.14 | 107 | 3 | 9 | K.AGFAGDDAPR.A | Cytoskeleton/mobility | |
| R.GYSFTTTAER.E | |||||||||
| K.SYELPDGQVITIGNER.F | |||||||||
| K1C18_HUMAN | Keratin, type I cytoskeletal 18 | 48.03/5.16 | 40 | 1 | 3 | R.AQIFANTVDNAR.I | Cytoskeleton/mobility | ||
| 25 | ACTS_HUMAN | Actin, alpha skeletal muscle | √ | 42.02/5.23 | 123 | 7 | 14 | K.AGFAGDDAPR.A | Cytoskeleton/mobility |
| R.AVFPSIVGRPR.H | |||||||||
| K.RGILTLK.Y | |||||||||
| R.GILTLK.Y | |||||||||
| K.IWHHTFYNELR.V | |||||||||
| R.LDLAGR.D | |||||||||
| K.EITALAPSTMK.I | |||||||||
| ACTB_HUMAN | Actin, cytoplasmic 1 | 41.71/5.29 | 162 | 7 | 22 | K.AGFAGDDAPR.A | Cytoskeleton/mobility | ||
| R.AVFPSIVGRPR.H | |||||||||
| K.RGILTLK.Y | |||||||||
| K.IWHHTFYNELR.V | |||||||||
| R.VAPEEHPVLLTEAP | |||||||||
| LNPK.A | |||||||||
| R.LDLAGR.D | |||||||||
| R.GYSFTTTAER.E | |||||||||
| K.EITALAPSTMK.I | |||||||||
| 26 | SH2D7_HUMAN | SH2 domain-containing protein 7 | √ | 49.78/5.96 | 35 | 1 | 1 | R.MNQAR.L | Transport/binding proteins |
| K2C8_HUMAN | Keratin, type II cytoskeletal 8 | 30.84/ 4.91 | 37 | 1 | 4 | R.LEGLTDEINFLR.Q | Cytoskeleton/mobility | ||
| 27 | DJB11_HUMAN | DnaJ homolog subfamily B member 11 | √ | 40.49/ 5.74 | 36 | 1 | 3 | R.TLEVEIEPGVR.D | Chaperone/stress response |
| 28 | gi 10567590 | Sodium bicarbonate cotransporter-like protein | √ | 118.58/8.00 | 45 | 2 | 4 | R.LCHAQSRSMNDISLT | Transport/binding proteins |
| PNTDQR.K | |||||||||
| R.LLGNSCKFIPDLALM | |||||||||
| SFILFFGTYSMTLTLK.K | |||||||||
| ALDOA_HUMAN | Fructose-bisphosphate aldolase A | 39.40/8.30 | 52 | 5 | 20 | M.PYQYPALTPEQK.K | Metabolism | ||
| K.ELSDIAHR.I | |||||||||
| K.GILAADESTGSIAK.R | |||||||||
| R.QLLLTADDR.V | |||||||||
| R.TVPPAVTGITFLSGG | |||||||||
| QSEEEASINLNAINK.C | |||||||||
| 29 | AFF4_HUMAN | AF4/FMR2 family member 4 | 69.38/9.45 | 38 | 1 | 1 | K.NSSSTSKQK.K | DNA replication/gene regulation | |
| ANXA2_HUMAN | Annexin A2 | 38.55/7.57 | 72 | 7 | 24 | R.DALNIETAIK.T | Signal transduction | ||
| K.GVDEVTIVNILTNR.S | |||||||||
| R.QDIAFAYQR.R | |||||||||
| K.SALSGHLETVILGLLK.T | |||||||||
| K.TPAQYDASELK.A | |||||||||
| R.TNQELQEINR.V | |||||||||
| K.DIISDTSGDFRK.L | |||||||||
| ANXA1_HUMAN | Annexin A1 | 38.69/6.63 | 36 | 1 | 3 | K.TPAQFDADELR.A | Signal transduction | ||
| gi 182073 | Erythroid protein 4.1 isoform A | √ | 86.55/5.31 | 42 | 2 | 2 | K.APIAAPEPELK.T | Metabolism | |
| K.TVKGGISETR.I | |||||||||
| 30 | ANXA2_HUMAN | Annexin A2 | 38.55/7.57 | 70 | 7 | 21 | K.AYTNFDAER.D | Signal transduction | |
| R.DALNIETAIK.T | |||||||||
| K.GVDEVTIVNILTNR.S | |||||||||
| R.QDIAFAYQR.R | |||||||||
| K.TPAQYDASELK.A | |||||||||
| R.TNQELQEINR.V | |||||||||
| K.SLYYYIQQDTK.G | |||||||||
| ANXA1_HUMAN | Annexin A1 | 38.69/6.63 | 52 | 4 | 15 | K.GVDEATIIDILTK.R | Signal transduction | ||
| K.TPAQFDADELR.A | |||||||||
| R.SEDFGVNEDLADSDAR.A | |||||||||
| K.GTDVNVFNTILTTR.S | |||||||||
| G3P_HUMAN | Glyceraldehyde-3-phosphate dehydrogenase | 36.03/ | 36 | 2 | 6 | K.VGVNGFGR.I | Metabolism | ||
| 8.26 | R.GALQNIIPASTGAAK.A | ||||||||
| K1C9_HUMAN | Keratin, type I cytoskeletal 9 | 62.09/5.19 | 35 | 2 | 4 | R.SGGGGGGGLGSGGSIR.S | Cytoskeleton/mobility | ||
| K.STMQELNSR.L | |||||||||
| 31 | ANXA1_HUMAN | Annexin A1 | 38.69/6.63 | 47 | 3 | 14 | K.GVDEATIIDILTK.R | Signal transduction | |
| K.GGPGSAVSPYPTFNP | |||||||||
| SSDVAALHK.A | |||||||||
| K.TPAQFDADELR.A | |||||||||
| ANXA5_HUMAN | Annexin A5 | √ | 35.91/4.73 | 35 | 1 | 4 | R.GTVTDFPGFDER.A | Signal transduction | |
| 32 | HRX_HUMAN | Histone-lysine N-methyltransferase HRX | √ | 431.68/9.23 | 36 | 3 | 0 | R.GRPPSTERIK.T | DNA replication/gene regulation |
| R.EPTFR.W | |||||||||
| R.GPRIK.H | |||||||||
| 33 | A8K1N0_HUMAN | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, isoform CRA_a | √ | 27.73/4.73 | 77 | 5 | 22 | K.SVTEQGAELSNEER.N | Metabolism |
| R.NLLSVAYK.N | |||||||||
| K.VFYLK.M | |||||||||
| K.GIVDQSQQAYQ | |||||||||
| EAFEISK.K | |||||||||
| K.DSTLIMQLLR.D | |||||||||
| 34 | gi 119610955 | TSR1 20S rRNA accumulation homolog yeast isoform CRA b | √ | 50.28/9.11 | 38 | 1 | 2 | K.LLLLDTQQEAGMLLR.Q | DNA replication/gene regulation |
| K1C18_HUMAN | Keratin, type I cytoskeletal 18 | 47.31/5.27 | 40 | 2 | 4 | R.AQIFANTVDNAR.I | Cytoskeleton/mobility | ||
| R.IVLQIDNAR.L | |||||||||
| 35 | TPIS_HUMAN | Triosephosphate isomerase | 26.65/6.50 | 47 | 2 | 11 | R.HVFGESDELIGQK.V | Metabolism | |
| K.VVLAYEPVWAIGTGK.T | |||||||||
| 36 | FLNA_HUMAN | Filamin-A isoform CRA_e | √ | 263.75/5.79 | 35 | 2 | 0 | K.DGTVTVR.Y | Cytoskeleton/mobility |
| K.RDSLPR.L | |||||||||
| Z804A_HUMAN | Zinc finger protein 804A | √ | 136.80/8.19 | 49 | 2 | 2 | M.ECYYIVISSTHLSNG | DNA replication/gene regulation | |
| HFRNIK.G | |||||||||
| R.SLVLQNDMK.H | |||||||||
| A0N4V7_HUMAN | HCG2039797 | √ | 2.21/9.79 | 36 | 1 | 38 | K.GITLSVRP.- | Unknown | |
| gi 119569964 | HCG2040487 | √ | 21.65/8.60 | 39 | 1 | 10 | R.IQGKDLTVWTTQDV | Unknown | |
| EGILGAK.G | |||||||||
| 37 | PRDX1_HUMAN | Peroxiredoxin-1 | 22.10/8.27 | 45 | 3 | 15 | R.TIAQDYGVLK.A | Protection and detoxification | |
| R.QITVNDLPVGR.S | |||||||||
| R.LVQAFQFTDK.H | |||||||||
| gi 16075531 | Immunoglobulin kappa chain variable region | √ | 9.11/9.13 | 36 | 1 | 13 | R.SGTDFTLKISR.V | Immunological response | |
| 38 | NOV_HUMAN | Nephroblastoma overexpressed precursor | √ | 39.14/8.12 | 39 | 2 | 7 | K.QTRLCMVRPCEQEP | DNA replication/gene regulation |
| EQPTDK.K | |||||||||
| K.AIHLQFK.N | |||||||||
| NGAL_HUMAN | Neutrophil gelatinase-associated lipocalin, NGAL, Lipocalin 2 | √ | 20.54/8.87 | 38 | 1 | 6 | K.SYPGLTSYLVR.V | Transport/binding proteins | |
| PRDX1_HUMAN | Peroxiredoxin-1 | 22.10/8.27 | 39 | 3 | 15 | R.TIAQDYGVLK.A | Protection and detoxification | ||
| R.QITVNDLPVGR.S | |||||||||
| R.LVQAFQFTDK.H | |||||||||
| 39 | DESP_HUMAN | Desmoplakin | √ | 201.24/6.68 | 40 | 2 | 1 | K.EISMQKEDDSK.N | Transport/binding proteins |
| R.GIVDSITGQR.L | |||||||||
| NGAL_HUMAN | Neutrophil gelatinase-associated lipocalin, NGAL, Lipocalin 2 | √ | 20.54/8.87 | 42 | 2 | 11 | K.MYATIYELK.E | Transport/binding proteins | |
| K.SYPGLTSYLVR.V | |||||||||
| 40 | Q59FK3_HUMAN | Annexin IV variant | √ | 25.41/6.03 | 41 | 2 | 6 | -.AASISR.L | Signal transduction |
| K.GDTSGDYR.K | |||||||||
| FAM3C_HUMAN | Protein FAM3C | 24.67/8.52 | 37 | 1 | 7 | R.LIADLGSTSITNLGFR.D | Signal transduction | ||
| 41 | K1C16_HUMAN | Keratin, type I cytoskeletal 16 | 51.24/4.79 | 74 | 6 | 15 | R.APSTYGGGLSVSSR.F | Cytoskeleton/mobility | |
| R.LASYLDK.V | |||||||||
| R.ALEEANADLEVK.I | |||||||||
| K.IIAATIENAQPILQID | |||||||||
| NAR.L | |||||||||
| K.ASLENSLEETK.G | |||||||||
| R.LEQEIATYR.R | |||||||||
| K1C9_HUMAN | Keratin, type I cytoskeletal 9 | 62.09/5.19 | 68 | 5 | 8 | R.SGGGGGGGLGSGGSIR.S | Cytoskeleton/mobility | ||
| R.LASYLDK.V | |||||||||
| K.TLLDIDNTR.M | |||||||||
| R.QGVDADINGLR.Q | |||||||||
| R.QEYEQLIAK.N | |||||||||
| 42 | A8KAH9_HUMAN | Ras-related protein Rap-1A | √ | 20.97/6.38 | 42 | 2 | 14 | K.SALTVQFVQGIFVEK.Y | Signal transduction |
| K.INVNEIFYDLVR.Q | |||||||||
| 43 | gi 150023192 | NADH dehydrogenase subunit 5 | √ | 67.02/9.04 | 41 | 1 | 1 | R.KMVGLLK.T | Metabolism |
| gi 62906706 | Diacylglycerol kinase alpha | √ | 28.08/4.98 | 35 | 1 | 2 | K.KVSDVLK.L | Metabolism | |
| LIPH_HUMAN | Lipase member H | √ | 50.83/7.14 | 35 | 1 | 1 | R.KVAMVLK.E | Metabolism | |
| gi 119586924 | Rotatin isoform CRA_e | √ | 44.01/9.19 | 36 | 1 | 1 | K.KEDGVIK.E | Transport/binding proteins | |
| 44 | gi∣158430561 | Chain A, Crystal Structure Of The Bard1 Brct Repeat | √ | 24.10/8.62 | 37 | 1 | 3 | K.YEIPEGPR.R | DNA replication/gene regulation |
| 45 | PRDX5_HUMAN | Peroxiredoxin-5, mitochondrial | 22.01/8.75 | 38 | 1 | 5 | R.LLADPTGAFGK.E | Protection and detoxification | |
| Q05D03_HUMAN | RHOV protein | √ | 27.79/8.93 | 40 | 2 | 7 | R.AMPPR.E | Transport/binding proteins | |
| R.ACCYLECSALTQK.N | |||||||||
| 46 | PROF1_HUMAN | Profilin-1 | 15.04/8.45 | 78 | 4 | 41 | K.TFVNITPAEVGVLVGK.D | Cytoskeleton/mobility | |
| R.SSFYVNGLTLGGQK.C | |||||||||
| R.DSLLQDGEFSMDLR.T | |||||||||
| K.STGGAPTFNVTVTK.T | |||||||||
| 47 | HBB_HUMAN | Hemoglobin subunit beta | 15.99/6.87 | 39 | 2 | 18 | R.LLVVYPWTKR.F | Transport/binding proteins | |
| K.LHVDPENFR.- | |||||||||
| 48 | gi 119619136 | HCG1790904, isoform CRA_b | 37.27/9.67 | 40 | 2 | 4 | K.SSILYRHK.V | Unknown | |
| K.NEMLTFK.A | |||||||||
| CENPF_HUMAN | Centromere protein F | √ | 22.41/7.11 | 35 | 1 | 0 | K.YTALEQKLK.K | Cell cycle | |
When the HuCCA-1 cell line is compared to the 4 hepatocellular carcinoma cell lines, all cell lines secreted distinct protein profiles. We also found distinct secreted protein profiles when we compared the secreted proteins to different types of cell lines (liver, lung, breast, cervical and oral). Thus, some 30%–50% proteins were expressed at the same level in 5 cell lines, while 50%–70% proteins showed differences in expression. Forty-nine secreted proteins were found only in HuCCA-1 which was marked in Table 2, including laminin 5 beta 3, heat shock 90 kDa protein 1, heat shock 70 kDs protein 8 isoform 1 variant, GRP78 precursor, desmoglein-4 nephroblastoma overexpressed precursor, neutrophil gelatinase-associated lipocalin (lipocalin 2, NGAL), desmoplakin, cathepsin D, DnaJ homolog subfamily B member 11, annexin IV variant, annexin A5, Ras-related protein Rap-1A, RHOV protein, and rotatin isoform CRA_e.
4. Validation of Potential Biomarkers
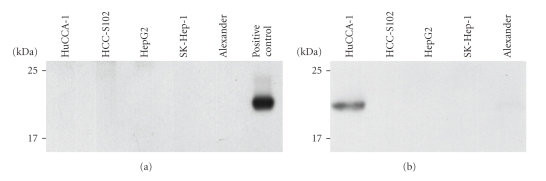
Of the overexpressed proteins found only in HuCCA-1, 15 appeared to be proteins related to cancer. 1-DE immmunoblotting was used to verify the presence of 4 proteins by using specific antibodies for nephroblastoma overexpressed precursor and neutrophil gelatinase-associated lipocalin (lipocalin 2, NGAL). Immunodetection only confirmed the presence of lipocalin 2 (Figure 3(b)), but not the presence of the other two proteins. Expression of lipocalin 2 was found only in the conditioned media of HuCCA-1, but not in the conditioned media of the other 4 cell lines (Figure 3(b)). Furthermore, there was no expression of lipocalin 2 in the cell lysate of any of the 5 cell lines, including HuCCA-1 (Figure 3(a)).
Figure 3.
The western blots for immunoreactive lipocalin 2 from the lysate of five cell lines (HuCCA-1, HCC-S102, HepG2, SK-Hep-1, and Alexander) (a) and five conditioned media samples (b).
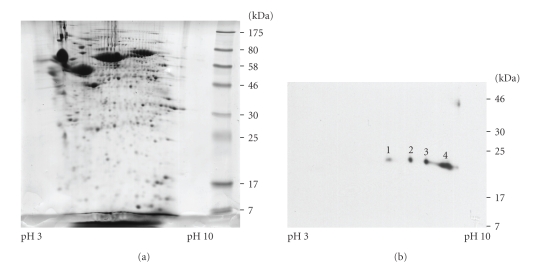
The 2-DE proteomic pattern of conditioned media of HuCCA-1 with Coomassie blue staining is shown in Figure 4(a) and 2-DE immunoblotting of the same sample was used to detect the lipocalin 2 forms (Figure 4(b)), revealing 4 spots with MW/pI 23.5/6.0, 23.5/6.5, 23.0/7.3, and 22.5/8.7. These different forms are likely to result from posttranslational modifications.
Figure 4.
Two-dimensional gel electrophoresis of conditioned media from HuCCA-1 stained with Coomassie Blue R-250 (a) and immunoblot labeled with the antibody to lipocalin 2 (b). The pI and MW of spot numbers 1, 2, 3, and 4 were approximately 6.0/23.5, 6.5/23.5, 7.3/23, and 8.7/22.5 respectively.
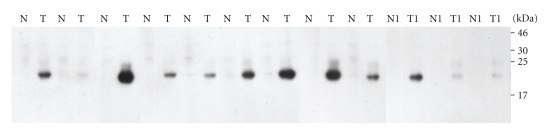
To confirm that lipocalin 2 is highly expressed in cholangiocarcinoma, the 12 homogenate samples from the pairs of normal and cancer of cholangiocarcinoma tissues described in Table 1 were resolved by SDS-PAGE gels and immunoblotted with monoclonal antibody to lipocalin 2. The results clearly showed a positive band of 23 kDa in all cancer tissues (Figure 5), with 9 out of the 12 cases showing high expression level of lipocalin 2, while the corresponding normal tissues were all negative.
Figure 5.
1-DE patterns of the pairs of normal and cancer tissues from 12 cholangiocarcinoma patients. Details of the patients are shown from Table 1. The proteins were separated in 15% SDS-PAGE and detected using by immunoblotting with antibody to lipocalin 2.
5. Discussion
The study of the cancer secretomes by using proteomic technology has greatly accelerated in recent years. With the rapid progress in mass spectrometry (MS), bioinformatics, and analytical techniques, cancer biomarker discovery has been greatly promoted by this approach. Our present work has investigated the cell secretomes of cholangiocarcinoma (CCA) compared to 4 hepatocellular carcinoma (HCC) cell lines. With each cell line, three repeat experiments were performed by collection of the conditioned media from CCA and HCC cell lines, followed by concentration and precipitation by TCA/acetone. The proteins were then run onto SDS-PAGE, gel bands excised consecutively, and proteins analyzed by LC/MS/MS. Some bands showed low levels of matching, some bands showed blue color, and some were almost clear. The pattern on SDS-PAGE of secreted proteins and the total identified proteins were differed with each cell line. When the protein expression of the HuCCA-1 cell line is compared to the HCC cell lines, only 14, 14, 19, and 6 proteins from Alexander, HCC-S102, HepG2, and SK-Hep-1, respectively, matched with HuCCA-1 (data not shown).
Proteins secreted into conditioned medium may be tumor specific and can represent potential biomarkers that may circulate in the blood stream. Our results showed 49 distinct proteins, expressed only in HuCCA-1. Some interesting secreted proteins expressed only in cholangiocarcinoma include laminin 5 beta 3, heat shock 90 kDa protein 1, heat shock 70 kDs protein 8 isoform 1 variant, GRP78 precursor, desmoglein-4 nephroblastoma overexpressed precursor, neutrophil gelatinase-associated lipocalin (lipocalin 2, NGAL), desmoplakin, cathepsin D, DnaJ homolog subfamily B member 11, annexin IV variant, annexin A5, Ras-related protein Rap-1A, RHOV protein, and rotatin isoform CRA_e.
Many of these proteins are related to cancer. For example, laminin-5 (LAMA3, LAMB3, and LAMC2) is a heterotrimeric glycosylated protein that belongs to th Ln family and is formed by α3β3γ2 chains assembled with disulfide bonds. Ln-5 is widely expressed in the human body but shows differential expression in metastatic and nonmetastatic hepatocellular carcinoma [21, 22]. Ln-5 also plays an important role in cell migration during tumor invasion and tissue remodeling [22], and laminin 5 γ2 chain and β3 chain have both been suggested to be important in the invasiveness of cancer cells. Unfortunately, no commercial antibody appears to be available for laminin 5 β3 chain; expression of the protein could not be validated in cholangiocarcinoma tissues. Laminin γ2 chain exhibits aberrant expression in a stepwise manner through different aggressive stages of tumor progression [23].
Nephroblastoma overexpressed (NOV) belongs to the CCN family of genes that encode secreted proteins associated with the extracellular matrix (ECM) and exert regulatory effects at the cellular level. NOV is likely to play a role in cell growth regulation, in the progression and in the metastatic potential of melanomas. The expression of this protein appears to be associated with a higher risk of developing metastases in Ewing's sarcoma [24]. We found the high expression of NOV secreted in the media of the cholangiocarcinoma cell line.
Neutrophil gelatinase-associated lipocalin (NGAL or lipocalin 2) is a prominent member of the lipocalin family. It was first isolated as a 25 kda glycoprotein covalently bound with matrix metalloproteinase-9 (MMP-9) in human neutrophil [25]. It is a secreted acute phase protein, which is also upregulated in multiple cancers, including breast, lung, and pancreas. Recently lipocalin 2 has been proposed as an early biomarker in pancreatic cancer [26].
Desmoplakin, a specialized adhesion junction protein and the principal plaque protein of desmosomes, has been found in various tissues including heart, skin, and meninges [27]. The biological significance of desmoplakin has been recently reported in both autosomal dominant and autosomal recessive disorders from naturally occurring human gene mutation. Desmoplakin was found to be abnormal in many conditions including autoimmune blistering diseases, epithelial malignancies, and blood vessel morphogenesis. Desmoplakin was also found to be secreted by the cholangiocarcinoma cell line in our study.
Cathepsin D, the aspartic protease, is an independent marker of poor prognosis in human breast cancer [28]. Cathepsin-D was reported to play an essential role in multiple tumor progression steps, affecting cell proliferation, angiogenesis, and apoptosis. Another report also suggested that cathepsin D was a key mediator in induced apoptosis [29]. Our results showed highly expression levels of cathepsin D in the cholangiocarcinoma cell line. A cathepsin D-like protein has also been reported from the gut and other tissues of the human liver fluke Opisthorchis viverrini where long-standing infection is associated with cancer of the bile ducts or cholangiocarcinoma [30]. This protein was also found in the excretory and secretory products of cultured adult flukes, indicating a role in host-parasite relationships.
The annexins are family of calcium-regulated phospholipid-binding proteins with diverse roles in cell biology. There are 12 human annexin subfamilies (A1–A11 and A13) that have been found to have various intra- and extracellular roles in a range of cellular processes such as cell signalling, ion transport, cell division, and apoptosis [31, 32]. Individual annexins have been implicated in tumour development and progression. Annexins A1, A2, and A5 interact with cytoskeletal proteins. Annexin A1 also has a role in controlling the inflammatory response while annexin A2 is present on the external surface of endothelial cells. Annexin A2 and annexin A4 appeared to be potential markers of interest for diagnosis of colorectal cancer [33]. The present studies showed that annexin A1, annexin A2, annexin IV variant, and annexin A5 were expressed in the cholangiocarcinoma cell line.
The presence of the proteins nephroblastoma overexpressed precursor and neutrophil gelatinase-associated lipocalin (lipocalin 2, NGAL) was validated in the cell lysates and conditioned media from all 5 cells by using specific antibodies. Lipocalin 2 was found to be clearly expressed in only conditioned media. Then 2-DE immunoblotting of the conditioned media showed the expression of lipocalin 2 as 4 spots at MW/pI 23.5/6.0, 23.5/6.5, 23.0/7.3, and 22.5/8.7 (Figure 4(b)). The posttranslation modification of the lipocalin 2 has not been reported yet and further studies will be needed to provide more information.
Finally, we verified the expression of lipocalin 2 in 12 homogenate samples from the pairs of normal and cancer tissues from cholangiocarcinoma patients. The lipocalin 2 antibody clearly detected a band of 23 kDa in all cancer tissues, while the pairs of normal tissues were all negative. Of the cancer tissues, 9 out of 12 cases showed high expression levels of lipocalin 2. However, since the cases studied here were in the late stage (grade 3-4) of cancer, it is of interest to see if tissues at the early stages of cholangiocarcinoma show lipoclain 2 expression. Further studies will investigate the expression of lipocalin 2 in different types and stages of cholangiocarcinoma in order to understand its relationship to grades and types of cancer.
6. Conclusion
Proteomic techniques have been used to compare the secretomes or proteins secreted into the external environment, cholangiocarcinoma, and hepatocellular carcinoma cell lines. Distinct proteins were found, in particular lipocalin 2, which may have potential value as a biomarker for cholangiocarcinoma.
Supplementary Material
List of secreted proteins identified by tryptic digest and LC/MS/MS from 4 hepatocellular carcinoma cell lines (Alexander, HepG2, SK-Hep-1 and HCC-S102).
Acknowledgments
The authors thank N. Monique Paricharttanakul and K. Lirdprapamongkol for helpful discussion. This investigation was also supported by the Chulabhorn Research Institute.
References
- 1.Sonakul D, Koompirochana C, Chinda K, Stitnimakarn T. Hepatic carcinoma with opisthorchiasis. The Southeast Asian Journal of Tropical Medicine and Public Health. 1978;9(2):215–219. [PubMed] [Google Scholar]
- 2.Reddy SB, Patel T. Current approaches to the diagnosis and treatment of cholangiocarcinoma. Current Gastroenterology Reports. 2006;8(1):30–37. doi: 10.1007/s11894-006-0061-1. [DOI] [PubMed] [Google Scholar]
- 3.Ong SL, Sachdeva A, Garcea G, et al. Elevation of carbohydrate antigen 19.9 in benign hepatobiliary conditions and its correlation with serum bilirubin concentration. Digestive Diseases and Sciences. 2008;53(12):3213–3217. doi: 10.1007/s10620-008-0289-8. [DOI] [PubMed] [Google Scholar]
- 4.Van Beers BE. Diagnosis of cholangiocarcinoma. HPB. 2008;10(2):87–93. doi: 10.1080/13651820801992716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sell S, Dunsford HA. Evidence for the stem cell origin of hepatocellular carcinoma and cholangiocarcinoma. American Journal of Pathology. 1989;134(6):1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 6.Srisomsap C, Sawangareetrakul P, Subhasitanont P, et al. Proteomic analysis of cholangiocarcinoma cell line. Proteomics. Clinical Applications. 2004;4(4):1135–1144. doi: 10.1002/pmic.200300651. [DOI] [PubMed] [Google Scholar]
- 7.Srisomsap C, Subhasitanont P, Sawangareetrakul P, et al. Comparison of membrane-associated proteins in human cholangiocarcinoma and hepatocellular carcinoma cell lines. Proteomics. Clinical Applications. 2007;1(1):89–106. doi: 10.1002/prca.200600168. [DOI] [PubMed] [Google Scholar]
- 8.Ladunga I. Large-scale predictions of secretory proteins from mammalian genomic and EST sequences. Current Opinion in Biotechnology. 2000;11(1):13–18. doi: 10.1016/s0958-1669(99)00048-8. [DOI] [PubMed] [Google Scholar]
- 9.Zwickl H, Traxler E, Staettner S, et al. A novel technique to specifically analyze the secretome of cells and tissues. Electrophoresis. 2005;26(14):2779–2785. doi: 10.1002/elps.200410387. [DOI] [PubMed] [Google Scholar]
- 10.Lou X, Xiao T, Zhao K, et al. Cathepsin D is secreted from M-BE cells: its potential role as a biomarker of lung cancer. Journal of Proteome Research. 2007;6(3):1083–1092. doi: 10.1021/pr060422t. [DOI] [PubMed] [Google Scholar]
- 11.Perera CN, Spalding HS, Mohammed SI, Camarillo IG. Identification of proteins secreted from leptin stimulated MCF-7 breast cancer cells: a dual proteomic approach. Experimental Biology and Medicine. 2008;233(6):708–720. doi: 10.3181/0710-RM-281. [DOI] [PubMed] [Google Scholar]
- 12.Mlynarek AM, Balys RL, Su J, Hier MP, Black MJ, Alaoui-Jamali MA. A cell proteomic approach for the detection of secretable biomarkers of invasiveness in oral squamous cell carcinoma. Archives of Otolaryngology. 2007;133(9):910–918. doi: 10.1001/archotol.133.9.910. [DOI] [PubMed] [Google Scholar]
- 13.Mbeunkui F, Fodstad O, Pannell LK. Secretory protein enrichment and analysis: an optimized approach applied on cancer cell lines using 2D LC-MS/MS. Journal of Proteome Research. 2006;5(4):899–906. doi: 10.1021/pr050375p. [DOI] [PubMed] [Google Scholar]
- 14.Kulasingam V, Diamandis EP. Proteomics analysis of conditioned media from three breast cancer cell lines: a mine for biomarkers and therapeutic targets. Molecular & Cellular Proteomics. 2007;6(11):1997–2011. doi: 10.1074/mcp.M600465-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Wu C-C, Chien K-Y, Tsang N-M, et al. Cancer cell-secreted proteomes as a basis for searching potential tumor markers: nasopharyngeal carcinoma as a model. Proteomics. Clinical Applications. 2005;5(12):3173–3182. doi: 10.1002/pmic.200401133. [DOI] [PubMed] [Google Scholar]
- 16.Wu H-Y, Chang Y-H, Chang Y-C, Liao P-C. Proteomics analysis of nasopharyngeal carcinoma cell secretome using a hollow fiber culture system and mass spectrometry. Journal of Proteome Research. 2009;8(1):380–389. doi: 10.1021/pr8006733. [DOI] [PubMed] [Google Scholar]
- 17.Sirisinha S, Tengchaisri T, Boonpucknavig S, et al. Identification and potential use of a soluble tumor antigen for the detection of liver-fluke-associated cholangiocarcinoma induced in a hamster model. Asian Pacific Journal of Allergy & Immunology. 1991;9:153–157. doi: 10.1002/ijc.2910570514. [DOI] [PubMed] [Google Scholar]
- 18.Laohathai K, Bhamarapravati N. Culturing of human hepatocellular carcinoma. A simple and reproducible method. American Journal of Pathology. 1985;118(2):203–208. [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Zakarias T, Bunkenborg J, Gronborg M, et al. A proteomic analysis of human bile. Molecular & Cellular Proteomics. 2004;3(7):715–728. doi: 10.1074/mcp.M400015-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Giannelli G, Fransvea E, Bergamini C, Marinosci F, Antonaci S. Laminin-5 chains are expressed differentially in metastatic and nonmetastatic hepatocellular carcinoma. Clinical Cancer Research. 2003;9(10):3684–3691. [PubMed] [Google Scholar]
- 22.Akimoto S, Nakanishi Y, Sakamoto M, Kanai Y, Hirohashi S. Laminin 5 β3 and γ 2 chains are frequently coexpressed in cancer cells. Pathology International. 2004;54(9):688–692. doi: 10.1111/j.1440-1827.2004.01681.x. [DOI] [PubMed] [Google Scholar]
- 23.Aishima S, Matsuura S, Terashi T, et al. Aberrant expression of laminin gamma 2 chain and its prognostic significance in intrahepatic cholangiocarcinoma according to growth morphology. Modern Pathology. 2004;17(8):938–945. doi: 10.1038/modpathol.3800143. [DOI] [PubMed] [Google Scholar]
- 24.Vallacchi V, Daniotti M, Ratti F, et al. CCN3/nephroblastoma overexpressed matricellular protein regulates integrin expression, adhesion, and dissemination in melanoma. Cancer Research. 2008;68(3):715–723. doi: 10.1158/0008-5472.CAN-07-2103. [DOI] [PubMed] [Google Scholar]
- 25.Triebel S, Bläser J, Reinke H, Tschesche H. A 25 kDa α2-microglobulin-related protein is a component of the 125 kDa form of human gelatinase. FEBS Letters. 1992;314(3):386–388. doi: 10.1016/0014-5793(92)81511-j. [DOI] [PubMed] [Google Scholar]
- 26.Tong Z, Kunnumakkara AB, Wang H, et al. Neutrophil gelatinase-associated lipocalin: a novel suppressor of invasion and angiogenesis in pancreatic cancer. Cancer Research. 2008;68(15):6100–6108. doi: 10.1158/0008-5472.CAN-08-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai Cheong JE, Wessagowit V, McGrath JA. Molecular abnormalities of the desmosomal protein desmoplakin in human disease. Clinical and Experimental Dermatology. 2005;30(3):261–266. doi: 10.1111/j.1365-2230.2005.01736.x. [DOI] [PubMed] [Google Scholar]
- 28.Berchem G, Glondu M, Gleizes M, et al. Cathepsin-D affects multiple tumor progression steps in vivo: proliferation, angiogenesis and apoptosis. Oncogene. 2002;21(38):5951–5955. doi: 10.1038/sj.onc.1205745. [DOI] [PubMed] [Google Scholar]
- 29.Beaujouin M, Liaudet-Coopman E. Cathepsin D overexpressed by cancer cells can enhance apoptosis-dependent chemo-sensitivity independently of its catalytic activity. Advances in Experimental Medicine and Biology. 2008;617:453–461. doi: 10.1007/978-0-387-69080-3_44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suttiprapa S, Mulvenna J, Huong NT, et al. Ov-APR-1, an aspartic protease from the carcinogenic liver fluke, Opisthorchis viverrini: functional expression, immunolocalization and subsite specificity. The International Journal of Biochemistry & Cell Biology. 2009;41(5):1148–1156. doi: 10.1016/j.biocel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerke V, Moss SE. Annexins: from structure to function. Physiological Reviews. 2002;82(2):331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 32.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nature Reviews Molecular Cell Biology. 2005;6(6):449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 33.Alfonso P, Cañamero M, Fernández-Carbonié F, Núñez A, Casal JI. Proteome analysis of membrane fractions in colorectal carcinomas by using 2D-DIGE saturation labeling. Journal of Proteome Research. 2008;7(10):4247–4255. doi: 10.1021/pr800152u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of secreted proteins identified by tryptic digest and LC/MS/MS from 4 hepatocellular carcinoma cell lines (Alexander, HepG2, SK-Hep-1 and HCC-S102).