Summary
Rho GTPases are key regulators of the actin-based cytoskeleton; Rab GTPases are key regulators of membrane traffic. We report here that the atypical Rho GTPase family member, RhoBTB3, binds directly to Rab9 GTPase, and functions with Rab9 in protein transport from endosomes to the trans Golgi network. Gene replacement experiments show that RhoBTB3 function in cultured cells requires both RhoBTB3’s N-terminal, Rho-related domain, and C-terminal sequences that are important for Rab9 interaction.9 Biochemical analysis reveals that RhoBTB3 binds and hydrolyzes ATP rather than GTP. Rab9 binding opens the auto-inhibited RhoBTB3 protein to permit maximal ATP hydroysis. Because RhoBTB3 interacts with TIP47 on membranes, we propose that it may function to release this cargo selection protein from vesicles to permit their efficient docking and fusion at the Golgi.
Introduction
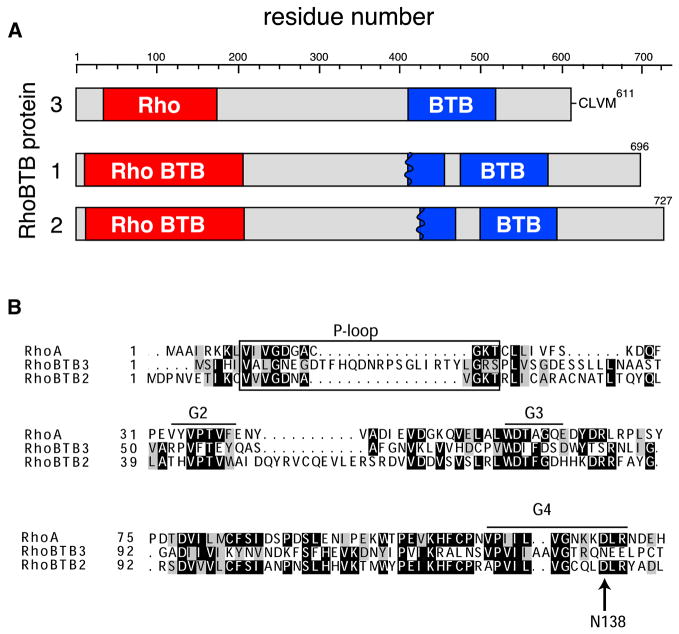
Rho GTPases are key regulators of the actin-based cytoskeleton and thus control most aspects of cell dynamics, including migration, division, and morphogenesis (Jaffe and Hall, 2005; Ridley, 2006). There are at least 20 members of this subfamily of Ras-like GTPases in humans, and although many have been studied in great detail, the so-called atypical Rho GTPase subset are much less well understood (Salas-Vidal et al., 2005; Aspenström et al., 2007). Three atypical Rho GTPases, RhoBTB1, 2 and 3, are much larger (67–83kD) than conventional Rho proteins and are comprised of N-terminal, Rho-related domains followed by one or more BTB (Bric-a-brac, Tramtrack, Broad-complex) domains (Fig. 1; Ramos et al., 2002; Berthold et al., 2008a). BTB domains are found in about 200 proteins and are thought to mediate protein:protein interactions.
Figure 1. Alignment of RhoBTB3 with RhoBTB 1 and 2 and RhoA.
(A) Amino acid residues are listed at the top; numbers at left indicate RhoBTB1, 2 or 3. Each of the proteins has an N-terminal Rho domain (red) and at least one intact BTB domain (blue). Because RhoBTB has a single ancestor closest to RhoBTB1 and 2, the conserved domain database identified these as “RhoBTB” domains, a subset of Rho domains (Marchler-Bauer et al., 2007). RhoBTB3 is unique with a C-terminal CAAX prenylation motif (CLVM shown). (B) Sequence alignment of RhoBTB3’s Rho domain with RhoA and the Rho domain of RhoBTB2. Canonical features of typical GTPases are noted (G1 P-loop, G2, G3 and G4). Arrow points to Asn138. In all GTPases, this residue is Asp and confers specificity for guanosine.
RhoBTB1 and RhoBTB2 show 79% similarity in sequence, and are likely orthologs of the single RhoBTB that is present in Dictyostelium, Drosophila and Anopheles (Ramos et al., 2002). RhoBTB2 has attracted attention as a tumor suppressor that is down regulated in several tumor types (Hamaguchi et al., 2002; Berthold et al., 2008a). RhoBTB2’s first BTB domain interacts with the Cullin3 ubiquitin ligase scaffold protein, and Wilkins et al. (2004) have proposed that RhoBTB2 acts as a tumor suppressor by recruiting substrates to a Cul3 ubiquitin ligase complex for degradation. Like most Rho proteins (Ridley, 2006), changes in RhoBTB2 levels also influence membrane trafficking pathways (Siripurapu et al., 2005; Chang et al., 2006), and both RhoBTB1 and 2 are associated with what appear to be late endosomes or lysosomes in cultured cells (Aspenstrom et al., 2004).
RhoBTB3 is likely to play a different role than RhoBTB1 and 2, as it is the most divergent of the three RhoBTB proteins (only ~48% similar to RhoBTB1 and 2), and residues that are key for RhoBTB2-Cul3 interaction (including Y248) are not present in RhoBTB3 (but see Berthold et al., 2008b). Moreover, RhoBTB3 is only found in vertebrates, but like RhoBTB1, appears to be expressed in all tissues analyzed (Ramos et al., 2002). Indeed, Boureux et al. (2007) do not classify RhoBTB3 as a Rho family member, but the conserved domain database identified these as “RhoBTB” domains, a subset of Rho domains (Marchler-Bauer et al., 2007).
We study the transport of mannose 6-phosphate receptors (MPRs) from late endosomes to the Golgi complex. This process requires Rab9 GTPase (Lombardi et al., 1993; Riederer et al., 1994), the cargo selection protein and Rab9-localizer, TIP47 (Diaz and Pfeffer, 1998; Aivazian et al., 2006), cytoplasmic dynein (Itin et al., 1999), the trans Golgi network (TGN)-localized, tethering protein, GCC185 (Reddy et al., 2006; Derby et al., 2007), Rab6 and Arl1 GTPases (Burguete et al., 2008), and a SNARE complex comprised of Syntaxin-10, Syntaxin-16, Vti1a and VAMP3 (Ganley et al., 2008). As part of our ongoing efforts to elucidate the molecular mechanism of this trafficking pathway, we carried out a yeast two hybrid screen to identify gene products that interact preferentially with Rab9-GTP (Reddy et al., 2006). We report here the characterization of a novel Rab9 effector identified in that screen: RhoBTB3. To our surprise, this protein turns out to be a Rab9-regulated ATPase, rather than a GTPase, that participates in transport vesicle docking at the Golgi complex.
Results
RhoBTB3 is a 611 amino acid protein that contains a Rho GTPase-related domain near its N-terminus and a BTB domain near its C-terminus, as annotated by the conserved domain database (Marchler-Bauer et al., 2007) (Fig. 1A). The related RhoBTB1 and RhoBTB2 proteins are shown for comparison (Fig. 1A, B). The Rho domains of these two proteins are more closely related to the canonical RhoA protein (37% and 39%, respectively). In addition, they appear to have a remnant of a second BTB domain, although sequence similarity algorithms that include structural information, such as the conserved domain architecture retrieval tool (Marchler-Bauer et al., 2007), indicate that each of these proteins contains only one intact BTB motif. Unlike RhoBTBs 1 and 2, RhoBTB3’s C-terminal sequence terminates in a consensus prenylation sequence, CAAX.
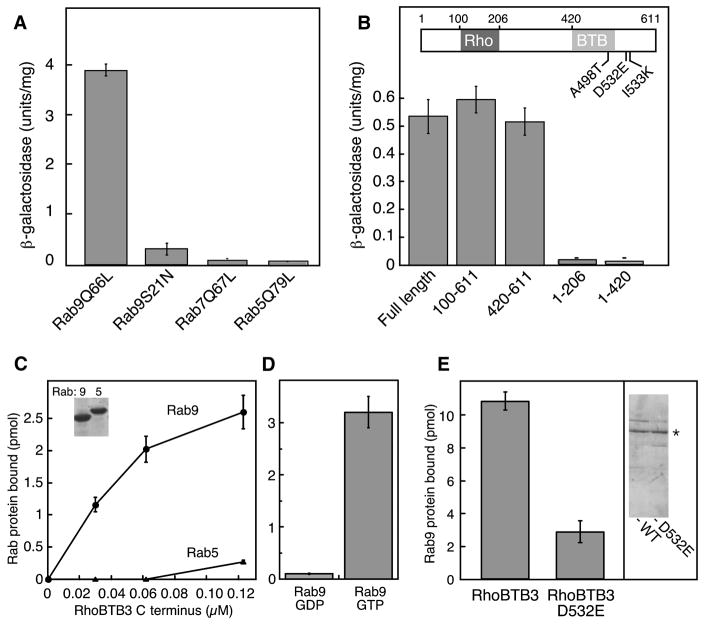
RhoBTB3 was identified in a yeast two hybrid screen for Rab9-GTP-interacting proteins. Binding was quantified by liquid culture analysis: full length RhoBTB3 interacted with the GTP conformation of Rab9 (Rab9Q66L) but not a mutant Rab9 that binds GDP with 50 fold preference to GTP (Rab9S21N; Riederer et al., 1994) (Fig. 2A). Binding was also specific, as no interaction was seen for the activated mutant forms of Rabs 5 (Rab5 Q79L) or 7 (Rab7 Q67L). Subsequent analysis of 54 different Rab GTPases confirmed binding specificity: only Rab9A and to a lesser extent, Rab9B showed interaction with RhoBTB3 (Supplemental Figure 1). We will focus here on Rab9A (“Rab9”).
Figure 2. RhoBTB3 interacts specifically with Rab9 GTPase via its C-terminus.
(A) Liquid culture β-galactosidase activity of yeast strains coexpressing the clone 8-GAL4 activation domain (RhoBTB3 131–607) hybrid and GAL4-DNA binding domain hybrids of either Rab9Q66L, Rab9S21N, Rab7Q67L, or Rab5Q79L. (B) Liquid culture of β-galactosidase activity of yeast strains coexpressing the indicated RhoBTB3 fragments with the GAL4 DNA binding domain of Rab9Q66L. Error bars in A and B represent standard deviation of duplicate determinations from a representative experiment. RhoBTB3 C-terminus (420–607) was subjected to PCR-based mutagenesis and assayed for subsequent Rab9 binding by two hybrid screening; point mutations that abolished Rab9 binding are indicated. (C) (Left panel) Increasing amounts of purified GST-RhoBTB3 C-terminus (AA 420-611) were incubated with purified Rab9 or Rab5 (600nM), preloaded with 35S-GTPγS. Inset, SDS-PAGE of proteins used for these experiments. (D) GST-RhoBTB3 C-terminus (AA 420-611; 180nM) binding to Rab9 bearing either 35S-GTPγS- or 3H-GDP as in C. Error bars represent standard deviation. (E) Binding of full length wildtype or RhoBTB3 D532E proteins (100nM) to Rab9-GTP (3μM). Inset at right shows Coomassie stained SDS-PAGE of the proteins used. Asterisk indicates His-RhoBTB3 full length.
Truncation constructs were generated to identify the precise Rab9 binding site within RhoBTB3. As shown in Figure 2B, the Rab9 binding site was localized to the C-terminal region of the protein, as residues 420-611 were sufficient to yield full β-galactosidase activation in the two hybrid system.
RhoBTB3 is a Rab9 effector and localized to the Golgi
To verify that the binding observed in yeast was due to a direct protein:protein interaction, we tested the ability of purified GST-RhoBTB3 C-terminal residues 420–611 to bind purified Rab proteins. Rabs were preloaded with GTPγ-[35S] or [3H]-GDP and then incubated with RhoBTB3; bound proteins were collected on glutathione beads and counted in a scintillation counter. As shown in Figure 2C, RhoBTB3 bound saturably to Rab9 but not Rab5, and binding showed strong preference for Rab9-GTP (Fig. 2D).
Error prone PCR mutagenesis was carried out to further define RhoBTB3 residues important for Rab9 binding; mutated constructs were screened for loss of interaction by two hybrid analysis, and then sequenced directly. These screens suggested that Rab9 interaction required A498, D532 or I533 (Fig. 1B) as their mutation abolished binding in the two hybrid system (not shown). The D532E mutation was of particular interest, as that residue is a glutamic acid in RhoBTB2. The importance of D532 for Rab9 binding was demonstrated directly: purified RhoBTB3 D532E protein (2E gel inset) showed significantly less binding to Rab9 than wild type protein, confirming the importance of this region of the protein in Rab9 interaction (Figure 2E).
RhoBTB3 is localized to the Golgi complex, as determined by indirect immunofluorescence of myc-RhoBTB3 transfected cells, using the proteins TGN46 or GCC185 as endogenous Golgi marker proteins (Supplemental Figure 2; see also Berthold et al., 2008b). Confirmation of this localization was obtained by disruption of the microtubule-based cytoskeleton using nocodazole. Under these conditions, the Golgi fragments into mini-stacks that distribute throughout the cytoplasm. The Golgi marker, TGN46, showed complete co-localization with RhoBTB3 under control and nocodazole conditions. Thus, myc-RhoBTB3 appears to be Golgi-associated. Unfortunately it was not possible to detect endogenous RhoBTB3 by light microscopy using two separate rabbit antisera raised against recombinant RhoBTB3 protein; these only detected endogenous protein by immunoblot.
The localization of RhoBTB3 to the Golgi did not require C-terminal residues that could potentially be prenylated, as a ΔCAAX mutant was not altered in its subcellular localization (see below). In addition, Rab9 does not determine RhoBTB3’s Golgi association as most Rab9 is not found on the Golgi, and a mutant form of RhoBTB3 (D532E) that cannot bind Rab9 is normally localized (see below).
RhoBTB3 is required for retrograde transport to the Golgi complex
Because of the close link between Rho GTPases and the actin cytoskeleton, we first tested whether depletion or overexpression of RhoBTB3 altered the appearance of either actin or microtubules. Superficial examination of light micrographs did not reveal any significant changes (not shown). To try to determine RhoBTB3’s function, we took advantage of the clue that it binds to Rab9 GTPase.
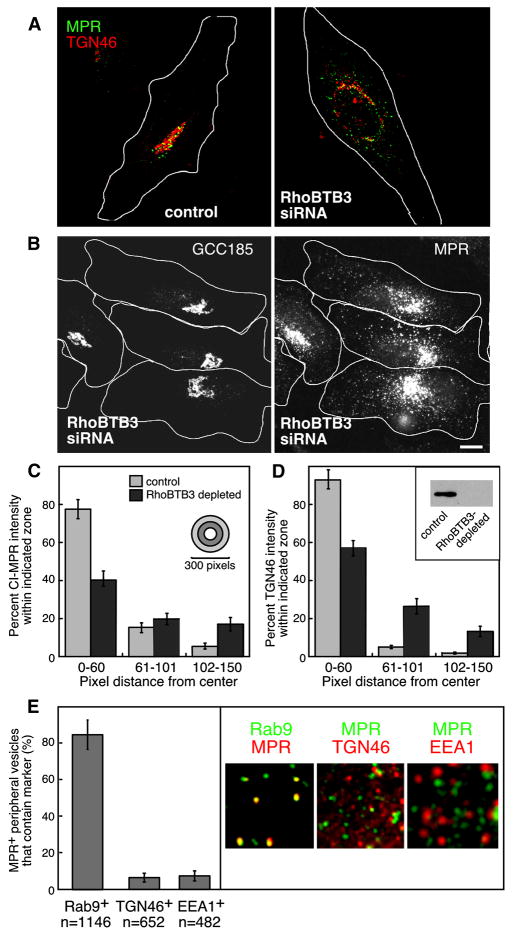
Rab9, and each of its effectors that have been studied to date (p40, GCC185, TIP47) are required for the recycling of MPRs from endosomes to the TGN. To test if RhoBTB3 is also involved in this process, we used siRNA-mediated protein depletion and monitored the distribution of MPRs in siRNA-transfected cells. MPRs normally display a tight perinuclear localization, adjacent to the Golgi complex (Fig. 3A, left panel). When another Golgi-localized Rab9 effector, GCC185, is depleted from cells, MPRs accumulate in peripheral, Rab9-positive transport intermediates (Reddy et al., 2006). This has implicated GCC185 as a tethering factor in this transport process.
Figure 3. Depletion of RhoBTB3 disperses MPRs into peripheral, Rab9-positive vesicles.
(A) Cells depleted of RhoBTB3 mis-sort the cycling protein TGN46 (red) as well as CI-MPR (green). White lines represent approximate cell outlines. (B) Endogenous GCC185 remains on the Golgi in RhoBTB3 depleted cells. Scale Bar is 10μm. (C) Fluorescence intensity distribution of CI-MPR in control (n=20) and RhoBTB3 depleted cells (n=23) was quantified using ImageJ. Circles of different radii, centered on the peak fluorescent signal, were drawn; the intensity within each ring of the bull’s eye was quantified relative to the total intensity in the cell. Shown is the percent CI-MPR intensity in the central 0–60 pixel region, the next region 61–101 pixels from the center, or 102–150 pixels from the center. (D) Intensity distribution of TGN46 was measured as in (C); (n=13 for control and RhoBTB3 depletion). Inset, western blot showing RhoBTB3 depletion upon siRNA treatment. (E) Peripheral MPR containing vesicles are Rab9 positive but do not contain TGN46 or EEA1. BsC-1 cells stably expressing GFP-Rab9 were depleted of RhoBTB3. Peripheral vesicles were scored for the presence of MPRs and Rab9 as in A and B. An example is shown at right. Hela cells were used to score TGN46 and EEA1 (examples at right). In all experiments, depletion was for 72 hours. Error bars represent standard deviation in C, D and E; data were from multiple experiments for each panel.
Upon siRNA depletion of RhoBTB3, MPRs appeared in small dispersed vesicles throughout the cytoplasm, reminiscent of the phenotype observed upon GCC185 depletion (Fig. 3A, B; Reddy et al., 2006). The phenotype was quantified by determining the percent of total MPR intensity detected at various distances from the center of intensity. Using this metric, the distribution of MPRs in depleted cells (Fig. 3C, dark bars) was demonstrably broader than in control cells (light bars, Fig. 3C). Although GCC185 was slightly altered in its appearance (see below), the Golgi remained intact in cells depleted of RhoBTB3 (Fig. 3B). In contrast to GCC185 and analogous to MPRs, TGN46 proteins were also broadly distributed (Fig. 3A and D). It is important to note that unlike GCC185, TGN46 cycles between endosomes and the Golgi. These data suggest that RhoBTB3 is needed for the retrograde transport of both MPRs and TGN46 to the Golgi complex; in its absence, cycling proteins are unable to return.
If RhoBTB3 functions in transport vesicle tethering, cells depleted of this protein should accumulate MPRs in Rab9-positive transport intermediates. To test this, cells stably expressing GFP-Rab9 were depleted of RhoBTB3 and the localization of MPRs and Rab9 GTPase in peripheral vesicles was quantified. As shown in Figure 3E, a large proportion of peripheral, small vesicles carrying MPRs were indeed Rab9-positive (>80%). These structures were not early or recycling endosomes as they did not contain fluorescent transferrin, endocytosed for 15 minutes (Supplemental Figure 3) or the early endosome marker, EEA1 protein (Fig. 3E). The accumulation of MPRs in Rab9-positive structures that are not early endosomes is consistent with a role for RhoBTB3 in transport vesicle tethering at the Golgi complex. In addition, ~95% of dispersed TGN46 was not in the same vesicles as MPRs, consistent with their accumulation in a different type of transport vesicle carrier (Fig. 3E). Unlike MPRs, TGN46 recycling requires Syntaxin 6 (Ganley et al., 2008). Given the distinct biochemical requirements for TGN46 recycling, its accumulation in a distinct type of vesicle is not unexpected, however both TGN46 and MPRs require RhoBTB3 function upon arrival at the Golgi complex.
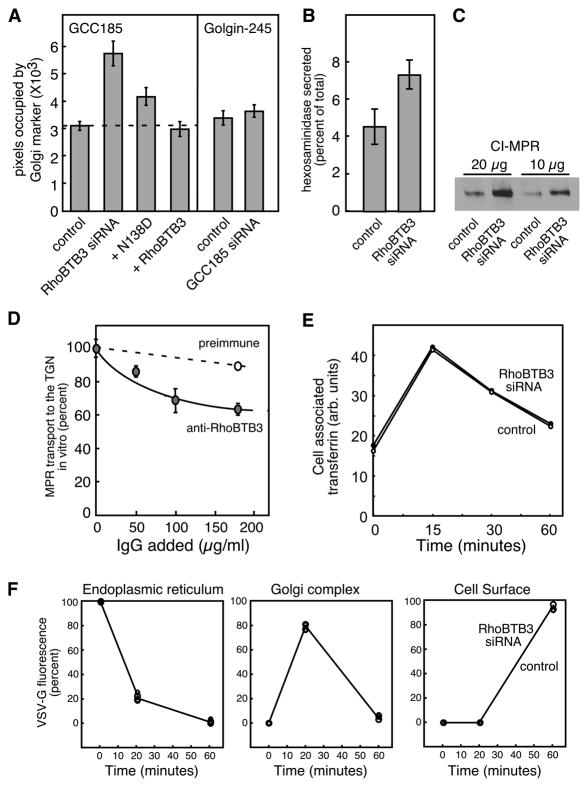
Careful examination of cells depleted of RhoBTB3 showed that the Golgi (monitored by GCC185 localization) appeared somewhat enlarged compared with control cells. This was quantified by determining the number of pixels occupied by the Golgi marker under various conditions (Figure 4A). In control cells, GCC185 occupied 2889 ± 109 pixels per cell (n=210). In depleted cells, this almost doubled to 5764 ± 453 pixels per cell (n=44), even though overall cell size remained unchanged. A similar expansion was seen for the Golgi marker, p115 (data not shown). Expression of the RhoBTB3 N138D mutant that cannot bind nucleotide (see below) caused a smaller increase in Golgi area (4195 ±323 (n=60)), but expression of wild type RhoBTB3 did not (Figure 4A). For comparison, depletion of GCC185, which causes the Golgi to fragment into mini-stacks (Reddy et al., 2006), did not change the overall area occupied by the Golgi complex, as monitored by the area occupied by Golgin-245 protein (Fig. 4A). Thus, RhoBTB3 also contributes to the overall structure of the Golgi complex.
Figure 4. RhoBTB3 depletion expands the Golgi and blocks MPR trafficking.
(A) Quantitation of Golgi area. Left, GCC185 area in control cells (n=210), cells depleted of RhoBTB3 (n=44), or in cells expressing wild type RhoBTB3 (n=42) or N138D RhoBTB3 (n=60), was quantified using a Matlab script. Right, Golgi area in control cells (n=48) or in cells depleted of GCC185 (n=67) using Golgin245 staining. (B) Lysosomal enzyme secretion upon depletion of RhoBTB3. Cells were treated for 72 hours with siRNA against RhoBTB3 and assayed for secretion of hexosaminidase. Both control and depleted cells contained 5.2 × 103 units/mg hexosaminidase. (C) Immunoblot analysis of CI-MPR levels in control or RhoBTB3 depleted cell extracts; 10 or 20μg of HeLa extracts were analyzed as indicated. (D) MPR transport from endosomes to the TGN (Reddy et al., 2006) was carried out for 90 min. in the presence or absence of the indicated amounts of non-immune or rabbit anti-RhoBTB3 IgG. In A, error bars represent standard error of the mean; in B and D, standard deviation of duplicate samples from a representative experiment are shown. (E) Alexa-594-Transferrin (50μg/ml) was prebound to cells on ice for 60 minutes; cells were warmed for various times and cell-associated transferrin was determined after washing with 20mM HOAC pH3, 500mM NaCl by immunofluorescence and MatLab analysis in control and RhoBTB3 depleted HeLa cells (>50 cells counted for each time point in two separate experiments; average of both is shown). (F) YFP-VSV-G-ts045 transport through the ER, Golgi and cell surface was carried out as described (Sklan et al., 2007) for control and RhoBTB3 depleted cells, with >150 cells counted at each time point, in two separate experiments, one of which is shown. The two data sets were identical.
Consistent with a role in MPR transport, cells depleted of RhoBTB3 showed a two fold increase in the secretion of the lysosomal hydrolase, hexosaminidase (Fig. 4B), despite comparable levels of total enzyme in controls and siRNA-depleted cells (both 5.2 × 103 units/mg hexosaminidase). In addition, the steady state levels of MPRs increased at least two fold (Fig. 4C). These are hallmarks of a block in MPR trafficking (Riederer et al., 1994), and are seen in cells expressing the GDP-preferring mutant of Rab9 (S21N), or depleted of Rab9 (Ganley et al., 2004), GCC185 (Reddy et al., 2006) or Syntaxin10 proteins (Ganley et al., 2008). Specifically, when MPRs are mis-sorted to lysosomes they are turned over more rapidly; in several cases, cells compensate for this loss by upregulating MPR synthesis.
To confirm a specific role for RhoBTB3 in MPR transport to the Golgi complex, we tested whether anti-RhoBTB3 antibodies could inhibit this transport process in an in vitro system that reconstitutes this transport event (Itin et al., 1999). Anti-RhoBTB3 IgG inhibited transport ~40% under conditions in which pre-immune IgG inhibited less than 10% (Figure 4D). Similarly, negative control anti-Syntaxin 6 antibodies inhibited ~10% while another positive control antibody, anti-Rab9, also inhibited 40% (Ganley et al., 2008). These data support the conclusion that RhoBTB3 function is needed for MPR transport to the Golgi complex both in vitro and in live cells. As mentioned earlier, the anti-RhoBTB3 antibody does not readily recognize the native protein upon immunofluorescence of fixed cells, which could easily explain the incomplete block of in vitro transport observed.
The specificity of RhoBTB3’s role in retrograde transport was confirmed by monitoring general endocytosis and exocytosis in RhoBTB3-depleted cells. Transferrin uptake and recycling in RhoBTB3-depleted cells was indistinguishable from that seen for control cells (Figure 4E). In addition, in RhoBTB3-depleted cells, the rate of export of VSV-G glycoprotein from the endoplasmic reticulum, its transit through the Golgi complex, and arrival at the cell surface was entirely unchanged (Figure 4F). In summary, these data show that RhoBTB3 functions specifically in retrograde transport from endosomes to the Golgi complex and in maintenance of the structure of the Golgi complex. Although the Golgi was slightly larger in area (Fig. 4A), it was nevertheless fully functional for G-glycoprotein export.
RhoBTB3 function requires its Rho related domain and Rab9 binding capacity
We identified two mutant proteins that blocked either RhoBTB3’s nucleotide-binding capacity (N138D, see below) or its ability to interact with Rab9 GTPase (D532E). Thus we could test the importance of these distinct domains on RhoBTB3’s role in MPR transport to the Golgi complex. For these experiments, we used fluorescent siRNA to deplete RhoBTB3 from cells and enable their identification. We then carried out gene rescue experiments with either myc-tagged, wild type RhoBTB3 or one of three mutant versions, each of which carried eight silent mutations to mask the rescuing construct from siRNA-mediated destruction.
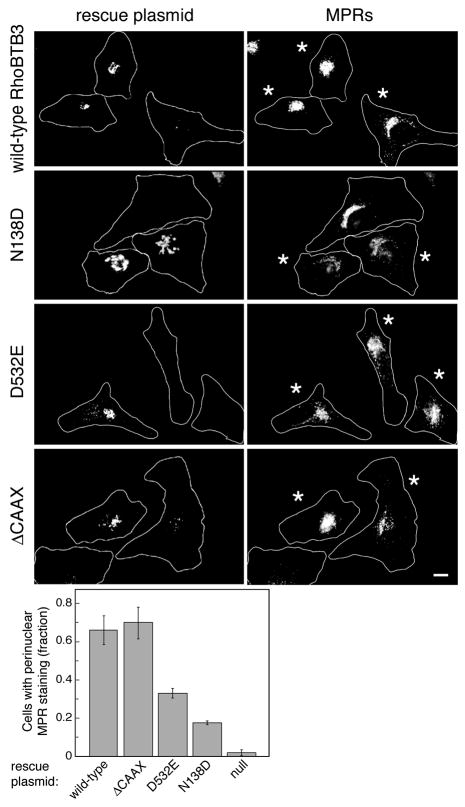
In Figure 5 (right column), fluorescent siRNA-containing cells are marked with an asterisk. They are depleted of endogenous RhoBTB3 based upon the high potency of the siRNA (Fig. 3D) and the dispersal of MPRs detected. In the top row, RhoBTB3-depleted cells that also received a wild type rescue plasmid (left column, 2 cells at left) showed a concentrated, perinuclear MPR distribution. In contrast, the right-most cell that expressed much less of the rescuing protein showed a dispersed MPR phenotype, as expected for RhoBTB3 depleted cells. Thus, the wild type construct can restore the normal, compact perinuclear MPR distribution. Similar data were obtained for the RhoBTB3 ΔCAAX mutant (Fig. 5, bottom row). Again, the presence of the rescue plasmid was sufficient to normalize MPR localization. It is important to note that the CAAX motif may not be modified in cells, which would represent a trivial explanation for its lack of importance for either RhoBTB3 localization or rescue.
Figure 5. RhoBTB3 function requires its ATPase activity and ability to bind Rab9.
RhoBTB3 was depleted by siRNA for 72 hours. Indicated myc-RhoBTB3 constructs that were masked by siRNA were expressed for 48 hours. Top, Expression of the wild type RhoBTB3 is sufficient to relocalize CI-MPR back to its tight perinuclear staining. Asterisked cells received fluorescent siRNA. Second, Expression of N138D, a mutant RhoBTB3 that cannot bind or hydrolyze nucleotide, cannot rescue CI-MPRs back to the perinuclear region. Third, Expression of D532E, a mutant RhoBTB3 that lacks capacity to bind Rab9, also does not rescue CI-MPR. Bottom, Expression of ΔCAAX, a mutant RhoBTB3 that lacks the C-terminal prenylation sequence, is capable of rescue. White lines are approximate cell outlines. Scale bar is 10μm. Quantitation at bottom was determined for >100 cells from multiple experiments under each condition; error bars represent standard deviation. Null samples were mock transfected.
This was in contrast to cells that were rescued with the ATPase-defective RhoBTB3 N138D construct (second row). Loss of RhoBTB3 in the two asterisked cells expanded the Golgi, as monitored by the localization of myc-N138D at left. Despite the presence of the mutant protein in the two lower cells, MPRs remained dispersed. Thus, RhoBTB3’s ATP binding and/or ATPase functionality is required for its ability to facilitate MPR recycling to the Golgi.
Similarly, the RhoBTB3 D532E rescue plasmid was also unable to correct a dispersed MPR phenotype in most cells analyzed (Fig. 5, third row). This suggests that either Rab9 or another interacting partner must be able to link to RhoBTB3 via this site for it to carry out its proper cellular function.
Quantitation of these data confirmed that rescue by the wild type or ΔCAAX RhoBTB3 proteins could restore a normal MPR phenotype to 66–70% of cells analyzed. In contrast, the D532E mutant was less able to rescue the phenotype (33%) and the N138D mutant was even less functional (17% rescue). Any rescue by the D532E or N138D mutants might be explained by retention of some Rab9 binding capacity and/or high enough expression to permit low basal ATP hydrolysis by RhoBTB3 protein.
RhoBTB3 is an ATPase
GTPases contain signature features that reflect the binding and hydrolysis capacities of these enzymes (Sprang, 1997). The phosphate binding or P-loop, and elements G2, G3 and G4 are indicated in Figure 1B. First, note that RhoBTB3 contains an unusual 16 residue insert in the middle of the P-loop. P-loops often contain multiple glycine residues that are thought to generate flexibility. In RhoBTB3, Pro-Ser-Gly-Leu occupies the middle of the extended loop and is predicted to create a tight, type II β-turn (Ybe and Hecht, 1996). This insert could permit extension of the P-loop without disrupting critical contacts with nucleotide. In addition, the canonical K(S/T) residues of the G1/Walker motif consensus sequence are replaced with RS in RhoBTB3, which will maintain charge at that position. A magnesium ion is coordinated by G2 sequences; in RhoBTB3, phenylalanine and glutamic acid replace proline and valine residues conserved between RhoA and RhoBTB2. Theoretically, these changes might still accommodate magnesium binding. RhoBTB3’s G3 residues are also poorly conserved. Most small GTPases contain a glycine residue immediately before the catalytic glutamine in G3; in RhoA, the sequence is WDTAGQ. RhoBTB3 instead contains WDIFDS. The significance of these changes is unclear, but as shown below, RhoBTB3 is capable of nucleotide hydrolysis, despite the divergence of G3 sequences.
The G4 region determines nucleotide specificity: small GTPases usually contain the sequence, NKXD. Mutation of this aspartic acid dramatically decreases the ability of GTPases to bind guanosine nucleotides, and conversion of this residue to asparagine strongly reduces GTP binding, while significantly enhancing xanthosine triphosphate (XTP) binding to elongation factor Ef-Tu (Hwang and Miller, 1987) and other small GTPases (Zhong et al., 1995; Rybin et al., 1996). Because the cell contains very little, if any, XTP, it was unclear whether RhoBTB3 (with the sequence TRQN instead of NKXD) could even bind nucleotide; hydrolysis seemed unlikely, as well. Indeed, Berthold et al. (2008b) reported that RhoBTB3 cannot hydrolyze GTP. Finally, G5 domain residues S160, A161 and K162 in RhoA interact with the carbonyl oxygen on the guanosine base. In RhoBTB3, these SAK residues are replaced with LDD, which could preclude guanosine interaction. To test these possibilities directly, we expressed a GST tagged RhoBTB3 Rho domain (residues 1–220) and examined the biochemical characteristics of this domain in vitro.
When small GTPases are expressed in bacteria, they carry with them bound nucleotide from the bacterial cytoplasm. Thus, after purification, Ras and Rab GTPases have bound GDP because GTP is slowly hydrolyzed during protein isolation (cf. Tucker et al., 1986). Reverse phase HPLC of the purified protein can be used to both detect and identify bound nucleotide. We first tested for the presence of bound nucleotide on RhoBTB3.
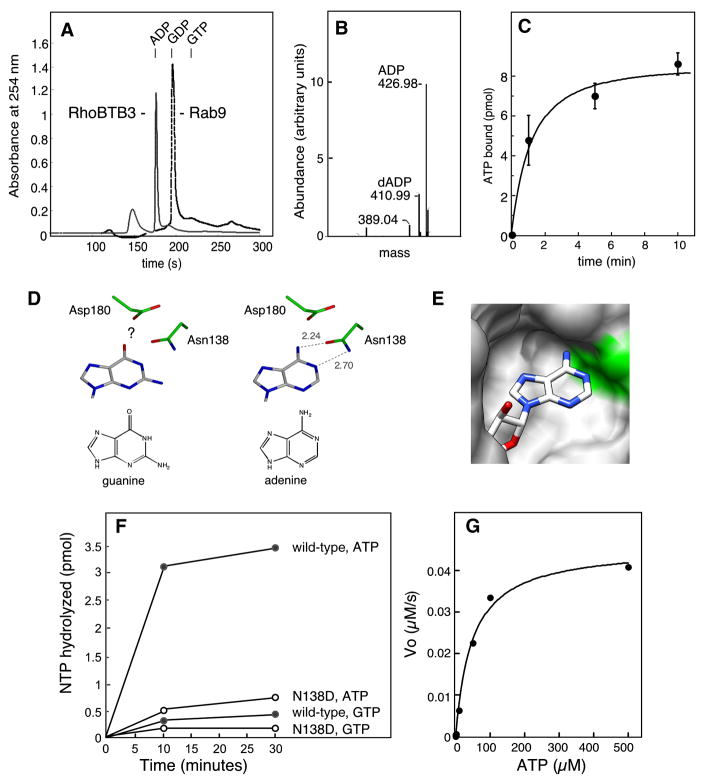
As expected, reverse phase HPLC confirmed that bacterially expressed control Rab9 protein carried GDP upon isolation (dashed profile, Fig. 6A). To our surprise, RhoBTB3’s Rho domain carried a substituent that showed absorbance at 254nm and eluted before GDP; subsequent chromatography showed precise co-elution with an ADP marker. Mass spectroscopic analysis of the bound material revealed it to be a mixture of 80% ADP and 20% dADP (Fig. 6B), a ratio of nucleotide to deoxynucleotide similar to that seen previously for p21 Ras (Tucker et al., 1986).
Figure 6. RhoBTB3 binds and hydrolyzes ATP.
(A) Purified proteins were injected onto a reverse phase HPLC column and nucleotide was eluted. Purified GST-RhoBTB3 Rho domain (residues 1–220, 5 nmols in 500μl) (solid profile) released bound ADP, eluting with a retention time of 177s. Rab9 (5 nmols in 500μl) released bound GDP (dashed profile), eluting with a retention time 196s on this system. (B) Mass spectroscopy analysis of the peak HPLC fraction revealed two major peaks at 427Da and 411Da. These correspond to the masses of ADP and dADP, respectively. (C) RhoBTB3 Rho domain (10pmol in 100μl) binds [α-32P]ATP to saturation; reactions were at 37°C with 100μM ATP. Error bars represent standard deviation of duplicates from a representative experiment. (D) RhoBTB3 was threaded onto the structure of RhoA-GMPPNP (pdb 1KMQ) using Modeller 9V3; steric clash of Van der Waals radii was determined using UCSF Chimera software. The adenine or guanine base was rotated 10° outward to relieve steric clash, and possible hydrogen bonding to N138 was modeled using Coot software. Left column, the position of guanine in relation to the catalytically important N138 and the nearby D180; right column, adenine. The question mark indicates possible negative charge repulsion. Hypothetical hydrogen bond lengths are indicated (dashed lines). (E). The 10°rotated, adenine base of ATP is shown modeled in the threaded structure; N138 is shadowed in green in the space-fill representation. (F) RhoBTB3 GST-Rho domain binds and hydrolyzes ATP and not GTP. Nucleotide free RhoBTB3 GST-Rho domain (20nM) was incubated with different concentrations of [γ-32P]ATP or [γ-32P]GTP and MgCl2. A point mutant in the G4 region of RhoBTB3 (N138D) was also tested. (G) Initial velocity measurements of ATP hydrolysis. Reactions were as in (F) using 600nM GST-Rho domain. The first 10% of each reaction (initial velocity) was plotted as a function of ATP concentration.
The ability of RhoBTB3 to bind ATP was confirmed directly using the purified Rho domain. Briefly, any already bound nucleotide in the protein preparation was released by EDTA pretreatment. After desalting, [α32P]-ATP was added and allowed to bind. As shown in Figure 6C, RhoBTB3 showed saturable binding of ATP to >80% of molecules added to the reaction.
To gain insight into ATP binding by RhoBTB3, we threaded the sequence of its Rho domain onto that of RhoA-GMPPNP (PDB 1KMQ). If we used RhoA’s precise orientation of bound GTP or ATP, the imido-ring of either base showed steric clash within the nucleotide binding pocket (not shown). However, a small (~10°), outward rotation of the glycosidic linkage was all that was required to position the adenosine at a position that enables optimal hydrogen binding to the catalytically important N138 (Fig. 6D right column; 6E). Indeed, ATP would fit snugly and favorably into the binding pocket under this hypothetical scenario. Modeling can only offer predictions about residues that might influence nucleotide specificity: in this light, it is noteworthy that D180 would be adjacent to the amino group on the adenine base. Perhaps negative charge repulsion by D180 on the partial negative charge on the guanine carbonyl oxygen makes guanine nucleotide binding less favorable.
In addition to binding ATP, RhoBTB3 can hydrolyze this nucleotide. RhoBTB3 readily hydrolyzed [γ32P]-ATP (Fig. 6F filled circles) at a rate of 4.5 per minute at 37°C but did not hydrolyze [γ32P]-GTP (Fig. 6F). For comparison, Rab5 hydrolyzes GTP with a single turnover rate constant of 0.12 min−1 at 37°C (Simon et al., 1996). The calculated Kcat/Km was 93,750 M−1 min−1 (Fig. 6G), comparable to that of the human Hsp70 and Hsp90 chaperones which hydrolyze ATP with rate constants of 0.6–140 per minute (Nadeau et al., 1993). In addition, binding and hydrolysis required the unique N138 residue of RhoBTB3, as when it was reverted to the D found in most other small GTPases (N138D), significantly less binding (not shown) and hydrolysis were observed (Fig. 6F, open circles).
Unlike Ras-related GTPases that bind GTP very tightly (but similar to ATPases such as Hsp90 (Nadeau et al., 1993)), RhoBTB3 binds nucleotide more weakly: it’s KM for ATP is 48μM (Fig. 6G), compared with nanomolar KDs for GTPases (cf. Simon et al., 1996). ATP is more abundant than GTP in the cytosol, at millimolar levels that far exceed this value. In addition, cellular interactions can alter nucleotide binding affinity: for example, a load on Myosin VI has been shown to change the affinity of the motor for ADP (Altman et al., 2004). In summary, RhoBTB3’s nucleotide binding and hydrolysis properties are similar to other cellular ATPases; as in GTPases, intrinsically slow nucleotide hydrolysis is likely to be catalyzed by other components.
Rab9 relieves RhoBTB3 ATPase auto-inhibition
One role for Rab9 binding could be to regulate the ATPase activity of RhoBTB3. To test this, we purified full length, recombinant His-RhoBTB3 protein (Fig. 7A, right gel insert) and compared its ATPase activity with that of the Rho domain alone. For these experiments, we used equimolar amounts of active enzymes, quantified by monitoring the ability of each preparation to bind [α32P]-ATP.
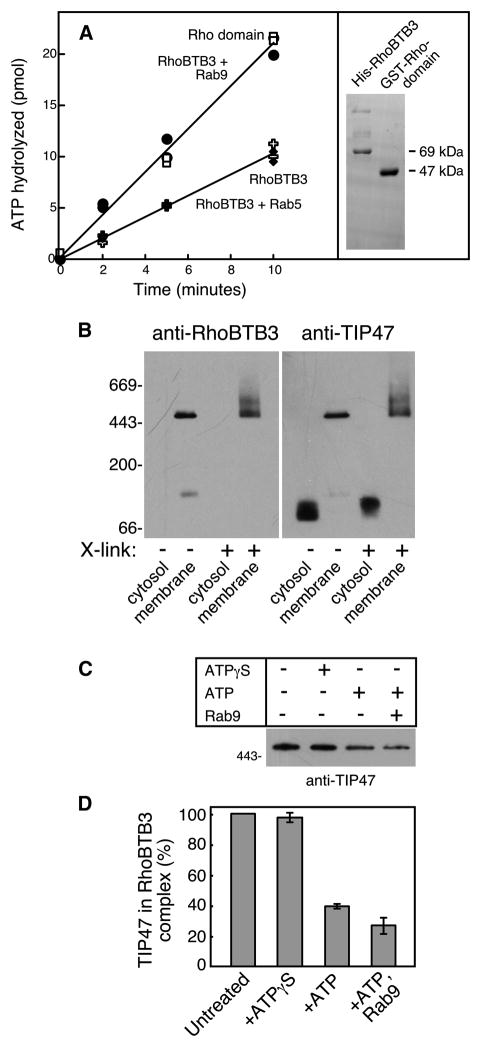
Figure 7. RhoBTB3 is a Rab9-regulated ATPase that binds TIP47.
(A). ATP hydrolysis was determined at the times indicated using 100nM active His-RhoBTB3 full length protein or GST-Rho domain, in the absence or presence of either Rab9 (7.6μM active protein) or Rab5 (8.25μM active protein) and 50μM ATP. Right inset, SDS-PAGE analysis of proteins used. (B). Immunoblot analysis of blue native gel electrophoresis of K562 Golgi/endosome-enriched membranes or K562 cytosol (10μg each). Native protein marker migration is shown at left (kD). Left panel, RhoBTB3; right panel, TIP47. X-link indicates samples that were reacted with DTSSP crosslinker. (C). Rab9 and ATP stimulate release of TIP47 from the RhoBTB3 complex. Top panel, representative immunoblot; below, quantitation of duplicate experiments, with standard deviation.
As shown in Figure 7A, full length RhoBTB3 protein (open crosses) displayed ~50% the rate of ATP hydrolysis seen for the Rho domain alone (open squares). Satisfyingly, addition of Rab9 GTPase (closed circles) but not Rab5 GTPase (closed diamonds) brought the hydrolysis rate of full length RhoBTB3 to that of the Rho domain alone. This suggests that the C-terminal portion of RhoBTB3 controls the N-terminal Rho domain; binding of Rab9 to the C-terminus releases this interaction, permitting ATP hydrolysis at an uninhibited, basal rate. These experiments demonstrate the ability of Rab9 to regulate RhoBTB3 ATPase.
RhoBTB3 binds TIP47 on membranes
In membrane traffic, ATPases are used to disassemble complexes. For example, the uncoating ATPase disassembles clathrin after endocytosis (Sousa and Lafer, 2006) and the Vps4 ATPase disassembles ESCRT complexes important for multivesicular endosome formation (Babst et al., 1998; Lata et al., 2008). To identify a substrate for RhoBTB3’s action, we attempted to isolate RhoBTB3 in complex with its substrate on native, cellular membranes. A human K562 cell membrane fraction enriched in Golgi and endosomes (by sucrose gradient flotation) was subjected to Blue Native gel electrophoresis (Swamy et al., 2006) in the presence of dodecyl maltoside, and the migration of RhoBTB3 was determined by immunoblot (Figure 7B, left panel lanes 2 and 4). Under these conditions, in the absence of SDS, RhoBTB3 migrated at ~475kD. RhoBTB3 was not detected in the same amount of cytosol analyzed in parallel (Fig. 7B lanes 1 and 3). When a chemical crosslinker (DTSSP) was added, RhoBTB3 also migrated as a larger form at ~550kD (7B lane 4).
Motivated by the analogy of the clathrin uncoating ATPase, we tested if RhoBTB3 bound to TIP47. TIP47 is recruited by Rab9 onto late endosomes and packages MPRs into nascent transport vesicles. TIP47 may also be present on the transport vesicles that carry MPRs to the Golgi complex. As shown in the right panel in Figure 7B, the membrane associated, RhoBTB3 complex contains the protein, TIP47. Cytosolic TIP47 gel filters as a hexamer (Sincock et al., 2003) but migrated as a dimer upon blue native gel electrophoresis in dodecyl maltoside (7B, right panel lane 1). On membranes, however, it perfectly co-migrated with RhoBTB3, in the absence or presence of crosslinker. The size of the RhoBTB3 complex (~475kD) could easily accommodate a TIP47 hexamer (282kD) plus a RhoBTB3 dimer (138kD).
IF RhoBTB3’s role is to release TIP47 from membranes, addition of ATP should release TIP47 from the RhoBTB3 complex identified here. As shown in Figure 7C and D, addition of the poorly hydrolysable ATP analog, ATPγS had no effect on the amount of TIP47 in the membrane complex. However, less TIP47 was recovered if membranes were incubated with ATP; still less was recovered if Rab9 GTPase was added to reaction mixtures. These data show that TIP47 is released from membranes by a process that is stimulated by Rab9 and ATP. Together these data implicate RhoBTB3 in TIP47 release as part of transport vesicle uncoating at the Golgi complex.
Discussion
We have revealed here the function of an orphan, distant Rho GTPase family member, RhoBTB3, and shown that this protein is an ATPase instead of a GTPase. RhoBTB3 is present at the exit face of the Golgi, and requires its intrinsic ability to bind (and likely hydrolyze) ATP to carry out its normal function. The lack of retention of nucleotide binding consensus sequences led several authors to assume that RhoBTB3 had lost its ability to bind nucleotide (cf. Ramos et al., 2002; Berthold et al., 2008a,b). Instead, the protein has diverged in a manner that enables it to accommodate a different nucleotide binding partner.
We identified RhoBTB3 as a protein that interacts specifically with Rab9 GTPase and not with 52 other Rab family members. RhoBTB3 appears to function in concert with Rab9--in facilitating the transport of MPRs from endosomes to the TGN. RhoBTB3 is also needed for normal Golgi morphology. Determinants in RhoBTB3 that are needed for Rab9 binding are also essential for RhoBTB3’s role in maintaining Golgi structure, as shown by gene rescue experiments. Indeed, Rab9 binding to the C-terminus enables RhoBTB3 to display its full, N-terminal, basal ATPase activity. Our data demonstrate that Rab9 regulates nucleotide hydrolysis by this enzyme. Such a scenario is reminiscent of the regulation of kinesin’s N-terminal ATPase activity by its C-terminus: in that case, cargo binding to kinesin’s C-terminus releases an inhibitory interaction that slows basal ATP hydrolysis (Coy et al., 1999).
A RhoATPase
RhoBTB3 is not the first GTPase family member to have ATPase activity. Careful evolutionary analysis of the structures of so-called P-loop GTPases segregates GTPases into two classes: TRAFAC and SIMIBI subgroups (Leipe et al., 2002). In the TRAFAC-category, Obg GTPases, YchF (or human OLA1) has an unusual G4 motif and was recently shown to be an ATPase (Koller-Eichhorn et al., 2007). The kinesin family of ATPases also lie in the TRAFAC family, and are entirely missing the conserved, nucleotide-specifying, G4 motif. In the SIMIBI group, MinD ATPase has diverged in its G4 motif to contain TRYN instead of the canonical NKXD. In comparison, RhoBTB3 uses the related G4 motif sequences, TRQN.
The OLA1 ATPase retains ATP binding preference even when its G4 motif is reverted to that of a typical GTPase (Koller-Eichhorn et al., 2007). In RhoBTB3, simple re-insertion of the key aspartic acid residue in the mutant, N138D decreased binding of ATP significantly, and decreased the low level of observed GTP binding even further. Thus, G4 sequences are important in RhoBTB3 for nucleotide binding and possibly also, hydrolysis.
Analogous to guanine nucleotide exchange factors, there may be proteins that enhance the rate of adenine nucleotide exchange: the GrpE subunit of the bacterial chaperone, DnaK functions precisely in this manner (Liberek et al., 1991); ATPase activating proteins are also anticipated (Lutkenhaus & Sundaramoorthy, 2003). Similarly, the motor ATPase, kinesin has a low intrinsic rate of ATP hydrolysis, and microtubules can stimulate kinesin’s ATP hydrolysis significantly (Kuznetsov & Gelfand, 1986). Because of RhoBTB3’s relatively low intrinsic rate of ATP hydrolysis, it is very likely to rely upon an ATPase activating protein to catalyze hydrolysis of ATP at the appropriate time and/or place. It will be of interest to try to determine whether membrane-associated, but not cytosolic TIP47 can activate RhoBTB3’s ATPase activity.
A model for RhoBTB3 in Rab9-dependent vesicle transport
Rab9 is present on late endosomes, and recruits TIP47 from the cytosol to that compartment, to package MPR cargo for export (Carroll et al., 2001; Ganley et al., 2004). Rab9 also decorates the transport vesicles that travel from late endosomes, along microtubule tracks towards the Golgi complex (Barbero et al., 2002). It has not been possible to determine if TIP47 remains on transport vesicles as they move to the Golgi, as N- or C-terminally tagged GFP proteins are non-functional, even with an additional linker included (Sincock et al., 2003). Also, >80% of TIP47 is in the cytosol (Diaz and Pfeffer, 1998), which interferes with live cell video microscopy. Nevertheless, it is likely that nascent vesicles will carry this cargo recognition protein.
Once Rab9+ vesicles reach the TGN, proteins coating the vesicle must be removed to permit subsequent membrane fusion. We favor a model in which Rab9, on the transport vesicle, activates RhoBTB3 when it arrives at the Golgi surface, to remove TIP47 from the vesicles to permit subsequent SNARE pairing and membrane fusion. RhoBTB3 was also needed for TGN46 retrograde transport. TIP47 does not bind to the cytoplasmic domain of TGN46 (Krise et al., 2000), thus RhoBTB3 would need another substrate for its action on this class of transport vesicle.
If TIP47 is on the vesicles, and RhoBTB3 is on the Golgi, do our isolated membrane complexes derive from docked vesicles? Future experiments will shed light on the full nature of this complex. It is of interest to note that Berthold et al. (2008b) reported myc-RhoBTB3 in a tight perinuclear pattern (like ours) while GFP-RhoBTB3 sometimes decorated peripheral vesicles. Perhaps the exogenous, GFP-RhoBTB3 protein is recruited onto TIP47-coated transport vesicles, en route to the Golgi complex. These workers also detected ubiquitylation of RhoBTB3 and interaction with exogenously expressed, dominant negative Cullin 3 ubiquitin ligase. The role of this interaction in membrane traffic is not yet known.
In summary, once again, the master Rab regulators of membrane traffic converge with the master Rho regulators of cell migration and motility, here by a direct interaction, to coordinate vesicle arrival at the Golgi complex. RhoBTB3 is a Rab9-regulated ATPase that may function in vesicle docking by uncoating transport vesicles at their target. How ATP hydrolysis is then coupled to the docking process represents an important area for future investigation.
Experimental Procedures
RhoBTB3 was identified in a Rab9Q66LΔCC two hybrid screen (Reddy et al, 2006). The two hybrid Rab collection was the generous gift of Dr. Francis Barr (Liverpool). Plasmid generation, protein production, binding, transport, enzyme assays and light microscopy were as described (Ganley et al., 2008; Burguete et al., 2008; Aivazian et al., 2006). Additional details can be found in the Supplemental Material.
Blue native PAGE--K562 membranes from the 0.8M/1.2M sucrose interface or K562 cytosol were prepared as described (Diaz and Pfeffer, 1998) and reacted with or without crosslinking with 1mM Dithiobis(sulfosuccinimidylproprionate (DTSSP, Pierce) by incubation of 10μg K562 membranes or cytosol in 50mM HEPES pH7.4, 150mM KCl, 2mM MgCl2 for 30 minutes at room temperature. The reaction was quenched with 20mM Tris for 10 minutes. Membranes were washed in BN-buffer (20mM Bis-Tris pH7.4, 500mM ε-aminocaproic acid, 20mM NaCl, 2mM EDTA, 10% glycerol) and solublized with 0.1% dodecyl maltoside in BN-buffer at 4°C for 1 hour. Coomassie G-250 was then added to 0.025%. Samples were loaded onto 3–12% polyacrylamide Bis-Tris gels (Invitrogen), transferred to PVDF, and probed with indicated antibodies.
For TIP47 release, 10μg K562 membranes were washed and incubated with 50mM HEPES pH7.4, 150mM KCl, 10mM EDTA for 15 minutes at 20°C. The membranes were then pelleted and resuspended in 50mM HEPES pH7.4, 150mM KCl, 5mM MgCl2. Either 100mM ATPγS, ATP regeneration system (Itin et al., 1997), or ATP regeneration system with Rab9 pre-loaded with GTPγS was added at 20°C for 30 minutes. Membranes were pelleted and resuspended in 0.1% dodecyl maltoside in BN-buffer.
Supplementary Material
Acknowledgments
This research was supported by a grant (NIDDK R37-37332) to SRP. K.S. was a fellow of the Leukemia and Lymphoma Society. We thank Dr. Jonathan Reddy for the original yeast two hybrid screen, Garret Hayes and Leonie Kastner for screening 54 Rab GTPases, and Margaret Cooke and Humsa Venkatesan for help with RhoBTB3 mutagenesis. Dr. Tim Fenn provided invaluable help with the structure modeling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aivazian D, Serrano RL, Pfeffer SR. TIP47 is a key effector for Rab9 Localization. J Cell Biol. 2006;173:917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman D, Sweeney HL, Spudich JA. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- Aspenström P, Fransson A, Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem J. 2004;377:327–337. doi: 10.1042/BJ20031041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenström P, Ruusala A, Pacholsky D. Taking Rho GTPases to the next level: the cellular functions of atypical Rho GTPases. Exp Cell Res. 2007;313:3673–3679. doi: 10.1016/j.yexcr.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbero P, Bittova L, Pfeffer SR. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J Cell Biol. 2002;156:511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold J, Schenkova K, Rivero F. Rho GTPases of the RhoBTB subfamily and tumorigenesis. Acta Pharmacol Sin. 2008a;29:285–295. doi: 10.1111/j.1745-7254.2008.00773.x. [DOI] [PubMed] [Google Scholar]
- Berthold J, Schenkova K, Ramos S, Miura Y, Furukawa M, Aspenström P, Rivero F. Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes-Evidence for an autoregulatory mechanism. Exp Cell Res. 2008b;314:3453–3465. doi: 10.1016/j.yexcr.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguete AS, Fenn TD, Brunger AT, Pfeffer SR. Dual GTPase regulation of the GCC185 Golgin: Communication between adjacent Rab6 and Arl1 binding sites. Cell. 2008;132:286–298. doi: 10.1016/j.cell.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KS, Hanna J, Simon I, Krise J, Barbero P, Pfeffer SR. Role of the Rab9 GTPase in facilitating receptor recruitment by TIP47. Science. 2001;292:1373–1377. doi: 10.1126/science.1056791. [DOI] [PubMed] [Google Scholar]
- Chang FK, Sato N, Kobayashi-Simorowski N, Yoshihara T, Meth JL, Hamaguchi M. DBC2 is essential for transporting vesicular stomatitis virus glycoprotein. J Mol Biol. 2006;364:302–8. doi: 10.1016/j.jmb.2006.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- Derby MC, Lieu ZZ, Brown D, Stow JL, Goud B, Gleeson PA. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 2007;8:758–773. doi: 10.1111/j.1600-0854.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- Diaz E, Pfeffer SR. TIP47: a cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Ganley I, Carroll K, Bittova L, Pfeffer SR. Rab9 regulates late endosome size and requires effector interaction for stability. Mol Biol Cell. 2004;15:5420–30. doi: 10.1091/mbc.E04-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Espinosa E, Pfeffer SR. A Syntaxin 10-containing SNARE complex distinguishes two distinct transport routes from endosomes to the trans Golgi network. J Cell Biol. 2008;180:159–172. doi: 10.1083/jcb.200707136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, Walsh T, Welcsh P, King MC, Wigler MH. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc Natl Acad Sci U S A. 2002;99:13647–13652. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang YW, Miller DL. A mutation that alters the nucleotide specificity of elongation factor Tu, a GTP regulatory protein. J Biol Chem. 1987;262:13081–13085. [PubMed] [Google Scholar]
- Itin C, Ulitzur N, Pfeffer SR. Mapmodulin, cytoplasmic dynein and microtubules enhance the transport of mannose 6-phosphate receptors from endosomes to the trans Golgi network. Mol Biol Cell. 1999;10:2191–2197. doi: 10.1091/mbc.10.7.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Koller-Eichhorn R, Marquardt T, Gail R, Wittinghofer A, Kostrewa D, Kutay U, Kambach C. Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J Biol Chem. 2007;282:19928–19937. doi: 10.1074/jbc.M700541200. [DOI] [PubMed] [Google Scholar]
- Krise JP, Sincock PM, Orsel JG, Pfeffer SR. Quantitative Analysis of TIP47 (Tail-interacting protein of 47kD)-receptor cytoplasmic domain interactions: implications for endosome-to-trans Golgi network trafficking. J Biol Chem. 2000;275:25188–25193. doi: 10.1074/jbc.M001138200. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Gelfand VI. Bovine brain kinesin is a microtubule-activated ATPase. Proc Natl Acad Sci U S A. 1986;83:8530–8534. doi: 10.1073/pnas.83.22.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. Rab9 functions in transport between late endosomes and the trans Golgi network in vitro. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J, Sundaramoorthy M. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol. 2003;48:295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: a conserved domain database for interactive domain family analysis. Nucl Acids Res. 2007;35:D237–240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau K, Das A, Walsh CT. Hsp90 chaperonins possess ATPase activity and bind heat shock transcription factors and peptidyl prolyl isomerases. J Biol Chem. 1993;268:1479–1487. [PubMed] [Google Scholar]
- Ramos S, Khademi F, Somesh BP, Rivero F. Genomic organization and expression profile of the small GTPases of the RhoBTB family in human and mouse. Gene. 2002;298:147–157. doi: 10.1016/s0378-1119(02)00980-0. [DOI] [PubMed] [Google Scholar]
- Reddy J, Burguete AS, Sridevi K, Ganley I, Nottingham RM, Pfeffer SR. A role for GCC185 in mannose 6-phosphate receptor recycling. Mol Biol Cell. 2006;17:4353–4363. doi: 10.1091/mbc.E06-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Riederer MA, Soldati T, Shapiro AD, Lin J, Pfeffer SR. Lysosome Biogenesis requires Mannose 6-Phosphate Receptor Recycling from Endosomes to the trans Golgi network. J Cell Biol. 1994;125:573–582. doi: 10.1083/jcb.125.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybin V, Ullrich O, Rubino M, Alexandrov K, Simon I, Seabra MC, Goody R, Zerial M. GTPase activity of Rab5 acts as a timer for endocytic membrane fusion. Nature. 1996;383:266–269. doi: 10.1038/383266a0. [DOI] [PubMed] [Google Scholar]
- Salas-Vidal E, Meijer AH, Cheng X, Spaink HP. Genomic annotation and expression analysis of the zebrafish Rho small GTPase family during development and bacterial infection. Genomics. 2005;86:25–37. doi: 10.1016/j.ygeno.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Simon I, Zerial M, Goody RS. Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J Biol Chem. 1996;271:20470–20478. doi: 10.1074/jbc.271.34.20470. [DOI] [PubMed] [Google Scholar]
- Sincock PM, Ganley IG, Krise J, Diederichs S, Sivars U, O’Connor B, Ding L, Pfeffer SR. Self-assembly is important for TIP47 function in mannose 6-phosphate receptor transport. Traffic. 2003;4:18–25. doi: 10.1034/j.1600-0854.2003.40104.x. [DOI] [PubMed] [Google Scholar]
- Siripurapu V, Meth J, Kobayashi N, Hamaguchi M. DBC2 significantly influences cell-cycle, apoptosis, cytoskeleton and membrane-trafficking pathways. J Mol Biol. 2005;346:83–89. doi: 10.1016/j.jmb.2004.11.043. [DOI] [PubMed] [Google Scholar]
- Sklan EH, Serrano RL, Einav S, Pfeffer SR, Lambright DG, Glenn JS. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem. 2007;282:36354–36361. doi: 10.1074/jbc.M705221200. [DOI] [PubMed] [Google Scholar]
- Sousa R, Lafer EM. Keep the traffic moving: mechanism of the Hsp70 motor. Traffic. 2006;7:1596–603. doi: 10.1111/j.1600-0854.2006.00497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang SR. G proteins, effectors and GAPs: structure and mechanism. Curr Opin Struct Biol. 1997;7:849–56. doi: 10.1016/s0959-440x(97)80157-1. [DOI] [PubMed] [Google Scholar]
- Swamy M, Siegers GM, Minguet S, Wollscheid B, Schamel WW. Blue native (BN-PAGE) for the identification and analysis of multiprotein complexes. Sci STKE. 2006;2006(345):p14. doi: 10.1126/stke.3452006pl4. [DOI] [PubMed] [Google Scholar]
- Tucker J, Sczakiel G, Feuerstein J, John J, Goody RS, Wittinghofer A. Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J. 1986;5:1351–1358. doi: 10.1002/j.1460-2075.1986.tb04366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins A, Ping Q, Carpenter CL. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev. 2004;18:856–861. doi: 10.1101/gad.1177904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ybe JA, Hecht MH. Sequence replacements in the central beta-turn of plastocyanin. Protein Sci. 1996;5:814–824. doi: 10.1002/pro.5560050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong JM, Chen-Hwang MC, Hwang YW. Switching nucleotide specificity of Ha-Ras p21 by a single amino acid substitution at aspartate 119. J Biol Chem. 1995;270:10002–10007. doi: 10.1074/jbc.270.17.10002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.