SUMMARY
Notch signaling plays multiple roles to direct diverse decisions regarding cell fate during T cell development. During helper T (Th) cell differentiation, Notch is involved in generating optimal Th2 cell responses. Here, we present data investigating how Notch mediates Th2 cell differentiation. Notch showed a CD4+ T cell intrinsic role in promoting IL-4 expression that required GATA-3. In the absence of Notch signals, Gata3 expression was markedly diminished. Introduction of an activated allele of Notch1 into CD4+ T cells led to the specific and direct upregulation of a developmentally regulated Gata3 transcript that included the exon 1a sequences. Furthermore, Notch acted in parallel with GATA-3 to synergistically activate IL-4 expression. Together, these data implicate Gata3 as a direct transcriptional Notch target that acts in concert with Notch signaling to generate optimal Th2 cell responses.
INTRODUCTION
Notch signaling initiates when a Notch receptor is engaged by a Jagged or Delta-like ligand (Maillard et al., 2005). This results in two proteolytic cleavages that release the intracellular domain of Notch (ICN) from the membrane, allowing it to translocate into the nucleus. In the nucleus, ICN binds the transcription factor CSL (gene name Rbpj) enabling Mastermind-like (MAML) (Nam et al., 2003; Wu et al., 2000) to bind to a groove at the interface between ICN and CSL (Nam et al., 2006). MAML in turn recruits critical coactivators, such as p300, that are required for transcriptional activation (Oswald et al., 2001; Wallberg et al., 2002; Wu et al., 2000).
During T cell development, Notch signals are critical for initial T cell specification from multipotent progenitors (Pui et al., 1999; Radtke et al., 1999; Sambandam et al., 2005) and development of the αβ T cell lineage (Maillard et al., 2006; Tanigaki et al., 2004; Wolfer et al., 2002). Outside of the thymus, Notch has roles in shaping peripheral T cell immunity (Adler et al., 2003; Amsen et al., 2004; Maekawa et al., 2003; Minter et al., 2005; Tanaka et al., 2006; Tu et al., 2005). Recent studies have focused on how Notch influences helper T (Th) cell differentiation. When naive CD4+ helper T cells encounter antigen, they differentiate into effector cells that secrete cytokines mediating immunity toward infectious organisms (Murphy and Reiner, 2002). Th1 cells secrete IFN-γ, express the transcription factor T-bet, and protect against intracellular microbes (Szabo et al., 2003). Th2 cells produce the cytokines interleukin-4 (IL-4), IL-5, and IL-13, express the transcription factor GATA-3, and are involved in immunity against extracellular parasites (Zheng and Flavell, 1997; Zhu et al., 2004).
Notch has been implicated in almost all aspects of helper T cell differentiation from negatively or positively regulating T cell activation to having a role in either Th1 or Th2 differentiation (Adler et al., 2003; Amsen et al., 2004; Eagar et al., 2004; Maekawa et al., 2003; Minter et al., 2005; Tanaka et al., 2006; Tu et al., 2005). Gain-of-function studies suggest that Notch has the capacity to drive either Th1 or Th2 development (Maekawa et al., 2003; Minter et al., 2005) or both (Amsen et al., 2004). In contrast, two different approaches that inhibit Notch signaling by specifically targeting CSL-dependent signaling via either conditional deletion of Rbpj or expression of a dominant-negative form of MAML (DNMAML) identified defects only in IL-4 production (Amsen et al., 2004; Tu et al., 2005). In addition, DNMAML mice failed to mount a protective Th2 response against Trichuris muris (Tu et al., 2005), revealing a physiologic function for Notch in Th2 differentiation. A recent report suggests that Notch is involved in promoting very rapid IL-4 production in cells such as memory-type Th2 cells or NKT cells (Tanaka et al., 2006). Although no defects were seen in IFN-γ production in either the conditional Rbpj-deficient or the DNMAML mice, pharmacological inhibition of the Notch pathway via γ-secretase inhibitors (GSIs) impaired IFN-γ secretion (Minter et al., 2005), suggesting a role for Notch in Th1 differentiation. Thus, the requirement for Notch signaling during Th1 or Th2 differentiation may depend on genetic background, disease model, or mode of Notch inhibition.
In this report, we focus on the mechanism underlying Notch-mediated IL-4 production. At the transcriptional level, Il4 is regulated by signals downstream of the T cell receptor as well as the transcription factors GATA-3, STAT6, and NFAT (Ansel et al., 2006). IL-4 transduces signals through the IL-4 receptor, which leads to phosphorylated STAT6 that activates transcription of both Il4 and Gata3 (Ouyang et al., 2000; Zhu et al., 2001). This positive reinforcement contributes to the increasingly stable expression of Th2 cytokines. Th2 differentiation is accompanied by progressive changes in chromatin structure; these changes include histone modifications and DNA methylation, which regulate accessibility across the Th2 gene cluster (Agarwal and Rao, 1998; Avni et al., 2002; Lee et al., 2002). In particular, DNase I hypersensitivity sites (HSs) have marked regions that possess regulatory function (Fields et al., 2004; Takemoto et al., 2000). HS V and HS VA of Il4 were deleted in tandem and resulted in impaired IL-4 expression in both CD4+ T cells and mast cells. This enhancer region contains a conserved CSL-binding site that is within HS V and that is responsive to Notch signals (Amsen et al., 2004; Tanaka et al., 2006). Therefore, Notch may induce IL-4 expression at least in part through this region.
GATA-3 exerts its effects as a key regulator of Th2 differentiation in several ways. GATA-3 is involved in acute transcription of Il5 and Il13 and binds to these promoters (Kishikawa et al., 2001; Zhang et al., 1998; Zhu et al., 2004). GATA-3 is also sufficient to induce changes in chromatin structure at the Il4 locus, and these mondifications contribute to Th2 differentiation. (Lee et al., 2001; Lee et al., 2000; Yamashita et al., 2004). Known signals that activate Gata3 transcription during Th2 differentiation are IL-4, STAT6, and GATA-3 itself (Kurata et al., 1999; Ouyang et al., 2000; Zhu et al., 2001). Gata3 is expressed at various stages of T cell development, starting with the earliest steps of T cell commitment (Hoflinger et al., 2004; Sambandam et al., 2005). Similar to other GATA family members (Minegishi et al., 1998), Gata3 utilizes alternative promoters in different tissues (Asnagli et al., 2002). In developing Th2 cells, Gata3 transcripts include the proximal exon 1a, whereas in thymocytes, distal exon 1b predominates. However, the mechanism by which these alternative exons are regulated has not been previously addressed.
Here, we present data demonstrating that in addition to a defect in IL-4 expression, Gata3 expression was also impaired in the absence of Notch signals. Notch associated with Gata3 regions that contained CSL-binding sites, and this was accompanied by changes in histone acetylation. In addition, Notch preferentially induced expression of the developmentally regulated Gata3 transcript that included exon 1a sequences. Furthermore, GATA-3 activity was required for Notch induction of IL-4. The dependence of Notch on GATA-3 was consistent with a synergistic effect observed between Notch and GATA-3 in promoting IL-4 expression. Together, these data implicate Gata3 as a direct downstream Notch target that cooperates with Notch signaling to generate an optimal Th2 response.
RESULTS
Notch Has a Cell-Intrinsic Role in Regulating IL-4 in Peripheral CD4+ T Cells
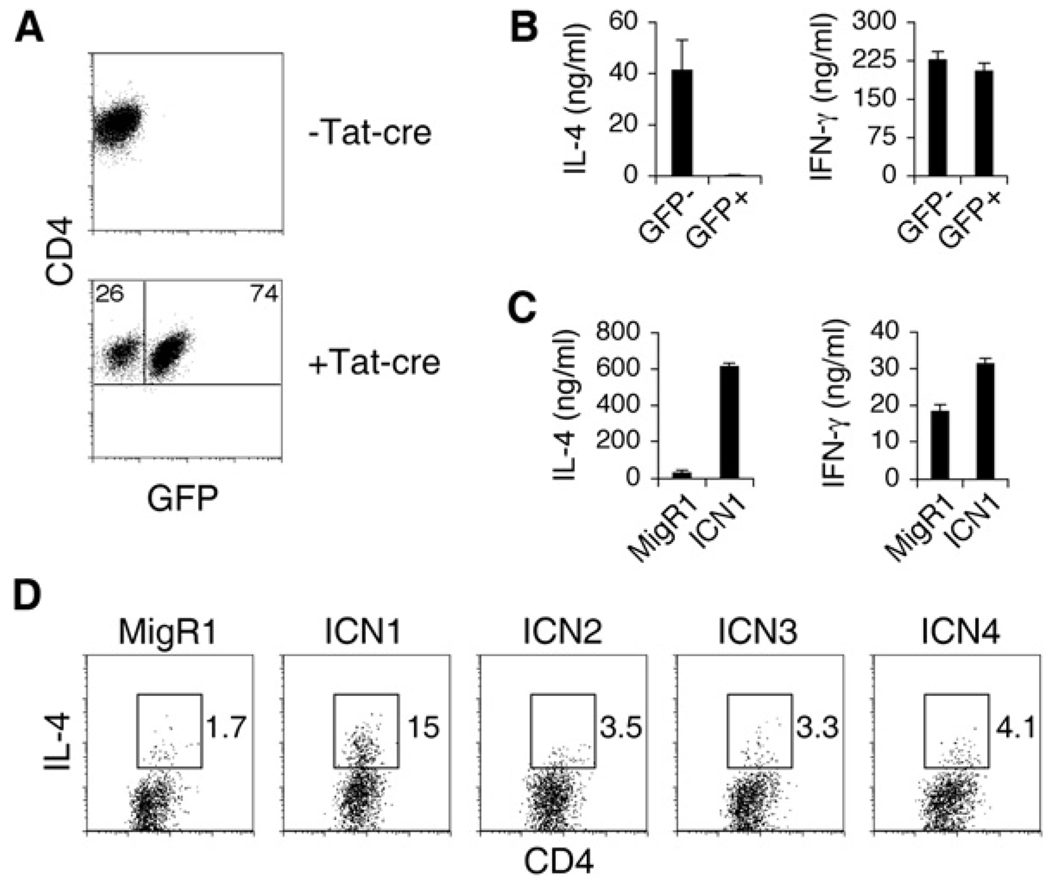
We previously found that breeding ROSA26-DNMAML (dominant-negative MAML) mice with CD4-cre mice resulted in normal lymphocyte development (Tu et al., 2005). However, IL-4 production by peripheral CD4+ T cells was defective. Although T cell development appeared normal in these mice, we wanted to confirm that the resulting IL-4 defect was not due to a defect acquired when the DNMAML transgene was turned on at the double-positive stage in the thymus. To address this, we took an approach to specifically activate DNMAML expression in peripheral naive CD4+ T cells. DNMAML mice were bred with DO11.10 mice that have TCRα and β transgenes specific for ovalbumin (OVA). CD4+ T cells from these mice do not express GFP because there was no Cre recombinase to turn on the DNMAML-GFP protein (Figure 1A). CD4+ T cells were sorted and treated with TAT-cre peptide, which is cell permeable (Peitz et al., 2002). The cells were then stimulated with irradiated antigen presenting cells (APCs) and OVA peptide. After 7 days, cultures contained a mixture of GFP− and GFP+ (expressing DNMAML) CD4+ T cells that were then sorted and restimulated. There was no difference in IFN-γ production between GFP− and GFP+ cells (Figure 1B). However, GFP+ cells produced markedly less IL-4, in concordance with previous results when DNMAML was expressed in the thymus (Tu et al., 2005). In the reciprocal experiment, retroviral expression of an activated allele of Notch1 (ICN1) potently induced (>50-fold) IL-4 when introduced into CD4+ T cells (Figure 1C), whereas its effect on IFN-γ production was modest (~1.5-fold increase). Activated alleles of other Notch family members induced smaller amounts of IL-4 compared to the effect of ICN1 (Figure 1D). Thus, Notch has a CD4+ T cell-intrinsic role in inducing IL-4 expression, and Notch1 is the strongest inducer of IL-4.
Figure 1. Notch Regulates IL-4 in CD4+ T Cells.
(A) Tat-cre induces expression of DNMAML. DO11.10 DNMAMLf/+ CD4+ T cells were treated with Tat-Cre to induce expression of DNMAML. After 24 h, cells were stimulated with irradiated APCs and OVA peptide in the presence of IL-2. GFP− and GFP+ CD4+ T cells were sorted on day 7. Flow-cytometry plots display GFP expression prior to Tat-cre treatment and 7 days after culture.
(B) DNMAML expression in peripheral CD4+ T cells results in impaired IL-4 expression. Equal numbers of GFP− and GFP+ CD4+ cells from (A) were restimulated with plate-bound α-CD3, and supernatants were collected after 48 hr to determine quantities of IL-4 or IFN-γ by ELISA. Graphs represent the mean of values of triplicate wells ±SD. Data are representative of three independent experiments.
(C) Notch1 induces IL-4 expression. Wild-type Balb/C CD4+ T cells were retrovirally transduced with either the control MigR1 or ICN1 (activated Notch1). GFP− and GFP+ CD4+ T cells were sorted after 6 days. Equal numbers of GFP− and GFP+ cells were restimulated with plate-bound α-CD3, and supernatants were collected after 48 hr for ELISA. Graphs represent the mean of values of triplicate wells ±SD. Data are representative of six independent experiments.
(D) Weak induction of IL-4 by other Notch family members. Wild-type Balb/C CD4+ T cells were retrovirally transduced with either the control MigR1 or various activated Notch alleles. Cells were restimulated with PMA and Ionomycin on day 6. Flow-cytometry plots represent GFP+ cells. Numbers indicate the percentage of CD4+IL-4+ cells within the gates. Data are representative of three independent experiments.
Notch Regulates Gata3
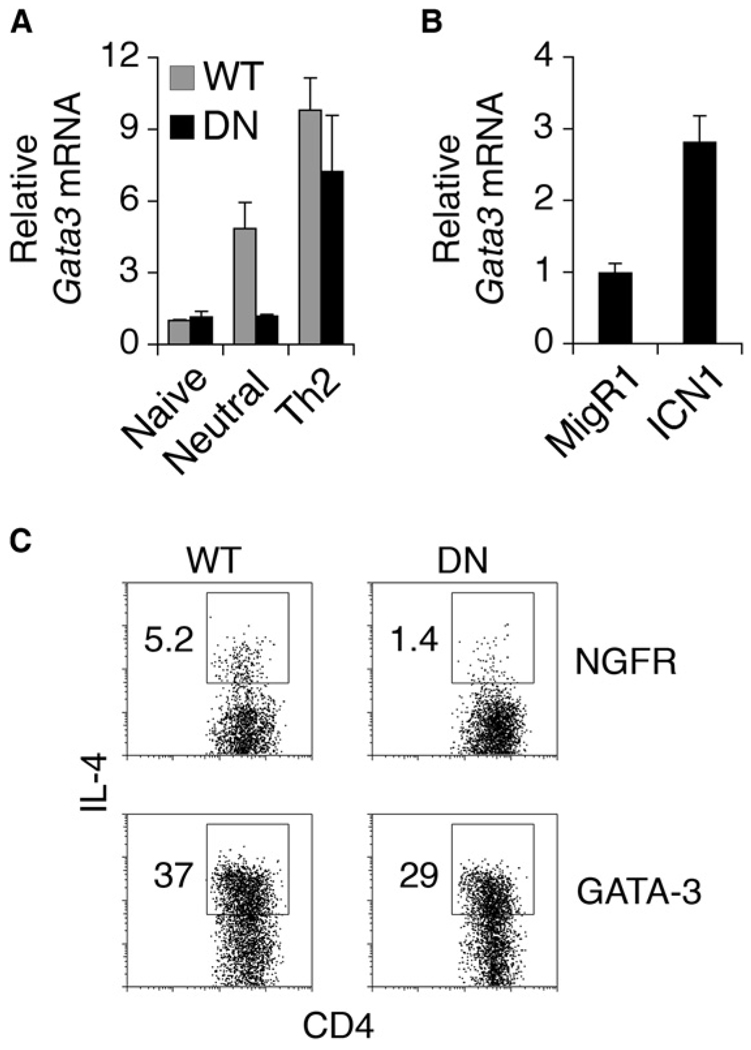
Because GATA-3 is a major factor involved in Th2 differentiation and there had been suggestions that Gata3 was a Notch target in CD4+ T cells (Amsen et al., 2004), Gata3 expression was examined in the absence of Notch signals (Figure 2A). After 7 days of culture, Gata3 expression was markedly reduced in DNMAML CD4+ T cells compared to the wild-type (WT) controls. Culturing cells in Th2 conditions (IL-4 and α-IL-12) restored Gata3 expression to that of wild-type (Figure 2A), probably because of the combined effects of inducing Gata3 expression via IL-4 and blocking IL-12 suppression of Gata3. In addition, ICN1 upregulated Gata3 expression compared to the vector control (Figure 2B). To determine whether restoring GATA-3 expression would restore IL-4 production in the absence of Notch signals, we transduced wild-type or DNMAML CD4+ T cells with either a control virus expressing nerve growth factor receptor (NGFR) or GATA-3-NGFR (Figure 2C). With the control NGFR virus, DNMAML CD4+ T cells exhibited decreased IL-4 production compared to the control. Expression of GATA-3 induced abundant amounts of IL-4 in both wild-type and DNMAML CD4+ T cells. Therefore, expression of GATA-3 rescued IL-4 expression in the absence of Notch signaling.
Figure 2. Notch Signaling Regulates Gata3 Expression.
(A) Diminished Gata3 expression in the absence of Notch. Wild-type or DNMAML CD4+ T cells that were naive and cultured in neutral or Th2 conditions for 7 days were purified. Abundance of Gata3 mRNA was determined by quantitative RT-PCR. Gata3 expression relative to Hprt is shown as a mean of values from duplicate wells ±SD. Data are representative of three independent experiments.
(B) Notch1 induces Gata3 expression. Wild-type Balb/C CD4+ T cells were retrovirally transduced with either the control MigR1 or ICN1. CD4+GFP+ T cells were sorted by flow cytometry. Gata3 expression relative to Hprt is shown as a mean of values from duplicate wells ±SD. Data are representative of three independent experiments.
(C) GATA-3 rescues the IL-4 defect in DNMAML CD4+ T cells. Wild-type or DNMAML CD4+ T cells were retrovirally transduced with either the control NGFR or GATA-3-NGFR. Cells were restimulated with PMA and Ionomycin on day 6. Plots show NGFR+ cells. Numbers indicate the percentage of CD4+IL-4+ cells within the gates. Data are representative of three independent experiments.
Notch Associates with Gata3 Regions that Contain CSL-Binding Sites
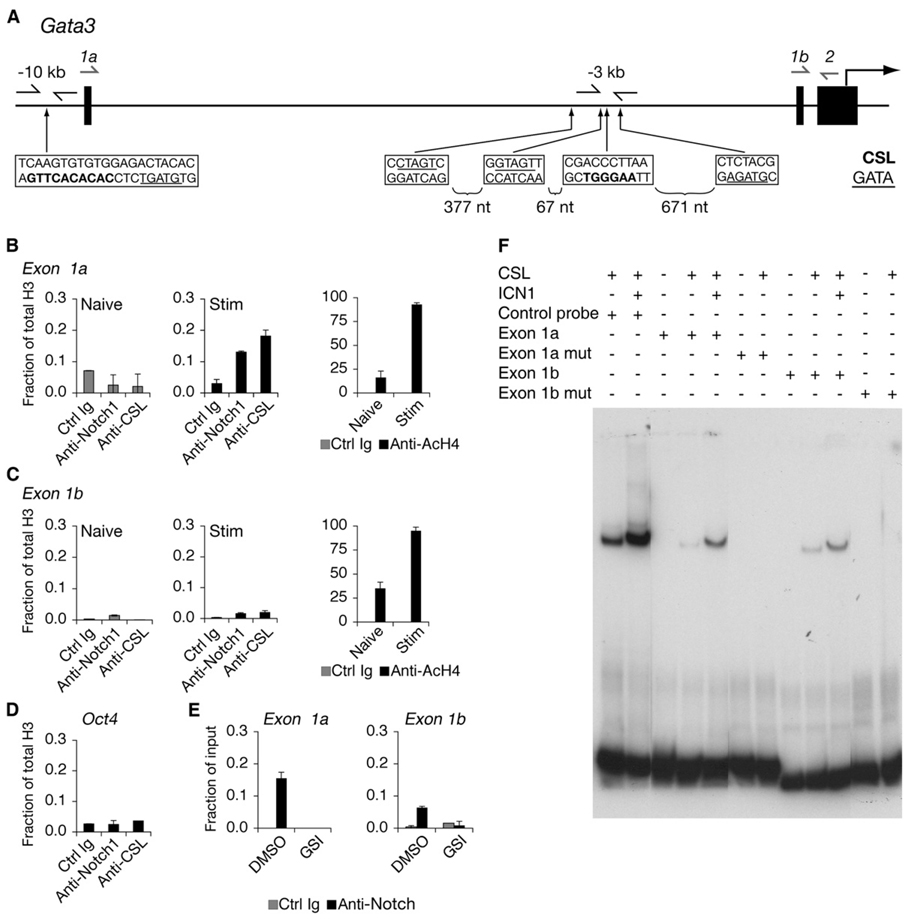
To further investigate the possibility that Notch may regulate Gata3, we searched for putative CSL-binding sites within the Gata3 gene locus. We identified multiple CSL sites and focused our attention on sites that were conserved between mouse and human Gata3. Conserved CSL-binding sites were found in regions of high DNA homology −3, −5, and −10 kb upstream of the translational start site (Figure 3A). Primers flanking these regions were designed to perform chromatin immunoprecipitation (ChIP). The −10 kb site was ~ 200 bp upstream of the alternative exon 1a and was denoted the “1a site.” The −3 kb site was −2.5 kb upstream of exon 1b and was referred to as the “1b site.” We compared naive CD4+ T cells, which do not contain activated Notch, to cells that were stimulated for 40 hr, a time point shown to have activated Notch protein (Adler et al., 2003; Amsen et al., 2004; Palaga et al., 2003). Naive cells served as a negative control, and PCR of α-Notch1 ChIP samples did not amplify DNA sequences corresponding to the CSL-binding sites of Gata3 (Figure 3B). Forty hours after stimulation, Notch1 associated with the −10 kb exon 1a CSL-binding site of Gata3. Very low binding of Notch1 occurred at the −3 kb exon 1b site of Gata3 (Figure 3C) that was similar to binding of an irrelevant control (Figure 3D). However, we consistently observed association at the 1b site, which could be inhibited by addition of GSI, suggesting that this was a specific interaction (Figure 3E). Like Notch1, CSL preferentially bound the exon 1a site in activated CD4+ T cells. In addition, histone 4 (H4) became hyperacetylated at both the 1a and 1b sites after stimulation, indicating that these are regions of open chromatin and are possible regulatory regions of Gata3 (Figures 3B and 3C). Notch binding did not occur at the −5 kb site (data not shown). An electrophoretic mobility shift assay (EMSA) was performed with probes corresponding to the 1a or 1b sites and CSL or CSL-ICN1 complexes (Figure 3F). No intrinsic difference was observed between the 1a or 1b probes. Binding was abrogated with mutated probes, indicating that the association was specific for the CSL-binding site. The lack of difference in binding with bare DNA templates suggests that differences in chromatin structure or presence of other factors might influence association of endogenous Notch with putative target sites. Thus, Notch has the potential to regulate Gata3 through either of these sites.
Figure 3. Notch Binds to Regions of Gata3 Containing CSL-Binding Sites.
(A) Schematic of the 5′ region of the Gata3 locus. Gata3 transcription can initiate from either exon 1a or exon 1b. The translational start site begins within exon 2. Locations of nucleotides matching the CSL-binding sites are shown in bolded sequences. GATA sites are underlined. Arrows immediately flanking these sites are primers used to amplify ChIP DNA. Arrows drawn above exon 1a or exon 1b were used as forward primers coupled with a reverse primer that annealed to exon 2 to amplify mRNA transcripts. The bottom strand in each box is indicated 5′ to 3′.
(B) Notch1 binds to the exon 1a CSL-binding site. Naive or CD4+ T cells stimulated for 40 hr were purified and fixed for ChIP. Antibodies used for IP are a control IgG, α-Notch1, α-CSL, α-acetylated H4 (AcH4), and α-H3. Total histone H3 was used for normalization. Graphs represent quantitative PCR presented as a mean of the ratios of the amount of IP DNA in each sample relative to total H3 from values of duplicate wells ±SD. Data are representative of at least three independent experiments.
(C) Notch1 binds weakly to the exon 1b CSL-binding site. The description is the same as (B), except the primers amplified the CSL-binding site upstream of exon 1b.
(D) ChIP from CD4+ T cells stimulated for 40 hr and quantitative PCR for an irrelevant gene control (Oct4). Data are representative of at least three independent experiments.
(E) GSI treatment blocks association of Notch1 with Gata3 CSL-binding sites. CD4+ T cells were treated with either DMSO (vehicle control) or GSI and stimulated for 40 hr, and ChIP was performed. Data are representative of three independent experiments.
(F) CSL binds to putative target sequences within the Gata3 locus. Wild-type or mutant oligonucleotides corresponding to exon 1a or exon 1b CSL-binding sites were labeled with 32P and mixed with buffer alone or buffer containing purified CSL or Notch1 polypeptides, or both, and electrophoresed. Gels were dried, and probes were detected by autoradiography. The control probe contained a consensus CSL-binding site.
Notch Regulates Exon 1a of Gata3
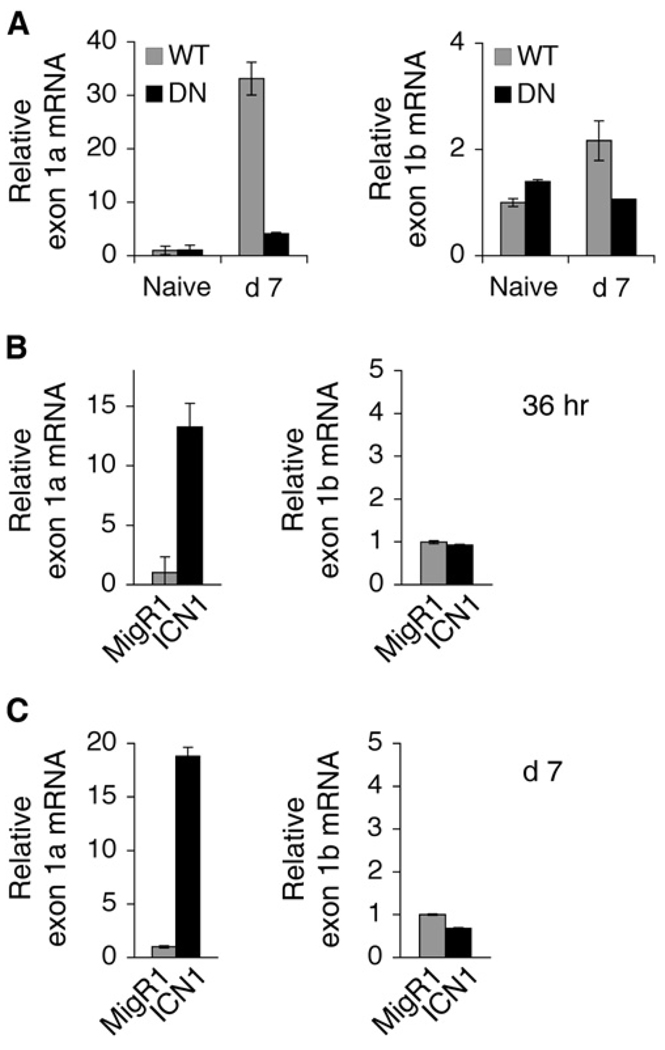
A previous report demonstrated that Gata3 transcripts including exon 1a were preferentially expressed in developing Th2 cells (Asnagli et al., 2002). We designed primers to distinguish between the 1a and 1b transcripts (Figure 3A). DNMAML CD4+ T cells that were stimulated with α-CD3 and α-CD28 and cultured for 7 days in the presence of IL-2 displayed a relative decrease in both transcripts compared to the wild-type control (Figure 4A). Enforced expression of ICN1 in CD4+ T cells induced exon 1a transcripts but had no effect on exon 1b expression (Figures 4B and 4C). Exon 1a transcripts were augmented as early as 36 hr after transduction, reducing the likelihood that the change was due to indirect effects, and persisted at day 7 after transduction, whereas there was still no alteration in exon 1b expression at this time point (Figure 4B). These data reveal that Notch regulates Gata3 expression by augmenting amounts of the alternative exon 1a transcript in CD4+ T cells.
Figure 4. Notch Preferentially Induces Expression of Gata3 Transcripts Including exon 1a.
(A) Expression of exon 1a and 1b transcripts is diminished in the absence of Notch signals. Wild-type or DNMAML CD4+ T cells that were naive or cultured in neutral conditions for 7 days were purified. mRNA expression of exon 1a or exon 1b of Gata3 were determined by quantitative RT-PCR. Primers used are illustrated in Figure 3A. Amounts of transcript compared to Hprt are shown as means of values from duplicate wells ±SD. Data are representative of three independent experiments.
(B) Rapid induction of exon 1a expression by Notch1. Wild-type Balb/C CD4+ T cells were retrovirally transduced with either the control MigR1 or ICN1. CD4+GFP+ T cells were sorted 36 hr after transduction. Relative abundance of exon 1a or exon 1b transcripts compared to Hprt is shown as a mean of values from duplicate wells ±SD. Data represent two independent experiments.
(C) Notch1 has no effect on exon 1b expression. The description is the same as (B), except cells were sorted 7 days after transduction. Data represent three independent experiments.
Induction of IL-4 by Notch Requires GATA-3
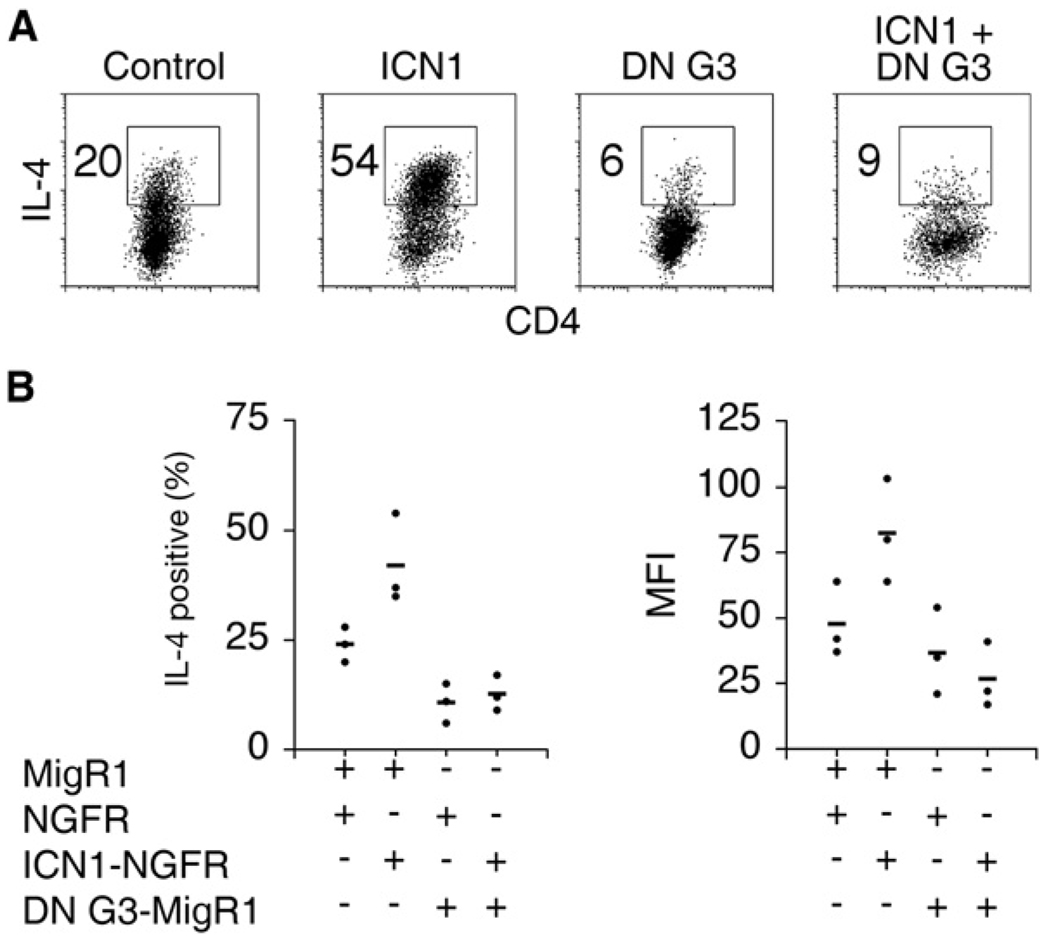
GATA-3 is essential for expression of Th2 cytokines (Pai et al., 2003; Zhu et al., 2004). A previous report showed that the distal 3′ enhancer of Il4 was directly responsive to Notch signals (Amsen et al., 2004). Therefore, we sought to determine whether Notch could induce IL-4 independently of GATA-3. To block GATA-3 activity, we used a dominant-negative GATA-3 allele (DN G3) in which the DNA-binding domain of GATA-3 is fused to the Drosophila Engrailed repression domain. This construct was shown to inhibit IL-4 production in developing Th2 cells (Martins et al., 2005). CD4+ T cells were cotransduced with a retrovirus expressing DN G3 (GFP) and ICN1 (NGFR) and cultured in Th2 conditions to achieve high IL-4 expression. Even in Th2 conditions, ICN1 enhanced the abundant amounts of IL-4 production in CD4+ T cells. Expression of DN G3 significantly decreased both the percentage of IL-4+ cells (Figures 5A and 5B) and the amount of IL-4 produced per cell in comparison to controls (Figures 5A and 5B). Furthermore, ICN1 failed to augment IL-4 production in the presence of DN G3. Therefore, the ability of Notch to induce IL-4 depends on GATA-3 activity.
Figure 5. IL-4 Induction by Notch Is Dependent on GATA-3 Activity.
(A) DN G3 antagonizes Notch1 induced IL-4 expression. Wild-type CD4+ T cells were cotransduced with ICN1 (NGFR), DN G3 (GFP), or control retroviruses and cultured under Th2 conditions. Cells were restimulated with PMA and Ionomycin on day 6 and stained for intracellular IL-4. Flow-cytometry plots show GFP+NGFR+ cells. Numbers represent the percentage of IL-4+ cells within the gates.
(B) Summary of independent experiments. Individual points on graphs represent results from separate experiments. The mean of the points within each group is shown as a horizontal line. The left plot displays the percentage of IL-4 cells as shown in (A). The right graph shows the mean fluorescence intensity of IL-4 as an indication of the amount of IL-4 expressed per cell.
Notch and GATA-3 Synergize to Promote IL-4 Expression
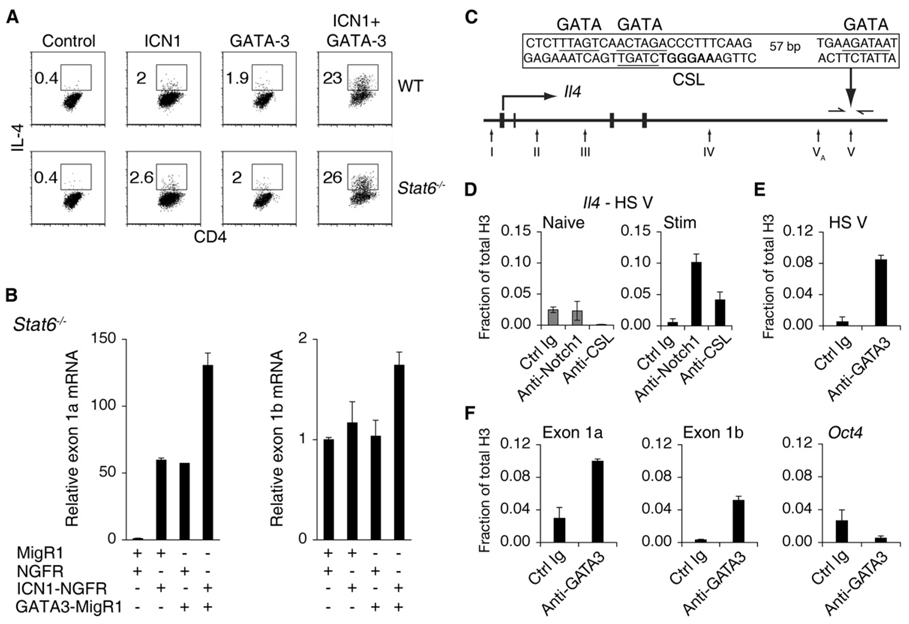
The preceding results demonstrated that Notch-mediated IL-4 expression is dependent on GATA-3. These data also show that one mechanism by which Notch influences IL-4 expression is via direct transcriptional control of Gata3. In addition, the existence of both GATA-3- (Agarwal et al., 2000; Avni et al., 2002) and CSL- (Amsen et al., 2004) binding sites in the Il4 locus suggests that Notch and GATA-3 may coordinately regulate the Il4 locus. To address this issue, we transduced CD4+ T cells with retroviruses expressing ICN1 and GATA-3 and cultured the cells in Th1 conditions, which include factors (recombinant IL-12 and α-IL-4) that strongly suppress both Gata3 expression and Th2 differentiation. Nevertheless, ICN1, by itself, induced IL-4 expression to a similar extent as GATA-3 alone (Figure 6A). Strikingly, coexpression of ICN1 and GATA-3 resulted in an even more potent effect on IL-4 expression than either alone. This synergistic effect of Notch and GATA-3 suggests that these two factors collaborate to induce IL-4 expression. Moreover, this effect was independent of STAT6 and any further positive feedback from IL-4, thereby demonstrating the robust effect that Notch and GATA-3 have together (Figure 6A).
Figure 6. Notch and GATA-3 Synergize to Induce IL-4 Expression Independently of STAT6.
(A) Notch1 and GATA-3 synergize to induce IL-4 expression. Wild-type or Stat6−/− CD4+ T cells were cotransduced with ICN1 (NGFR), GATA-3 (GFP), or control retroviruses and cultured under Th1 conditions. Cells were restimulated with PMA and Ionomycin on day 6 and stained for intracellular IL-4. Flow-cytometry plots show GFP+NGFR+ cells. Numbers represent the percentage of IL-4+ cells within the gates. Data are representative of three independent experiments.
(B) Notch1 relieves suppression of Gata3 expression independently of STAT6. Stat6−/− CD4+ T cells were cotransduced with ICN1 (NGFR), GATA-3 (GFP), or control retroviruses and cultured under Th1 conditions. GFP+NGFR+CD4+ T cells were sorted 6 days after transduction. Relative abundance of exon 1a or exon 1b transcripts compared to Hprt is shown as a mean of values from duplicate wells ±SD. Data represent three independent experiments.
(C) Schematic of the Il4 locus. HS sites are indicated by arrows below the locus. A conserved CSL site (nucleotides matching the consensus CSL-binding sites are shown in bold) is located within HS V. Conserved GATA sites are underlined. Arrows flanking HS V were used to amplify ChIP sequences. The bottom strand in each box is indicated 5′ to 3′.
(D) Notch1 binds to the CSL-binding site in HS V of Il4. Naive or CD4+ T cells stimulated for 40 hr were purified and fixed for ChIP. Antibodies used for IP are a control IgG, α-Notch1, α-CSL, α-acetylated H4 (AcH4), and α-H3. Graphs represent quantitative PCR presented as a mean of the ratios of the amount of IP DNA in each sample relative to total H3 from values of duplicate wells ±SD. Data are representative of at least three independent experiments.
(E) GATA-3 associates with HS V of Il4. CD4+ T cells stimulated for 40 hr were purified and fixed for ChIP. Antibodies used for IP are a control IgG, α-GATA-3, and α-H3. Data are representative of at least three independent experiments.
(F) GATA-3 associate with regions upstream of exon 1a and 1b of Gata3. The description is same as that of (E). Oct4 was used as a negative control for GATA-3 binding.
GATA-3 autoregulates its own expression in CD4+ T cells (Ouyang et al., 2000). This occurs in the absence of STAT6 and therefore independently of IL-4 signaling. In addition, retrovirally expressed GATA-3 induces expression of endogenous Gata3 even in the presence of IL-12, a potent Gata3 repressor. Therefore, we investigated whether Notch could regulate expression of exon 1a-derived transcripts in a similar fashion. Stat6−/− CD4+ T cells were used to distinguish direct effects from effects mediated through IL-4, a known mechanism of Gata3 induction, and cultured in Th1 conditions to suppress endogenous Gata3 expression. Seven days after retroviral transduction by ICN1 (NGFR), GATA-3 (GFP), or both, GFP+NGFR+ cells were sorted for analysis of the abundance of Gata3 mRNA. ICN1 or GATA-3 preferentially upregulated exon 1a-derived transcripts and when expressed together had an additive effect on exon 1a expression (Figure 6B). Neither ICN1 nor GATA-3 had an effect on the exon 1b transcript when expressed alone. However, there was a modest effect when ICN1 and GATA-3 were expressed together (Figure 6B). These data reveal that the effect of Notch in promoting Gata3 expression is sufficiently potent to oppose IL-12-mediated repression of Gata3 and occurs independently of signaling through IL-4. Furthermore, these findings suggest that Notch and GATA-3 coordinately regulate preferential induction of Gata3 exon 1a expression.
The 3′ Il4 enhancer contains a CSL-binding site within HS V (Figure 6C). This site mediates responsiveness of the Il4 locus to Notch1 (Amsen et al., 2004). Consistent with these findings, α-Notch1 ChIP demonstrated that Notch1 binds directly to HS V (Figure 6D). In addition, conserved GATA-binding sites were found within HS V (Figure 6C), and ChIP showed GATA-3 binding in this region (Figure 6E). The synergistic effect of Notch1 and GATA-3 may involve cooperation at these sites within HS V.
An additive effect was also seen with combined expression of Notch1 and GATA-3 at the Gata3 locus. A conserved GATA-binding site is located immediately adjacent to the exon 1a CSL-binding site and GATA-3 bound to this region (Figure 3A and Figure 6F). GATA-3 also associated with the region surrounding the exon 1b CSL site (Figure 6F). Conserved GATA-binding sites are located within the region amplified by the exon 1b primers (Figure 3A). However, their locations are not immediately adjacent to the CSL site. These data suggest that there may also be direct cooperation of Notch1 and GATA-3 at the Gata3 locus.
DISCUSSION
Recent data identified important roles for Notch in generating Th2 immunity (Amsen et al., 2004; Tu et al., 2005). Inhibiting Notch signaling by either expression of DNMAML or deletion of Rbpj at the double positive (DP) stage did not perturb T cell development but impaired IL-4 production in peripheral CD4+ T cells. We now demonstrate that blocking Notch signaling specifically in peripheral naive CD4+ T cells also results in loss of IL-4 production. Furthermore, introduction of an activated allele of Notch1 into naive CD4+ T cells results in robust IL-4 induction. Together, these data demonstrate that Notch regulates IL-4 production in a peripheral CD4+ T cell-intrinsic manner. Of the four mammalian Notch receptors, retroviral expression of the intracellular domain of Notch1 in CD4+ T cells strongly induces IL-4 production. However, the intracellular domains of Notch2–Notch4 weakly induce IL-4 expression. Because IL-4 production was not impaired in the absence of Notch1 (Tacchini-Cottier et al., 2004), signaling by one or more of the other Notch receptors may rescue Notch1 deficiency.
Previous reports suggested a relationship between Notch and GATA-3 and that GATA-3 may be downstream of Notch (Amsen et al., 2004; Hoflinger et al., 2004; Sambandam et al., 2005). We have now utilized a loss-of-function approach with DNMAML to show that the absence of CSL-dependent Notch signaling correlates with decreased Gata3 expression. We have further shown that Notch directly regulates Gata3 expression. Located ~10 kb apart are two alternative noncoding first exons of Gata3, termed exon 1a and exon 1b. Expression of Gata3 from exon 1a- or exon 1b-derived transcripts appears to vary in different contexts. For example, Gata3 transcripts including exon 1a are expressed in developing Th2 and neural cells, whereas transcripts including exon 1b predominate in thymocytes (Asnagli et al., 2002). The mechanism that influences alternative exon selection has not been previously addressed. We now show that Notch preferentially influences expression of Gata3 transcripts including exon 1a. Although both ChIP and EMSA experiments showed association of Notch1 with conserved CSL sites that are proximal to both exon 1a and exon 1b, gain-of-function experiments demonstrated a relatively large change in transcripts from exon 1a, whereas there was no effect on exon 1b-derived transcripts. This suggests that the major effect of Notch signaling at the Gata3 locus is to regulate exon 1a transcription. This preferential activation of the exon 1a transcript may have broad implications for the way Notch activates other genes and how Gata3 is developmentally regulated. For example, Gata3 may be a Notch target during the earliest stages of intrathymic T cell development, and these stages are Notch dependent (Sambandam et al., 2005). In contrast, Gata3 is likely to be expressed independently of Notch in double-positive and single-positive thymocytes, which display minimal Notch dependence (Tanigaki et al., 2004; Tu et al., 2005; Wolfer et al., 2001).
Binding of Notch1 to the 1a and 1b sites was accompanied by H4 hyperacetylation, suggesting that Notch affects the chromatin structure of Gata3. We also detected Notch1 binding at the conserved CSL-binding site in Il4 HS V, which was associated with acetylated H4 and p300 recruitment (data not shown), consistent with a direct role for Notch signaling in Il4 activation. In contrast, Notch was reported to regulate IFN-γ expression via direct Notch1 regulation of Tbx21, the gene encoding the Th1 master regulator, T-bet (Minter et al., 2005). However, in the same samples in which we found Notch bound to Gata3 or Il4, we failed to detect endogenous Notch residing at the Tbx21 promoter (data not shown). This is consistent with data obtained from two different loss-of-function approaches that showed in vitro and in vivo defects in Th2 differentiation in the absence of CSL-dependent Notch signaling (Amsen et al., 2004; Tu et al., 2005). Defects in IFN-γ expression or Th1 differentiation have not yet been observed in either of the models that specifically ablate CSL-dependent Notch signaling. These data suggest that the Notch signals that regulate IFN-γ or Th1 differentiation are likely to involve CSL-independent functions of Notch.
GATA-3, IL-4, and STAT6 participate in an autocrine signaling loop that acts positively on Th2 differentiation. In addition to activating transcription of Gata3, Notch also participates in transactivation of the Il4 gene (Amsen et al., 2004). In the presence of DN G3, Notch was unable to induce IL-4 expression, indicating that Gata3 is an obligatory downstream target of Notch for IL-4 expression and Th2 differentiation. Furthermore, our studies suggest that the dependence of Notch on GATA-3 activity is due to a cooperative action of Notch and GATA-3. Although either Notch1 or GATA-3 could lead to IL-4 expression in the absence of STAT6, the combined effect of coexpressing ICN1 and GATA-3 far exceeded their additive effects. Culturing cells in Th1 conditions suppresses the endogenous Gata3 gene. However, expression of the endogenous gene was induced by retroviral GATA-3 even in the absence of STAT6 (Ouyang et al., 2000). Similarly, Notch could relieve repression Gata3 repression independently of STAT6. Either ICN1 or GATA-3 induced a similar and marked upregulation of exon 1a expression without influencing exon 1b expression. Combined expression of ICN1 and GATA-3 further enhanced exon 1a expression and also enhanced (although to a lesser extent) exon 1b expression. Sequence analysis revealed adjacent GATA and CSL-binding sites immediately upstream of exon 1a. The configuration of the GATA-CSL-binding sites is conserved between humans and rodents, suggesting that evolutionary pressure was exerted to maintain these sequences. In contrast, the GATA sites upstream of exon 1b are not immediately adjacent to the CSL site. Together, our data support a model in which either Notch1 or GATA-3 can bind sequences immediately upstream of exon 1a leading to increased exon 1a Gata3 expression; however, the presence of both GATA-3 and Notch1 leads to coordinate binding and much greater quantities of both exon 1a Gata3 transcripts and IL-4.
Tandem deletion of both HS VA and HS V of Il4 impairs IL-4 expression in both CD4+ T cells and mast cells (Solymar et al., 2002). The CSL-binding site resides in HS V, whereas GATA and NFAT sites are located within HS VA. Whether deletion of HS V alone will have a similar effect as inactivation of Notch signals and whether Notch can still induce IL-4 expression in the absence of HS V awaits further studies. GATA-3 binds to HS VA, which is proximal to HS V (Agarwal et al., 2000; Avni et al., 2002). In addition, GATA-3 is sufficient to induce Th2-cell-specific patterns of DNaseI hypersensitivity independently of STAT6 at the Il4 locus (Ouyang et al., 2000). Notch may also participate in the chromatin-remodeling function of GATA-3. The synergistic effect of Notch and GATA-3 could reflect cooperative recruitment of transcriptional activators and histone acetyltransferases, such p300, leading to a transcriptionally permissive chromatin state. Alternatively, chromatin remodeling of Il4 by GATA-3 may be a prerequisite to a separate function of Notch.
In response to microbial stimuli, Notch ligands are upregulated on dendritic cells (Amsen et al., 2004). Activation of Notch signals during the initial events of T cell activation may serve to induce early Gata3 expression. Alternatively, Notch could function later in differentiation to establish stable Gata3 expression. Regardless of its temporal requirement, Notch seems to serve two important roles during Th2 cell fate determination. Notch regulates expression of the alternative exon 1a of Gata3 and induces IL-4 production in a GATA-3-dependent and STAT6-independent mechanism. The CSL-binding sites at the Gata3 locus reside within T cell-specific DNase I hypersensitivity sites that were previously identified (Lieuw et al., 1997). Preliminary data from our laboratory suggest that H4 hyperacetylation at these sites are Notch dependent. The effect of Notch on chromatin structure of Gata3 may also be important for regulating memory Th2 development. The polycomb group protein MLL influences memory Th2 cell development and promotes H3-K9 acetylation at the Gata3 locus (Yamashita et al., 2006). In addition, recent data showed that Notch could regulate IL-4 in NKT cells and in a subset of CD4+ cells that have a memory-like phenotype through a mechanism involving HS V of the Il4 locus (Tanaka et al., 2006). We demonstrated that Notch is required for development of the primary Th2 response during Trichuris muris infection (Tu et al., 2005). Whether Notch is also required for the maintenance of IL-4 or development of memory Th2 cells in vivo awaits further studies. A role for Notch in Th2 memory or the maintenance of IL-4 would suggest that therapeutic approaches that block Notch signaling may benefit diseases associated with excessive production of Th2 cytokines, such as asthma or allergy.
EXPERIMENTAL PROCEDURES
Mice
ROSA26-DNMAML mice were previously described (Maillard et al., 2006; Tu et al., 2005). C57Bl/6, Balb/C, and B10.D2 DO11.10 mice were obtained from Taconic Farms (Taconic, NY). STAT6−/− mice were obtained from Jackson Labs (Bar Harbor, ME). All mice were housed in specific pathogen-free facilities at the University of Pennsylvania. Experiments were performed according to guidelines from the National Institutes of Health with approved protocols from the University of Pennsylvania Animal Care and Use Committee.
TAT-Cre
Expression of TAT-cre was induced with IPTG in bacteria during log phase of growth at 37°C in the presence of chlorampenicol and carbenicillin. Purification of TAT-Cre protein was carried out by the Children’s Hospital of Pennsylvania Protein Core. 107 CD4+ T cells obtained from DNMAMLf/fDO11.10 (B10.D2) mice were washed, resuspended in 1 ml serum-free OPTI-MEM (Life Technologies), and incubated with 1 ml of 100 µg/ml of TAT-Cre in OPTI-MEM for 45 min at 37°C. Cells were then washed and cultured in IMDM (Life Technologies) supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 2 mM L-glutamine, and 50 µM 2-mercaptoethanol.
Quantitative Transcript Analysis
CD4+ T cells were isolated by positive selection with CD4 Microbeads (Miltenyi). RNA was purified with QIAGEN RNeasy Mini Kit. cDNA was synthesized with Superscript II kit (Invitrogen). Real-time quantitative RT-PCR was carried out on an ABI 7900. Transcripts were amplified with either Taqman Universal PCR Master Mix (ABI) or Sybr Green PCR Master Mix (ABI). Primers included the following: Hprt forward 5′-CTCCTCAGACCGCTTTTTGC-3′, Hprt reverse 5′-TAACCTGGTTCATCATCGCTAATC-3′, Hprt probe 5′-VIC-CCGTCATGCCGACCCGCAG-TAMRA-3′, Gata3 forward 5′-CGAGATGGTACCGGGCACTA-3′, Gata3 reverse 5′-GACAGTTCGCGCAGGATGT-3′, Gata3 probe 5′-6FAM-CTGCCGACAGCCTTCGCTTGG-TAMRA-3′. Exon 1a forward 5′-CTGGCTGAGATGCAGTGAAG-3′, Exon 1b forward 5′-AGCTGTCTGCGAACACTGAG-3′, and Exon 2 reverse 5′-GCTCAGAGACGGTTGCTCTT-3′.
Chromatin Immunoprecipitation
ChIP was performed with ChIP assay kits (Upstate Biotechnology). Antibodies used were normal rabbit IgG (Upstate Biotechnology), antiserum specific for Notch1 TAD domain (Maillard et al., 2006), α-CSL (Chemicon), α-GATA3 (Santa Cruz), α-histone 3 (Abcam), or α-acetylated histone 4 (Upstate Biotechnology). DNA was purified with Qiaquick PCR purification kit (QIAGEN). DNA sequences were quantified by real-time quantitative PCR with SYBR Green (ABI) performed on an ABI 7900HT Sequence Detection System (Applied Biosystems). Input DNA that was not immunoprecipitated was serially diluted and used as a standard curve to quantify levels of DNA recovered after IP. The amount of DNA recovered from each ChIP sample was presented as a relative value to either the input of that sample or the material recovered from α-H3 IP to correct for differences in starting material. Primers used were as follows: exon 1a forward 5′-TTCCACAGGGCAGTGTCATT-3′, exon 1a reverse 5′-CACACAAACCGCACATCAGA-3′, exon 1b forward 5′-CTCCCCTGCTCTGTGTTTCT-3′, exon 1b reverse 5′-CCAGCCTGTAGGGGGTATTT-3′, Il4 HS V forward 5′-TTGAAGTAGCCCTCCTCACGATCA-3′, and Il4 HS V reverse 5′-AGCCTCCAGACAAATTGGTGAGTG-3′.
Retroviral Constructs and Transduction and T cell Differentiation
All Notch-related cDNAs have been described (Maillard et al., 2004) and were cloned into retroviral vectors carrying either GFP (MigR1) or a truncated nerve growth factor receptor (NGFR) as a surrogate marker. GATA-3-MigR1, GATA-3-NGFR, and DN G3-MigR1 were provided by Steve Reiner. Transduced DNMAML CD4+ T cells were from a C57Bl/6 background. All other transductions were carried out in cells from wild-type Balb/C mice. Cell culture and transductions were carried out as described (Martins et al., 2005).
ELISA
Equal numbers of CD4+ T cells were plated in triplicate and restimulated in wells coated with 2.5 µg/ml α-CD3. After 48 hr, standard sandwich ELISA protocols were used as described (Tu et al., 2005).
Flow Cytometry
Cells were restimulated, fixed, and stained as described (Tu et al., 2005). Antibodies were peridinin chlorophyll-a protein-α-CD4, phycoerythrin-α-IL-4, and biotin-α-NGFR. Allophycyanin-conjugated streptavidin was used as a second step for detection of NGFR. Cells were acquired on a FACS Calibur (Becton Dickinson), and data was analyzed with FlowJo (TreeStar).
EMSA
Oligonucleotides with 5′-overhangs were labeled with 32P-α-dCTP (Perkin-Elmer) by incubation with the Klenow fragment of Escherichia coli DNA polymerase I (New England Biolabs) for 15 min at room temperature. DNA samples were purified with Microspin G-50 Columns (Amersham Biosciences). Sequences of the oligonucleotides (Integrated DNA technologies) were as follows:
exon 1a 5′-TCCAACTCAGTTTCACACACCTCTGAT-3′,
mutated exon 1a 5′-TCCAACTCAGTTAAAAACACCTCTGAT-3′,
exon 1b 5′-GCGGACCGGCTGGGAATTACATGTTA-3′,
mutated exon 1b 5′-GCGGACCGGCTGAAAATTACATGTTA-3′,
consensus CSL-binding site 5′-TCCAAATTTTTTCCCACGGCGTGT-3′.
CSL and the RAM-ANK domains of Notch1 were prepared as described (Nam et al., 2003). EMSAs were performed by incubation of 0.2 pmol of probes for 30 min at 30°C in binding buffer (10% glycerol, 20 mM HEPES [pH 7.9], 60 mM KCl, 10 mM DTT, 5 mM MgCl2, 250 ng dGdC, and 0.2 mg/ml bovine serum albumin) in the presence or absence of 300 ng of CSL and 800 ng of RAM-ANK of ICN1.
Sequence Analysis
ECR browser is located at http://ecrbrowser.dcode.org.
ACKNOWLEDGMENTS
We are grateful to Steve Reiner for advice, reagents, and critical reading of the manuscript; Bill Demuth for cell sorting; Hong Sai for retroviral production; and members of the Pear laboratory for critical reading of the manuscript. This work was supported by a predoctoral fellowship from the Cancer Research Institute to T.C.F., a predoctoral fellowship T32GM007229 to D.M.K., a long-term fellowship from the Human Frontier Science Program to C.D.B., and grants from the National Institutes of Health to S.C.B. and W.S.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adler SH, Chiffoleau E, Xu L, Dalton NM, Burg JM, Wells AD, Wolfe MS, Turka LA, Pear WS. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J. Immunol. 2003;171:2896–2903. doi: 10.4049/jimmunol.171.6.2896. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–652. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Asnagli H, Afkarian M, Murphy KM. Cutting edge: Identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J. Immunol. 2002;168:4268–4271. doi: 10.4049/jimmunol.168.9.4268. [DOI] [PubMed] [Google Scholar]
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- Eagar TN, Tang Q, Wolfe M, He Y, Pear WS, Bluestone JA. Notch 1 signaling regulates peripheral T cell activation. Immunity. 2004;20:407–415. doi: 10.1016/s1074-7613(04)00081-0. [DOI] [PubMed] [Google Scholar]
- Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Hoflinger S, Kesavan K, Fuxa M, Hutter C, Heavey B, Radtke F, Busslinger M. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J. Immunol. 2004;173:3935–3944. doi: 10.4049/jimmunol.173.6.3935. [DOI] [PubMed] [Google Scholar]
- Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J. Immunol. 2001;167:4414–4420. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- Kurata H, Lee HJ, O’Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11:677–688. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- Lee GR, Fields PE, Flavell RA. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity. 2001;14:447–459. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O’Garra A, Arai N. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J. Exp. Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieuw KH, Li G, Zhou Y, Grosveld F, Engel JD. Temporal and spatial control of murine GATA-3 transcription by promoter-proximal regulatory elements. Dev. Biol. 1997;188:1–16. doi: 10.1006/dbio.1997.8575. [DOI] [PubMed] [Google Scholar]
- Maekawa Y, Tsukumo S, Chiba S, Hirai H, Hayashi Y, Okada H, Kishihara K, Yasutomo K. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity. 2003;19:549–559. doi: 10.1016/s1074-7613(03)00270-x. [DOI] [PubMed] [Google Scholar]
- Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, Allman D, Aster JC, Pear WS. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- Martins GA, Hutchins AS, Reiner SL. Transcriptional activators of helper T cell fate are required for establishment but not maintenance of signature cytokine expression. J. Immunol. 2005;175:5981–5985. doi: 10.4049/jimmunol.175.9.5981. [DOI] [PubMed] [Google Scholar]
- Minegishi N, Ohta J, Suwabe N, Nakauchi H, Ishihara H, Hayashi N, Yamamoto M. Alternative promoters regulate transcription of the mouse GATA-2 gene. J. Biol. Chem. 1998;273:3625–3634. doi: 10.1074/jbc.273.6.3625. [DOI] [PubMed] [Google Scholar]
- Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, et al. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat. Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 2003;278:21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- Palaga T, Miele L, Golde TE, Osborne BA. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J. Immunol. 2003;171:3019–3024. doi: 10.4049/jimmunol.171.6.3019. [DOI] [PubMed] [Google Scholar]
- Peitz M, Pfannkuche K, Rajewsky K, Edenhofer F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: A tool for efficient genetic engineering of Mamm. Genomes. Proc. Natl. Acad. Sci. USA. 2002;99:4489–4494. doi: 10.1073/pnas.032068699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, Pear WS. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat. Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- Solymar DC, Agarwal S, Bassing CH, Alt FW, Rao A. A 3′ enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity. 2002;17:41–50. doi: 10.1016/s1074-7613(02)00334-5. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu. Rev. Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- Tacchini-Cottier F, Allenbach C, Otten LA, Radtke F. Notch1 expression on T cells is not required for CD4+ T helper differentiation. Eur. J. Immunol. 2004;34:1588–1596. doi: 10.1002/eji.200324337. [DOI] [PubMed] [Google Scholar]
- Takemoto N, Kamogawa Y, Jun Lee H, Kurata H, Arai KI, O’Garra A, Arai N, Miyatake S. Cutting edge: Chromatin remodeling at the IL-4/IL-13 intergenic regulatory region for Th2-specific cytokine gene cluster. J. Immunol. 2000;165:6687–6691. doi: 10.4049/jimmunol.165.12.6687. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Tsukada J, Suzuki W, Hayashi K, Tanigaki K, Tsuji M, Inoue H, Honjo T, Kubo M. The interleukin-4 enhancer CNS-2 is regulated by Notch signals and controls initial expression in NKT cells and memory-type CD4 T cells. Immunity. 2006;24:689–701. doi: 10.1016/j.immuni.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- Tu L, Fang TC, Artis D, Shestova O, Pross SE, Maillard I, Pear WS. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell. Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A, Bakker T, Wilson A, Nicolas M, Ioannidis V, Littman DR, Lee PP, Wilson CB, Held W, MacDonald HR, Radtke F. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta lineage thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, Kimura MY, Hasegawa A, Nakayama T. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24:611–622. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Shinnakasu R, Nigo Y, Kimura M, Hasegawa A, Taniguchi M, Nakayama T. Interleukin (IL)-4-independent maintenance of histone modification of the IL-4 gene loci in memory Th2 cells. J. Biol. Chem. 2004;279:39454–39464. doi: 10.1074/jbc.M405989200. [DOI] [PubMed] [Google Scholar]
- Zhang DH, Yang L, Ray A. Differential responsiveness of the IL-5 and IL-4 genes to transcription factor GATA-3. J. Immunol. 1998;161:3817–3821. [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. Stat6 is necessary and sufficient for IL-4’ role in Th2 differentiation and cell expansion. J. Immunol. 2001;166:7276–7281. doi: 10.4049/jimmunol.166.12.7276. [DOI] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr, Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]