SUMMARY
Hemoglobin digestion is an essential process for blood-feeding parasites. Using chemical tools, we deconvoluted the intracellular hemoglobinolytic cascade in the tick Ixodes ricinus, a vector of Lyme disease and tick-borne encephalitis. In tick gut tissue, a network of peptidases was demonstrated through imaging with specific activity-based probes and activity profiling with peptidic substrates/inhibitors. This peptidase network is induced upon blood feeding and degrades hemoglobin at acidic pH. Selective inhibitors were applied to dissect the roles of the individual peptidases and determine the peptidase-specific cleavage map of the hemoglobin molecule. The degradation pathway is initiated by endopeptidases of aspartic and cysteine class (cathepsin D supported by cathepsin L and legumain) and continued by cysteine amino- and carboxy-dipeptidases (cathepsins C and B). The identified enzymes are potential targets to developing novel anti-tick vaccines.
INTRODUCTION
Ticks are important ectoparasites that transmit a wide range of infectious agents including arboviruses, rickettsiae, spirochetes and protozoa causing diseases in humans and domestic animals (for review, see Jongejan and Uilenberg, 2004). The hard ticks of the genus Ixodes (e.g. I. ricinus and I. scapularis, in Europe and North America, respectively) are important vectors for Lyme disease (borreliosis) and tick-borne encephalitis. In addition to the threat to public health, a number of the hard tick species such as Boophilus microplus negatively impact on livestock productivity in tropical and subtropical areas.
A primary interface for vector-pathogen interaction is the tick gut, hence pathogen transmission is linked to the physiology of blood feeding and digestion. Blood provides a rich source of proteins for anabolic processes such as vitellogenesis and egg production. Hemoglobin released from erythrocytes is a primary nutritive component. Unlike other blood-feeding arthropods, ticks digest hemoglobin and other proteins intracellularly in the acidic endosomal/lysosomal vesicles of gut cells (Coons and Alberti, 1999; Sonenshine, 1991) at pH values well below the pH 6.3–6.5 of the gut contents (Mendiola et al., 1996). The absence of extracellular digestion enables the gut lumen to serve as a storage organ where hemoglobin may form crystalline deposits. Lara et al. (2005) showed that the digestive cells of the hard tick Boophilus microplus have specific receptor-mediated endocytic pathways for hemoglobin. The intracellular digestion of hemoglobin is linked to the detoxification of released heme, which forms aggregates accumulated inside specialized organelles called hemosomes (Lara et al., 2003).
Our understanding of the proteolytic arsenal in the tick gut tissue is fragmented. Previous studies have focused on individual enzymes in particular species and identified mainly cysteine peptidases (e.g. Renard et al., 2000; Sojka et al., 2007; Tsuji et al., 2008) and aspartic peptidases (Boldbaatar et al., 2006). Peptidases of other classes are also expressed in the tick gut, e.g. a leucine metallo-aminopeptidase and a hemolytic serine endopeptidase found in cytoplasm and lumen, respectively (e.g. Hatta et al., 2006; Miyoshi et al., 2007). Recently, we undertook a more systematic approach and determined a set of cDNA sequences for cysteine and aspartic peptidases from the gut of I. ricinus (Sojka et al., 2008). The midgut transcriptome (mialome) from the hard tick Dermacentor variabilis has now been reported that contains a panel of proteolytic enzymes from four main classes (Anderson et al., 2008).
At the protein level, however, there is a lack of information regarding the molecular proteolytic machinery in the tick gut tissue that degrades host proteins. The present work provides biochemical insight into the question using a proteomics platform that incorporates functional genomics approaches. This has only recently become feasible with the help of chemical tools such as activity-based probes for selectively imaging target peptidases (for review, see Fonovic and Bogyo, 2008). Herein, we uncover how hemoglobin is proteolytically digested in I. ricinus. First, we characterize a multienzyme network of peptidases that orchestrates intracellular hemoglobinolysis. Second, using cleavage mapping of the hemoglobin molecule, we define the contributions of individual peptidases to this process. Finally, we compare digestive proteolysis in ticks and other blood-feeding parasites, and discuss potential application of our results in anti-tick vaccine development.
RESULTS
Profiling of Peptidase Activities in the Tick Gut
The distribution of peptidases was analyzed by activity profiling in the extract from the gut tissue of partially engorged I. ricinus females. Table S1 summarizes the specific substrates and inhibitors used as diagnostic tools; the pH profiles and inhibitory sensitivity of the identified activities are presented in Figure 1. The major activity that cleaves the substrate Z-Phe-Arg-AMC was attributed to cathepsin B according to sensitivity to the inhibitor CA-074. A minor part of the activity to this substrate (insensitive to CA-074) was inhibited by Z-Phe-Phe-DMK, a preferential inhibitor of cathepsin L. In addition to endopeptidase activity (Z-Phe-Arg-AMC), mammalian cathepsin B can function as an exopeptidase, specifically as a peptidyl dipeptidase. We demonstrate that the same is also true for the I. ricinus cathepsin B by hydrolysis of the substrate Abz-Phe-Arg-Nph-Ser (sensitive to CA-074, see Table S1). Cathepsin C activity was assayed with the substrate Gly-Arg-AMC and was fully blocked by Gly-Phe-DMK. The legumain activity measured with Z-Ala-Ala-Asn-AMC was insensitive to E-64 (in contrast to cathepsins B, L and C) but sensitive to chicken cystatin as well as the specific inhibitor Aza-N-11a. Cathepsin D activity monitored with the substrate Abz-Lys-Pro-Ala-Glu-Phe-Nph-Ala-Leu was inhibited by the class-selective inhibitor pepstatin. In summary, five significant endo- and exopeptidase activities were profiled in the gut tissue: (i) cysteine peptidase class: cathepsins B and L, and dipeptidyl peptidase I (cathepsin C) of the CA clan (papain-type), and asparaginyl endopeptidase (legumain) of the CD clan, and (ii) aspartic peptidase class: cathepsin D of the AA clan. In addition, activities of two monopeptidases were detected: (iii) carboxypeptidase of serine peptidase class (sensitive to PMSF) most likely from the SC clan (Motobu et al., 2007), and (iv) aminopeptidase of metallo peptidase class (sensitive to bestatin) most likely a leucine aminopeptidase from the MF clan (Hatta et al., 2006). Importantly, no significant endopeptidase activities belonging to metallo or serine peptidases were identified in the gut tissue (Table S1).
Figure 1. Substrate and Inhibitor Profiling of Tick Gut Peptidases.
Proteolytic activities in the gut tissue extract from I. ricinus were measured with specific peptidase substrates (listed on the side of the bar graphs) using a continuous fluorimetric assay.
Right: The pH profiles of individual peptidase activities (RFU, relative fluorescence units).
Left: The inhibitory profiles of individual activities. Each activity was measured in the presence of selective peptidase inhibitors (bars) relevant to the target enzyme. The values are expressed as percent inhibition of the control activity. The inhibition was assayed at the pH optimum; three pH values were used for inhibition profiling of cathepsin B/L activities. The diagnostic activity and inhibitory responses allowed to classify the peptidase activities as indicated. For details, see Table S1 and Experimental procedures. The mean values ±SE are given.
The cysteine peptidases exhibited the greatest activity at the mildly acidic pH (from 4 to 6) and aspartic proteases at the pH range of 3 to 4 (Figure 1). The pH profile of legumain shows a sharp drop of the activity between pH 5.5 and 6.0, previously described for the recombinant legumain from I. ricinus (Sojka et al., 2007). A three-peak pH profile was recorded with the substrate Z-Phe-Arg-AMC; the inhibitory pattern measured along the profile suggests the presence of two distinct cathepsin B activities and a cathepsin L (Figure 1). The pH optimum (~6.5) of leucine aminopeptidase may suggest that this enzyme is functioning in a different compartment than the other, more acidic, peptidases.
Visualizing Tick Gut Peptidases with Active-Site Probes
The major identified peptidases were demonstrated in the tick gut extract by LC-MS/MS analysis (Table S2). For their visualization, a panel of specific activity-based probes (ABPs) was used (Figure 2). Three chemically reactive ABPs were employed for imaging cysteine peptidases, namely Green-DCG-04 (Greenbaum et al., 2002), FY01 (Yuan et al., 2006), and Fhex-PD-AOMK (Sexton et al., 2007), which selectively label cathepsin B/L, cathepsin C and legumain, respectively (Figure 2A). The visualized peptidase species were authenticated by quenching the signal when the active-site labeling was performed in competition with specific active-site inhibitors of the targeted enzymes. The competition experiments with CA-074 and Z-Phe-Phe-DMK clearly distinguished cathepsin B and cathepsin L. The molecular masses determined for cathepsin B and L (MW ~32 and 30 kDa) and legumain (MW ~38–40 kDa) are in accord with values calculated from the cDNA sequences of the respective mature enzymes (Sojka et al., 2008). The observed mass of cathepsin C (MW ~23–25 kDa) suggests that the enzyme precursor of MW ~50 kDa (Sojka et al., 2008) is proteolytically processed; the same chain pattern was found for mammalian cathepsin C (Horn et al., 2002).
Figure 2. Imaging of Tick Gut Peptidases with Active-Site Probes.
Extract from the gut tissue of I. ricinus was treated with a panel of selective activity-based probes (ABPs) that interact with the active site of the target peptidases.
(A) ABPs for labeling of cysteine peptidases: Green-DCG-04 for cathepsin B (CatB) and cathepsin L (CatL), FY01 for cathepsin C (CatC), and Fhex-PD-AOMK for legumain (AE). The ABP structures contain an irreversible active-site ligand as chemically reactive warhead (gray box) and fluorescent tag as a reporter group (open oval).
(B) ABP named bPSP-07 for labeling of aspartic peptidase cathepsin D (CatD). The bPSP-07 structure contains a reversible active-site ligand (gray box), which is cross-linked to the protein target by a photoreactive group, and biotin tag as a reporter group (open oval).
The reaction mixture was resolved by Laemmli-SDS-PAGE and the labeled peptidases were visualized in the gel (fluorescence signal) or on the chemiluminescent avidin blot (biotin signal). The competitive labeling was performed in the presence of selective active-site inhibitors: CA-074 (CatB inhibitor), Z-Phe-Phe-DMK (CatL inhibitor), Aza-N-11a (AE inhibitor), Gly-Phe-DMK (CatC inhibitor) and pepstatin (CatD inhibitor) (for details, see Experimental procedures).
For labeling of aspartic peptidases, we designed a novel photoreactive ABP, denoted bPSP-07 (Figure 2B) (see Materials). The probe selectively visualized a species of ~45 kDa, which accords with the cDNA-calculated mass of I. ricinus cathepsin D (Sojka et al., 2008). Labeling was quenched in the presence of pepstatin, an inhibitor of aspartic peptidases.
Quantification of Tick Gut Peptidases by Active-Site Titration
The molar content of effectively acting peptidases in the tick gut was determined. For this purpose, we employed the active-site titration of individual cysteine and aspartic peptidases in the gut tissue extract by their specific irreversible or tight binding inhibitors (see Experimental procedures). This profiling revealed a large abundance of cathepsins B and C at 202±22 and 194±19 fmol/ μg of gut tissue protein, respectively. The content of cathepsin D and legumain was much lower at the level 15±2 and 18±3 fmol/ μg of gut tissue protein, respectively. Cathepsin L was not directly quantified due to a lack of good titrant, however, its amount can be estimated from the active-site labeling experiment to be <5% of the cathepsins B amount (Figure 2A).
Hemoglobin Degradation in the Tick Gut Occurs at Acidic pH
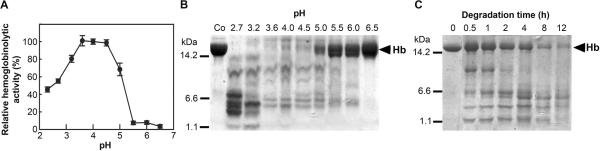
Degradation of hemoglobin by proteolytic activities of gut tissue extract was investigated in vitro using direct quantification of hemoglobin fragments and their SDS-PAGE analysis. The degradation was most efficient at acidic pH between 3.5 and 4.5, and was not observed above pH 5.5 (Figure 3A and 3B). The time course of hemoglobin degradation at the pH optimum demonstrates a gradual hydrolysis of the hemoglobin substrate (α-subunit: ~15 kDa, β-subunit: ~16 kDa) into initial fragments of 8–11 kDa, then 2–7 kDa peptidic fragments that are further processed (3–5 kDa species) and gradually removed (Figure 3C). The pattern of hemoglobin-derived fragments was pH dependent outside the pH optimum range; the large fragments (2–7 kDa) accumulated at pH below 3.5 (Figure 3B).
Figure 3. pH Profile of Hemoglobin Degradation by Tick Gut Peptidases.
Bovine hemoglobin was digested in vitro with the gut tissue extract from I. ricinus at various pH values.
(A) The degradation rate of hemoglobin was determined with the fluorescamine derivatization assay quantifying the liberated fragments. The mean values ±SE are expressed relatively to the maximum value.
(B) The hemoglobin digest (12 h) was subjected to Tricine-SDS-PAGE. The hemoglobin substrate (Hb mark) and its degradation products are visualized by protein staining.
(C) The time course of hemoglobin degradation was performed at the optimal pH 4.2.
A Network of Peptidases is responsible for Hemoglobinolysis in the Tick Gut
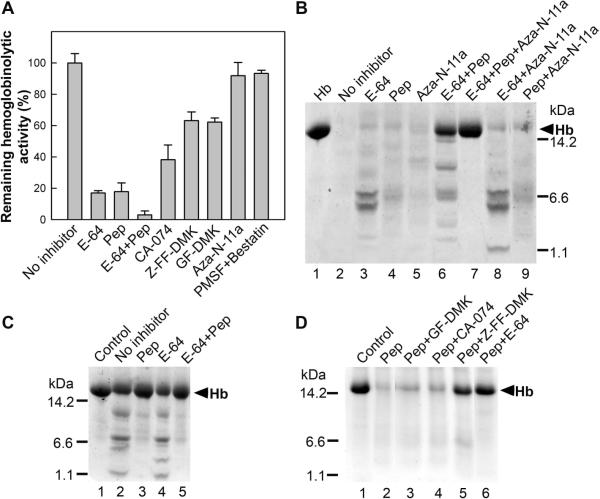
The contribution of individual peptidase activities to hemoglobin degradation was evaluated by analyzing the in vitro impact of selective peptidase inhibitors (Figure 4). Two different assays were employed, namely (i) quantification of hemoglobin fragments by fluorescamine derivatization to determine degradation rate, and (ii) SDS-PAGE visualization of the fragmentation pattern of hemoglobin.
Figure 4. Inhibition of Hemoglobin Degradation by Selective Peptidase Inhibitors.
Bovine hemoglobin was digested in vitro with the gut tissue extract at the optimal pH of 4.2. The extract was preincubated with the peptidase inhibitors (or their mixtures) prior to the initiation of digestion. A glossary of inhibitors (and target enzymes): pepstatin (Pep; aspartic peptidases), E-64 (papain-type cysteine peptidases), CA-074 (cathepsin B), Z-FF-DMK (cathepsin L), GF-DMK (cathepsin C), Aza-N-11a (legumain), PMSF (serine peptidases), bestatin (metallo peptidases).
(A) The degradation rate of hemoglobin was determined with the fluorescamine derivatization assay. The inhibition of the digestion is expressed as remaining degradation activity relative to the uninhibited control (100%). The mean values ±SE are given.
(B,C,D) The hemoglobin digest was subjected to Tricine-SDS-PAGE; the hemoglobin substrate (Hb mark) and its degradation products are visualized by protein staining.
(B) Three-inhibitor profile defines three groups of peptidases critical for hemoglobin degradation (a prolonged 16 h digest).
(C) Inhibitory profile of a short-term digest (30 min) shows critical importance of aspartic peptidases for the initial phase of degradation.
(D) Pepstatin inhibited profile demonstrates the supportive role of cathepsin L among papain-type peptidases in the initial fragmentation (16 h digest).
The fluorescamine derivatization assay showed that individual application of E-64 and pepstatin (targeting papain-type cysteine peptidases and aspartic peptidases, respectively) dramatically reduced the rate of hemoglobin digestion to about 20% (Figure 4A). Their combination resulted in a near total suppression of the hydrolysis rate to ~5%. Among cysteine peptidases, three papain-type peptidases, cathepsins B, C and L contribute to the degradation; their selective inhibition by CA-074, Gly-Phe-DMK and Z-Phe-Phe-DMK blocked about 60%, 40% and 40% of hemoglobinolysis rate, respectively. Inhibitors of serine and metallo peptidases did not significantly suppress the hemoglobin degradation process.
The SDS-PAGE visualization of the long-term digest further demonstrated that papain-type and aspartic peptidases act with some redundancy (Figure 4B, lines 2 and 3). The complete blockage of hemoglobinolysis was observed when E-64 and pepstatin were combined with Aza-N-11a, a legumain inhibitor (Figure 4B, line 7). Legumain alone can also degrade hemoglobin (Figure 4B, lines 6 and 7), however, its proteolytic contribution is much less than to either of the papain-type and aspartic peptidases (Figure 4B, lines 6, 8 and 9 ).The result demonstrates the critical involvement of peptidases of aspartic and cysteine classes in acidic hemoglobinolysis and suggests their cooperative action in vivo in the gut tissue.
Which peptidase(s) is responsible for the initial cleavage of the hemoglobin molecule? In Figure 4C and D, we show that both cathepsins D and L contribute, although legumain can also participate (see above). Figure 4C shows that the initial phase of the degradation process is specifically blocked by the cathepsin D inhibitor, pepstatin. However, cathepsin D can be substituted by cathepsin L (but not by cathepsin B or C) during prolonged digestion (Figure 4D). Hemoglobinolysis by the cathepsin L activity could be specifically inhibited with Z-Phe-Phe-DMK (Figure 4D, line 5) in which cathepsin D activity was also inhibited.
The papain-type peptidases were found to be important for the removal of large hemoglobin fragments. These species (5–7 kDa) accumulated in the experiments where the action of papain-type peptidases was suppressed by (i) inhibition with E-64 (Figure 4B), and (ii) extremely low pH that favors cathepsin D action (Figure 3B). Using two approaches, we demonstrated that papain-type peptidases are responsible for the processing of large fragments and the generation of small peptides and dipeptides. First, the pattern of hemoglobin-derived fragments on RP-HPLC profiles was compared to the digests inhibited by E-64 or pepstatin. There was a significant difference in the distribution of large fragments versus small peptides/dipeptides, the latter being absent in the digest treated by E-64 (Figure 5A). Second, we prepared a pool of large hemoglobin fragments produced by cathepsin D-driven digestion (see Experimental procedures). This material was found to be efficiently degraded by the tick gut extract, however, the degradation was blocked in the presence of E-64 (Figure 5B).
Figure 5. Papain-Type Peptidases Generate Small Fragments and Dipeptides from Hemoglobin.
(A) Bovine hemoglobin was digested (16 h) in vitro with extract from the gut tissue of I. ricinus at pH 4.2 in the presence of E-64 or pepstatin. The fragments derived from hemoglobin were resolved using C18 RP-HPLC. The E-64-inhibited elution profile was subtracted from the pepstatin-inhibited profile: Section 1 (majority in pepstatin-treated digest) contains small fragments and dipeptides, Section 2 (majority in E-64-treated digest) contains large fragments. The position of the intact hemoglobin is indicated (subunits αHb, βHb) in the control profile (dashed line).
(B) The large hemoglobin fragments of Section 2 were pooled (see inset for Tricine-SDS-PAGE pattern) and digested in vitro with the gut tissue extract at pH 4.2 in the presence of E-64 and/or pepstatin. The degradation rate of the large fragments was determined with the fluorescamine derivatization assay. The inhibition of the digestion is expressed as remaining degradation activity relative to the uninhibited control (100%). The mean values ±SE are given.
A Proteolytic Cleavage Map of the Hemoglobin Molecule
A detailed proteomic study was aimed at elucidating the cleavage sites in the hemoglobin molecule attacked by the key peptidases. The tick gut tissue extract was treated with combinations of three selective peptidase inhibitors (E-64, pepstatin, Aza-N-11a) to obtain the peptidase-specific digests of hemoglobin driven by papain-type cysteine peptidases, aspartic peptidases (namely cathepsin D) and legumains, respectively. The resulting fragments were characterized by mass spectrometry (see Experimental procedures). The identified cleavage sites in the sequence of α- and β-subunit of hemoglobin are shown in Figure 6A. In addition, these positions are presented in the 3D model of the hemoglobin αβ-dimer (Figure 6B). The αβ-dimer is the major oligomeric form of hemoglobin at a pH of 4.2 (Boys et al., 2007) that we found to be the optimal pH for hemoglobin degradation by the tick peptidase cocktail (Figure 3A).
Figure 6. Proteolytic Cleavage Map of the Hemoglobin Molecule.
(A) Bovine hemoglobin was digested in vitro with the gut tissue extract from I. ricinus at pH 4.2. The extract was preincubated with the selective inhibitors of key hemoglobinolytic peptidases to obtain peptidase-specific digests. These digests were driven by papain-type peptidases (pepstatin and Aza-N-11a treated extract), legumains (pepstatin and E-64 treated extract), and cathepsin D (E-64 and Aza-N-11a treated extract). The fragments were identified by mass spectrometry and the corresponding cleavage sites are indicated in the hemoglobin sequence: cleavage by papain-type peptidases (▲); cleavage by legumain (▲); cleavage by cathepsin D (▼) with the initial cleavage sites marked ( ). Unambiguously assigned dipeptides produced by the action of exopeptidases are underlined.
). Unambiguously assigned dipeptides produced by the action of exopeptidases are underlined.
(B) The map of cleavage sites in (A) is presented in the 3D ribbon model of the hemoglobin αβ-dimer. The digested peptide bonds are highlighted according to the color coding for individual peptidase-specific digests (see A).
(C) The size distribution of hemoglobin fragments in individual peptidase-specific digests. The fragments are represented by circles (for color coding see (A)); relative size of the circles corresponds to the number of assigned peptides of the same length.
(D) The P1-P1' subsite specificities of the key hemoglobinolytic peptidases were inferred from the cleavage site map (A). The plots generated by WebLogo (Crooks et al., 2004) depict relative entropy between the observed and background distributions of amino acids at each subsite. The overall height of each letter stack indicates the sequence conservation at that position, whereas the height of amino acid symbols within the stack reflects the relative frequency of the corresponding residue at that position. Amino acid color coding: acidic (red), basic (blue), neutral (purple), polar (green), hydrophobic (black).
The data were used for analysis of the cleavage site specificity and fragmentation pattern (Figure 6C and 6D). The cleavage sites of cathepsin D (~26% of cuts) contain hydrophobic residues in the P1/P1' position with a preference for Leu and Phe in P1. Three of these sites, Phe-Leu(35α), Leu-Ala(111α) and Phe-Phe(41β) are most likely responsible for the initial fragmentation of the hemoglobin molecule since these site are only found in the short-term digest by cathepsin D (Figure 6A) as well as in the non-inhibited short-term digest (data not shown). These sites are located on the exposed termini of helices of the α-subunit and a loop at the heme-binding pocket of the β-subunit, which might have an important impact on unfolding of the hemoglobin substrate. The legumain cleavage (~11% of cuts) was targeted strictly to Asn and Asp residues in the P1 position. Although legumains are generally highly specific for hydrolysis of asparaginyl bonds, thay can also cleave aspartyl bonds; this secondary activity increases towards lower pH (Rotari et al., 2001). Thus the pH of the digest (~4.2 corresponding to the hemoglobinolytic optimum) most likely explains the observed considerable cleavage at Asp residues. A dense series of cleavage sites (~64% of cuts) was found for papain-type peptidases, which exhibited less defined specificity requirements. This reflects the fact that several peptidases participated in this digest. An involvement of cathepsins B, C and L is indicated by the prevalent cleavage after Lys (Figure 6D) which agrees with the general preference of these enzymes for basic residues at P1 (Choe et al., 2006; McGuire et al., 1997). Figure 6C shows that the fragments produced by papain-type peptidases are predominantly small peptides and dipeptides, supporting the proposed function that these enzymes complete the degradation of hemoglobin which involves less specific exopeptidase trimming. This fragmentation pattern significantly differs from that found for aspartic peptidases and legumains which produce generally longer peptides.
Dynamics of the Hemoglobinolytic System during Bloodfeeding by I. ricinus
In order to explore the dynamics of digestive proteolysis, we investigated changes in the hemoglobinolytic capacity of ticks during the blood-feeding process (Figure 7). It typically takes ~8 days in I. ricinus and contains the slow feeding period (days 1–6) and the rapid engorgement period (days 6–8) (Coons and Alberti, 1999; Sonenshine, 1991). There is low hemoglobinolytic activity (a lag phase) during the first 4 days of blood-feeding. A dramatic 10-fold boost in hemoglobinolysis occurs between days 4 and 6, towards the end of the slow feeding period. The activity further increased during the rapid engorgement period to full repletion and detachment from the host. The result demonstrates the correlative upregulation of the hemoglobinolytic system in the tick gut tissue in response to bloodfeeding.
Figure 7. Dynamics of Hemoglobinolytic System during Bloodfeeding by I. ricinus.

Time course of changes of hemoglobinolytic activity in the gut tissue of I. ricinus females during blood-meal uptake. The activity was measured at pH 4.2 in the hemoglobinolytic fluorescamine assay; the mean values ±SE are given. The data are normalized to one tick, and the maximum is set to 100%. The insets show the size of a feeding tick at the indicated time points from attachment to the host to repletion and detachment on the 8th day (scale bar: 1 cm).
In this work, we used the 6-day-fed tick as a representative time point for detailed analysis of digestive system to ensure high proteolytic activity, easy manipulation with the gut tissue, and for comparison with a preferred time-point in analyses of other hard tick species (Anderson et al., 2008).
DISCUSSION
In the present work, we identify and unravel a proteolytic system functioning in intracellular protein digestion in the gut tissue of the hard tick I. ricinus. The focus was on the mechanism of the degradation of hemoglobin, a major host blood protein and a key nutrient for ticks (Coons and Alberti, 1999; Sonenshine, 1991).
In the first step, we biochemically dissected a number of peptidase activities in tick gut tissue. For the activity profiling, we employed a battery of specific substrates and inhibitors. The peptidases were further authenticated by imaging using a unique panel of selective activity-based probes. This combined approach enabled the classification of the main component peptidase activities: (i) clan CA cathepsins B, C and L (ii) clan CD asparaginyl endopeptidase (legumain) and (iii) clan AA cathepsin D. Other activities detected were attributed to monopeptidases, namely a serine carboxypeptidase and a leucine metallo-aminopeptidase.
In the second step, we simulated in vitro the degradation of hemoglobin with gut tissue extracts. Hydrolysis is optimal at acidic pH (3.5 to 4.5) which corresponds to the pH environment of gut digestive vesicles (Lara et al., 2005). The contribution of each peptidase activity to degradation of the hemoglobin substrate was dissected by selective inhibitors. The subsequent data on degradation potency and the fragment profile enabled us to trace the degradation pathway and propose the following hemoglobinolytic model (Figure 8).
Figure 8. The Hemoglobinolytic Pathway in I. ricinus.
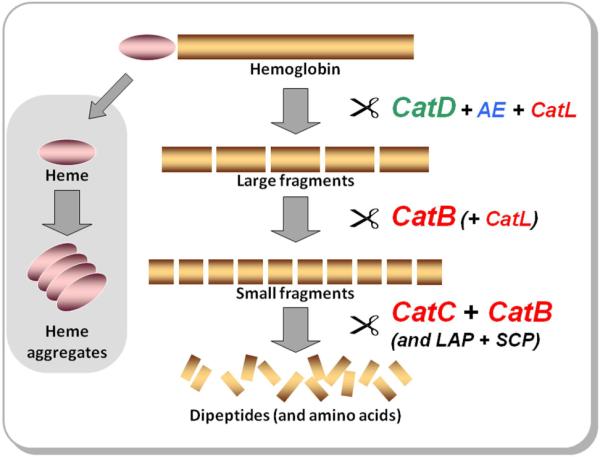
A mechanistic model for the proteolytic pathway of hemoglobin degradation in the digestive vesicles of I. ricinus midgut cells. The endopeptidases, cathepsins D (CatD) supported by cathepsin L (CatL) and legumain (AE), are responsible for primary cleavage of hemoglobin. The production of secondary small fragments is dominated by the endopeptidase activity of cathepsin B (CatB). Exopeptidases act on the peptides released by the action of the endopeptidases through the carboxy-dipeptidase activity of CatB and the amino-dipeptidase activity of cathepsin C (CatC). Monopeptidases, including serine carboxypeptidase (SCP) and leucine aminopeptidase (LAP), might participate in the liberation of free amino acids. The enzymes are color-coded according to clan membership; AA clan aspartic peptidases (green), CD clan cysteine peptidases (blue), CA clan (papain-type) cysteine peptidases (red), and serine and metallo peptidases (black). The heme moiety forms aggregates that accumulate inside the hemosome, a specialized organelle of the digestive cell (Lara et al., 2003).
Three endopeptidases could perform the initial cleavage of the hemoglobin molecule, namely, cathepsin D, cathepsin L and legumain. Among them, cathepsin D is the most dynamic enzyme due to its high turnover efficiency. Cathepsin L has lower turnover efficiency, and hence its role can be supportive. Legumain is less effective regarding the extent of fragmentation, however it plays a specific role in completion of the digestion. The papain-type peptidases cathepsin B and cathepsin C are dominant in the next phases of the hemoglobinolysis that lead from the level of large fragments to dipeptides. We show that these two enzymes, executing massive decomposition of the gross fragments, are ~10 fold more abundant in the tick gut tissue than those endopeptidases involved in the first step of the pathway. Cathepsin B exerts its dual endopeptidase and peptidyl dipeptidase activity to generate small peptides and remove carboxy-terminal dipeptides, while cathepsin C is functioning as dipeptidyl peptidase responsible for production of dipeptides by amino-terminal trimming.
We suppose that the in vivo pathway might also incorporate the action of monopeptidases in the liberation of free amino acids from hemoglobin-derived peptides. However, although we detected the activity of serine carboxypeptidase and leucine aminopeptidase in the gut tissue extract, their participation in hemoglobinolysis was not proven by our in vitro experiments. This might be linked to other factors like compartmentation that can influence the reconstituted pathway; for example, an alkaline leucine aminopeptidase was reported to be a cytosolic enzyme in the gut cells of the hard tick Haemaphysalis longicornis (Hatta et al., 2006).
Hemoglobin degradation with the native cocktail of the gut tissue peptidases occurs at acidic pH (up to pH ~5.5), which complies with the identified activity range of the peptidases that constitute the hemoglobinolytic network. Interestingly, we observed that the pH optimum of the individual enzymes apparently increases along their hypothesized position downstream the pathway (Figure 1). Thus, cathepsins D and L are the most acidic enzymes: the pH optimum of the I. ricinus cathepsin L is unusually low in comparison with mammalian homologs but similar to that of a cathepsin L in B. microplus larvae (Estrela et al., 2007). We hypothesize that such a gradient of pH optima might be associated with a physiological regulation mechanism of hemoglobinolysis controlled by pH in the digestive organelle.
An analysis of cleavage sites in the hemoglobin molecule revealed a distinct pattern of subsite specificities of the component peptidases (Figure 6D). The cleavage sites cover hydrophobic residues (P1-P1') for cathepsin D, highly restricted hydrophilic neutral/acidic residues (Asn/Asp in P1) for legumain, and a promiscuous pattern with a basic residue prevalent in P1 for papain-type peptidases. Further, each of these three clans of peptidases digested hemoglobin with markedly different distributions of peptide fragment lengths (Figure 6C). This demonstrates that the peptidase clans participating in hemoglobinolysis are generally complementary in their substrate site preferences.
Hemoglobin-derived peptides possessing antimicrobial activity have been identified in the guts of several hard and soft tick species. The best characterized is a peptide ranging from Phe(34α) to Lys(62α) from B. microplus (Sforca et al., 2005). The comprehensive cleavage map of the hemoglobin molecule we have generated (Figure 6A) allows us to predict the possible origin of these peptides. For I. ricinus, we found that the N-terminal Met-Phe(34α) bond is susceptible to hydrolysis by cathepsin D, and that the C-terminal Lys-Val(63α) bond is cleaved by a papain-type peptidase. The N-terminal cleavage site is located in the proteolytically accessible hinge region of the hemoglobin α-subunit, which is frequently recognized for initial cleavage on the hemoglobin molecule. This has been demonstrated for digestive aspartic peptidases of several other blood-feeding parasites, e.g. cathepsin D from Schistosoma japonicum and plasmepsins from Plasmodium falciparum (Brinkworth et al., 2001; Goldberg et al., 1991).
The hemoglobinolytic pathway herein identified for I. ricinus allows us to compare the digestive proteolytic system of this tick with those of other blood-feeding parasites. In general, there is a remarkable similarity in the pathway and its component enzymes to platyhelminths and nematodes, as represented by Schistosoma and hookworms (Caffrey et al., 2004; Delcroix et al., 2006; Ranjit et al., 2009; Williamson et al., 2004) and protozoa such as Plasmodium (Goldberg, 2005). In these phylogenetically diverse organisms, digestive proteolysis is likewise based on cooperating aspartic and cysteine peptidases. Thus, this acidic proteolytic system clearly differs from alkaline digestive proteolysis that utilizes serine peptidases, e.g., in hematophagous insects.
In summary, our study identifies and dissects a network of aspartic and cysteine peptidases that orchestrates the intracellular digestion of hemoglobin in the I. ricinus gut at acidic pH. The process is partially ordered in that some enzymes act to initially cleave the substrate and that these and others then cooperate (with some redundancy) to complete substrate hydrolysis. The hemoglobinolytic system is dynamic being upregulated during blood-feeding by the tick. Considering the complexity of digestive proteolysis in ticks uncovered here, new avenues for further research are offered. Thus, we are engaged in addressing the following. (i) Does the identified hemoglobinolytic pathway operate in a similar manner to degrade other host blood proteins? (ii) What is the contribution of peptidase isoenzymes, the existence of which is suggested in this study? (iii) What mechanisms regulate the pathway e.g. trans-activation by other proteases? These studies will be the subjects of future reports.
SIGNIFICANCE
Ticks are vectors for a number of viral and bacterial diseases in humans and domestic animals. To survive and reproduce, ticks feed on host blood and digest hemoglobin. This critical process is still poorly understood. Using Ixodes ricinus as a model hard tick, our work fills the gap in tick biology and provides biochemical insight into the mechanism of hemoglobin peptidolysis in the tick gut. We pursue functional proteomic approaches to identify a suite of gut peptidases that operate in an ordered pathway to complete the hydrolysis of hemoglobin. Because of their central function in nutrition of the parasite, one or more of the digestive peptidases may prove useful as a molecular vaccine to limit parasite survival and transmission of associated diseases. The vaccination efficacy of proteins from the tick gut (`concealed' antigens) in controlling tick infestations has already been successfully demonstrated (for review, see de la Fuente and Kocan, 2006), and new candidate antigens are increasingly in demand to combat the spread of tick-borne diseases.
EXPERIMENTAL PROCEDURES
Materials
The fluorogenic AMC-substrates (AMC, 7-amino-4-methylcoumarin) were from Bachem, fluorescence resonance energy transfer (FRET) substrates Abz-Lys-Pro-Ala-Glu-Phe-Nph-Ala-Leu and Abz-Phe-Arg-Nph-Ser (Abz, aminobenzoic acid; Nph, 4-nitrophenylalanine) was prepared as described previously ((Masa et al., 2006; Pohl et al., 1987)). Peptidase inhibitors were from Bachem, bestatin from Sigma (Sigma), Gly-Phe-diazomethyl ketone (DMK) was prepared as described in Green and Shaw (1981), and chicken egg white cystatin was isolated according to Nicklin and Barrett (1984). The aza-peptide Michael acceptor CBz-Ala-Ala-(aza-Asn)-CH=CH-COOEt (Aza-N-11a) (Ekici et al., 2004) and vinyl sulfone Ala-Hph-VS-Ph (Kam et al., 2004) were kindly donated by Dr. J.C. Powers. The probes Green-DCG-04, FY01 and Fhex-PD-AOMK were synthesized as described previously (Greenbaum et al., 2002; Kato et al., 2005; Yuan et al., 2006). The bPSP-07 probe was designed based on the pepstatin structure optimized for cathepsin D inhibition (Majer et al., 1997); it was synthesized by Fmoc solid phase chemistry and postsynthetic modification by sulfosuccinimidyl-6-(4'-azido-2'-nitrophenylamino)hexanoate (Pierce); bPSP-07 was demonstrated to label human cathepsin D (not shown).
Ticks and the Tick Gut Tissue Extract
I. ricinus ticks were collected by flagging in localities around the town of Ceske Budejovice (Czech Republic). The guts were dissected from 10 I. ricinus females fed for 6 days (if not otherwise stated) on laboratory guinea pigs (Sojka et al., 2007). The gut contents were carefully removed without disrupting the epithelium, and the gut tissue was washed from the host blood in phosphate buffered saline. Gut tissue extract (150 μg protein/ml) was prepared by homogenization of the pooled gut tissue in 0.1 M Na-acetate, pH 4.5, 1% CHAPS, 2.5 mM DTT on ice. The extract was cleared by centrifugation (16000 g, 10 min, 4°C), filtered with Ultrafree-MC 0.22 μm (Millipore) and stored at −80 °C.
Profiling of Tick Peptidases with Substrates and Inhibitors
Proteolytic activities were identified and characterized by hydrolysis of the following substrates: 25 μM Z-Arg-Arg-AMC or Abz-Phe-Arg-Nph-Ser for cathepsin B (Barrett and Kirschke, 1981; Pohl et al., 1987), 25 μM Z-Phe-Arg-AMC for cathepsin L/B or trypsin (Barrett and Kirschke, 1981), 30 μM Gly-Arg-AMC or Gly-Phe-AMC for cathepsin C (McGuire et al., 1997), 40 μM Abz-Lys-Pro-Ala-Glu-Phe-Nph-Ala-Leu for cathepsin D (Masa et al., 2006), 30 μM Z-Ala-Ala-Asn-AMC for legumain (Kembhavi et al., 1993), 30 μM Leu-AMC for leucine aminopeptidase (Hatta et al., 2006), 1 mM Z-Phe-Leu for serine carboxypeptidase (Motobu et al., 2007), 50 μM Ac-Asp-Glu-Val-Asp-AMC for caspases (Garcia-Calvo et al., 1999), 50 μM Z-Gly-Gly-Leu-AMC, Suc-Ala-Ala-Pro-Phe-AMC and MeOSuc-Ala-Ala-Pro-Val-AMC for subtilisin-, chymotrypsin- and elastase-like peptidases (Barrett et al., 2004).
Activity measurements were performed at 35 °C using an aliquot of the gut tissue extract (20 to 200-fold diluted stock solution) in 0.1 M Na-citrate-phosphate, pH 2.3–8.0 including 2.5 mM DTT (for cysteine peptidases) and 25 mM NaCl (for cathepsin C). For activity assays in the presence of peptidase inhibitors, an aliquot of the extract was preincubated (15 min at 35 °C) in the assay buffer of the pH optimum value (Table S1) with the inhibitor: 10 mM E-64 for papain-type cysteine peptidases (Barrett et al., 1982), 20 nM cystatin for cysteine peptidases (Turk et al., 2008), 10 μM CA-074 for cathepsin B (Murata et al., 1991), 1 μM Z-Phe-Phe-DMK for cathepsin L (Caffrey and Ruppel, 1997), 1 μM Gly-Phe-DMK or Ala-Hph-VS-Ph for cathepsin C (Green and Shaw, 1981; Kam et al., 2004), 1 μM Aza-N-11a for legumain (Ekici et al., 2004), 10 μM pepstatin for cathepsin D (Knight and Barrett, 1976), 1 μM PMSF or Pefabloc for serine peptidases (James, 1978; Motobu et al., 2007), 10 mM EDTA for metallo peptidases (Auld, 1988), 0.1 μM bestatin for leucine aminopeptidase (Hatta et al., 2006). Proteolytic activity was continuously measured after addition of substrate in a fluorescence reader GENios Plus at 320 nm excitation and 420 nm emission wavelengths (for Abz-containing substrate) or at 360 nm excitation and 465 nm emission wavelengths (for AMC-containing substrates). Carboxypeptidase activity was monitored after derivatization of liberated product with fluorescamine (Sorgine et al., 2000). Assays of legumain and cathepsin L were measured in the presence of 10 μM CA-074 to prevent confounding proteolysis by cathepsin B (Delcroix et al., 2006; Grunclova et al., 2006). All measurements were performed in triplicate.
Active-Site Titration of Tick Peptidases
The absolute molarity of individual peptidases in the gut tissue extract was determined by stoichiometric titration according to Barrett and Kirschke (1981) and Knight and Barret (1976) with the following inhibitors as active-site titrants: CA-074 for cathepsin B, pepstatin for cathepsin D, Ala-Hph-VS-Ph for cathepsin C, and Aza-N-11a for legumain. An aliquot of the gut extract was incubated with various amounts of the titrant for 30 min at 35°C in 0.1 M Na-citrate-phos phate including 2.5 mM DTT (for cysteine peptidases) and 25 mM NaCl (for cathepsin C) at pH 3.5 (for cathepsin D) or pH 5.5 (for cysteine peptidases). Residual peptidase activities were measured using the assay systems described in the previous paragraph, and plotted in the titration curves.
Active-Site Labeling of Tick Peptidases
For cysteine peptidase labeling, an aliquot of the gut tissue extract (0.1–1.0 μg of protein) was incubated (1 h at 35 °C) with 1 μM of the active-site probe: Green-DCG-04 (Greenbaum et al., 2002), Fhex-PD-AOMK (Sexton et al., 2007), or FY01 (Yuan et al., 2006). The reaction was performed in 25 mM Na-acetate including 2.5 mM DTT and 25 mM NaCl (for cathepsin C) at pH 4.5 (for legumain) or pH 5.5 (for cathepsin B, L and C). The competitive labeling was performed after preincubation (15 min at 35 °C) with one or more of the following inhibitors: 1 or 10 μM CA-074, 10 μM Z-Phe-Phe-DMK, 1 μM Aza-N-11a or 1 μM Gly-Phe-DMK. The labeling reaction was stopped by heating to 70 °C in reducing Laemmli sample buffer. The reaction mixture was separated by 15% Laemmli-SDS-PAGE and visualized in a Typhoon 8600 Imager (GE Healthcare) using excitation at 532 nm (green laser) and a cut-off filters set to 526 nm for Fhex-PD-AOMK or 580 nm (bp 30 nm) for Green-DCG-04 and FY01.
For aspartic peptidases labeling, an aliquot of the gut tissue extract (10 μg protein) was incubated (15 min at 26 °C) with 0.5 μM bPSP-07 in 50 mM Na-acetate pH 4.0 containing 10 μM E-64. Afterwards, the reaction mixture was irradiated for 20 min on ice with a 125 W high-pressure mercury-vapor lamp to allow for photoactivated cross-linking. The competitive labeling was performed after preincubation (15 min at 26 °C) with 2 μM pepstatin. The mixture was separated by Laemmli-SDS-PAGE (see above), transferred to a PVDF membrane, and the biotin label visualized by chemiluminescence using VectaStain reagents (Vector Labs).
Quantification of Hemoglobin Degradation
Digestion of bovine hemoglobin (10 μg) was performed with gut tissue extract (0.5 μg of protein) in 25 mM Na-citrate-phosphate, pH 2.3–8.0 including 2.5 mM DTT and 25 mM NaCl, in a total volume of 35 μl for 30 min to 16 h at 35 °C. For hemoglobin degradation in the presence of peptidase inhibitors, an aliquot of the extract was preincubated (15 min at 35°C) in buffer at pH 4.2 with the inhibitor: 50 μM E-64, 10 μM pepstatin, 10 μM CA-074, 1 μM Gly-Phe-DMK, 1 μM Aza-N-11a, 1 μM Z-Phe-Phe-DMK, or 0.1 mM PMSF. For quantification, aliquots of the digest were subjected to derivatization with fluorescamine to quantify the newly formed amino-terminal ends; the degradation rate was determined using aliquots withdrawn at different time points (up to 1 h) (Sorgine et al., 2000). The fluorescence signal was measured using a GENios Plus reader at 370 nm excitation and 485 nm emission wavelengths. All measurements were performed in triplicate. For SDS-PAGE visualization of hydrolysis, hemoglobin digests were separated in Tricine gels (16% T/ 6% C) containing 6 M urea (Schagger, 2006).
Identification of Hemoglobin Fragments
Bovine hemoglobin (0.3 mg) was incubated with gut tissue extracts (10 μg of protein) in 50 mM Na-acetate, pH 4.2 including 2.5 mM DTT and 25 mM NaCl, in a total volume of 200 μl for 30 min to 16 h at 35 °C. For some experiments, extracts were preincub ated with different combinations of inhibitors (10 μM pepstatin, 50 μM E-64, 1 μM Aza-N-11a) for 15 min at 35 °C in the same buffer. The reaction mixture was treated with 10 μl of 10% trifluoracetic acid and separated by RP-HPLC on C4 or C18 Vydac columns (Vydac) equilibrated in 0.1% (v/v) TFA and eluted with a 1%/min gradient of a 99% (v/v) acetonitrile solution in 0.1% (v/v) TFA. The collected peak fractions were analyzed by mass spectrometry. Mass spectra of peptides were measured by FT/MS using an LTQ Orbitrap XL mass spectrometer (Thermo) operating in high-resolution mode (R ~105). Cleavage sites were searched by the MS-NonSpecific module of ProteinProspector software (University of California San Francisco) using a mass tolerance of 3 ppm. The cleavage map combines data from digests at three time points.
Preparation of Large Hemoglobin Fragments
Bovine hemoglobin (0.5 mg) was incubated for 16 h in 50 mM Na-acetate, pH 4.2 with gut tissue extracts (1.0 μg of protein) preincubated for 15 min with 10 μM E-64. The reaction mixture was separated by RP-HPLC on a C4 Vydac column as described above. The collected peak fractions containing fragments of 5–7 kDa were pooled and lyophilized. The experimental fragmentation of this material was performed with gut tissue extract at pH 4.2 as described for intact hemoglobin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants no. 206/06/0865 (GACR) and IAA600960910 (GAASCR), by research projects no. Z60220518 and Z40550506, and Research Center no. LC06009. M.H. was supported by grant no. KJB400550516 (GAASCR), C.R.C. by the Sandler Foundation, and M.B. by Center on Proteolytic Pathways NIH grant no. U54-RR020843. We thank J.C. Powers (Georgia Institute of Technology, Atlanta), L. Marešová and M. Hradilek (IOCB ASCR, Praha) for providing selected peptidase inhibitors, I. Pražáková for technical assistance, and J. Erhart a J. Kopecký (IP BC ASCR, Ceske Budejovice) for photographs.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA The Supplemental Data include two tables and can be found with this article online at http://www.cell.com/chemistry-biology/supplemental.
REFERENCES
- Anderson JM, Sonenshine DE, Valenzuela JG. Exploring the mialome of ticks: an annotated catalogue of midgut transcripts from the hard tick, Dermacentor variabilis (Acari: Ixodidae) BMC. Genomics. 2008;9:552. doi: 10.1186/1471-2164-9-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auld DS. Use of chelating agents to inhibit enzymes. Methods Enzymol. 1988;158:110–114. doi: 10.1016/0076-6879(88)58051-5. [DOI] [PubMed] [Google Scholar]
- Barrett AJ, Kembhavi AA, Brown MA, Kirschke H, Knight CG, Tamai M, Hanada K. L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem. J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ, Kirschke H. Cathepsin B, Cathepsin H, and cathepsin L. Methods Enzymol. 1981;80(Pt C):535–561. doi: 10.1016/s0076-6879(81)80043-2. [DOI] [PubMed] [Google Scholar]
- Barrett AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes. Elsevier; London: 2004. [Google Scholar]
- Boldbaatar D, Sikalizyo SC, Battsetseg B, Xuan X, Fujisaki K. Molecular cloning and functional characterization of an aspartic protease from the hard tick Haemaphysalis longicornis. Insect Biochem. Mol. Biol. 2006;36:25–36. doi: 10.1016/j.ibmb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Boys BL, Kuprowski MC, Konermann L. Symmetric behavior of hemoglobin alpha- and beta- subunits during acid-induced denaturation observed by electrospray mass spectrometry. Biochemistry. 2007;46:10675–10684. doi: 10.1021/bi701076q. [DOI] [PubMed] [Google Scholar]
- Brinkworth RI, Prociv P, Loukas A, Brindley PJ. Hemoglobin-degrading, aspartic proteases of blood-feeding parasites: substrate specificity revealed by homology models. J. Biol. Chem. 2001;276:38844–38851. doi: 10.1074/jbc.M101934200. [DOI] [PubMed] [Google Scholar]
- Caffrey CR, McKerrow JH, Salter JP, Sajid M. Blood `n' guts: an update on schistosome digestive peptidases. Trends Parasitol. 2004;20:241–248. doi: 10.1016/j.pt.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Caffrey CR, Ruppel A. Cathepsin B-like activity predominates over cathepsin L-like activity in adult Schistosoma mansoni and S. japonicum. Parasitol. Res. 1997;83:632–635. doi: 10.1007/s004360050310. [DOI] [PubMed] [Google Scholar]
- Choe Y, Leonetti F, Greenbaum DC, Lecaille F, Bogyo M, Bromme D, Ellman JA, Craik CS. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem. 2006;281:12824–12832. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- Coons LB, Alberti G. The Acari-Ticks. In Microscopic Anatomy of Invertebrates. In: Harrison FW, Foelix R, editors. Chelicerata Arthropoda. Vol. 8B. Wiley-Liss; New York: 1999. pp. 267–514. [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J, Kocan KM. Strategies for development of vaccines for control of ixodid tick species. Parasite Immunol. 2006;28:275–283. doi: 10.1111/j.1365-3024.2006.00828.x. [DOI] [PubMed] [Google Scholar]
- Delcroix M, Sajid M, Caffrey CR, Lim KC, Dvorak J, Hsieh I, Bahgat M, Dissous C, McKerrow JH. A multienzyme network functions in intestinal protein digestion by a platyhelminth parasite. J. Biol. Chem. 2006;281:39316–39329. doi: 10.1074/jbc.M607128200. [DOI] [PubMed] [Google Scholar]
- Ekici OD, Gotz MG, James KE, Li ZZ, Rukamp BJ, Asgian JL, Caffrey CR, Hansell E, Dvorak J, McKerrow JH, Potempa J, Travis J, Mikolajczyk J, Salvesen GS, Powers JC. Aza-peptide Michael acceptors: a new class of inhibitors specific for caspases and other clan CD cysteine proteases. J. Med. Chem. 2004;47:1889–1892. doi: 10.1021/jm049938j. [DOI] [PubMed] [Google Scholar]
- Estrela A, Seixas A, Termignoni C. A cysteine endopeptidase from tick (Rhipicephalus (Boophilus) microplus) larvae with vitellin digestion activity. Comp Biochem. Physiol B Biochem. Mol. Biol. 2007;148:410–416. doi: 10.1016/j.cbpb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Fonovic M, Bogyo M. Activity-based probes as a tool for functional proteomic analysis of proteases. Expert. Rev. Proteomics. 2008;5:721–730. doi: 10.1586/14789450.5.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Calvo M, Peterson EP, Rasper DM, Vaillancourt JP, Zamboni R, Nicholson DW, Thornberry NA. Purification and catalytic properties of human caspase family members. Cell Death. Differ. 1999;6:362–369. doi: 10.1038/sj.cdd.4400497. [DOI] [PubMed] [Google Scholar]
- Goldberg DE. Hemoglobin degradation. Curr. Top. Microbiol. Immunol. 2005;295:275–291. doi: 10.1007/3-540-29088-5_11. [DOI] [PubMed] [Google Scholar]
- Goldberg DE, Slater AF, Beavis R, Chait B, Cerami A, Henderson GB. Hemoglobin degradation in the human malaria pathogen Plasmodium falciparum: a catabolic pathway initiated by a specific aspartic protease. J. Exp. Med. 1991;173:961–969. doi: 10.1084/jem.173.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green GD, Shaw E. Peptidyl diazomethyl ketones are specific inactivators of thiol proteinases. J. Biol. Chem. 1981;256:1923–1928. [PubMed] [Google Scholar]
- Greenbaum D, Baruch A, Hayrapetian L, Darula Z, Burlingame A, Medzihradszky KF, Bogyo M. Chemical approaches for functionally probing the proteome. Mol. Cell Proteomics. 2002;1:60–68. doi: 10.1074/mcp.t100003-mcp200. [DOI] [PubMed] [Google Scholar]
- Grunclova L, Horn M, Vancova M, Sojka D, Franta Z, Mares M, Kopacek P. Two secreted cystatins of the soft tick Ornithodoros moubata: differential expression pattern and inhibitory specificity. Biol. Chem. 2006;387:1635–1644. doi: 10.1515/BC.2006.204. [DOI] [PubMed] [Google Scholar]
- Hatta T, Kazama K, Miyoshi T, Umemiya R, Liao M, Inoue N, Xuan X, Tsuji N, Fujisaki K. Identification and characterisation of a leucine aminopeptidase from the hard tick Haemaphysalis longicornis. Int. J. Parasitol. 2006;36:1123–1132. doi: 10.1016/j.ijpara.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Horn M, Baudys M, Voburka Z, Kluh I, Vondrasek J, Mares M. Free-thiol Cys331 exposed during activation process is critical for native tetramer structure of cathepsin C (dipeptidyl peptidase I) Protein Sci. 2002;11:933–943. doi: 10.1110/ps.2910102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James GT. Inactivation of the protease inhibitor phenylmethylsulfonyl fluoride in buffers. Anal. Biochem. 1978;86:574–579. doi: 10.1016/0003-2697(78)90784-4. [DOI] [PubMed] [Google Scholar]
- Jongejan F, Uilenberg G. The global importance of ticks. Parasitology. 2004;129(Suppl):S3–14. doi: 10.1017/s0031182004005967. [DOI] [PubMed] [Google Scholar]
- Kam CM, Gotz MG, Koot G, McGuire M, Thiele D, Hudig D, Powers JC. Design and evaluation of inhibitors for dipeptidyl peptidase I (Cathepsin C) Arch. Biochem. Biophys. 2004;427:123–134. doi: 10.1016/j.abb.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nat. Chem. Biol. 2005;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- Kembhavi AA, Buttle DJ, Knight CG, Barrett AJ. The two cysteine endopeptidases of legume seeds: purification and characterization by use of specific fluorometric assays. Arch. Biochem. Biophys. 1993;303:208–213. doi: 10.1006/abbi.1993.1274. [DOI] [PubMed] [Google Scholar]
- Knight CG, Barrett AJ. Interaction of human cathepsin D with the inhibitor pepstatin. Biochem. J. 1976;155:117–125. doi: 10.1042/bj1550117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara FA, Lins U, Bechara GH, Oliveira PL. Tracing heme in a living cell: hemoglobin degradation and heme traffic in digest cells of the cattle tick Boophilus microplus. J. Exp. Biol. 2005;208:3093–3101. doi: 10.1242/jeb.01749. [DOI] [PubMed] [Google Scholar]
- Lara FA, Lins U, Paiva-Silva G, Almeida IC, Braga CM, Miguens FC, Oliveira PL, nsa-Petretski M. A new intracellular pathway of haem detoxification in the midgut of the cattle tick Boophilus microplus: aggregation inside a specialized organelle, the hemosome. J. Exp. Biol. 2003;206:1707–1715. doi: 10.1242/jeb.00334. [DOI] [PubMed] [Google Scholar]
- Majer P, Collins JR, Gulnik SV, Erickson JW. Structure-based subsite specificity mapping of human cathepsin D using statine-based inhibitors. Protein Sci. 1997;6:1458–1466. doi: 10.1002/pro.5560060710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masa M, Maresova L, Vondrasek J, Horn M, Jezek J, Mares M. Cathepsin D propeptide: mechanism and regulation of its interaction with the catalytic core. Biochemistry. 2006;45:15474–15482. doi: 10.1021/bi0614986. [DOI] [PubMed] [Google Scholar]
- McGuire MJ, Lipsky PE, Thiele DL. Cloning and characterization of the cDNA encoding mouse dipeptidyl peptidase I (cathepsin C) Biochim. Biophys. Acta. 1997;1351:267–273. doi: 10.1016/s0167-4781(97)00021-3. [DOI] [PubMed] [Google Scholar]
- Mendiola J, Alonso M, Marquetti MC, Finlay C. Boophilus microplus: multiple proteolytic activities in the midgut. Exp. Parasitol. 1996;82:27–33. doi: 10.1006/expr.1996.0004. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji N, Islam MK, Huang X, Motobu M, Alim MA, Fujisaki K. Molecular and reverse genetic characterization of serine proteinase-induced hemolysis in the midgut of the ixodid tick Haemaphysalis longicornis. J. Insect Physiol. 2007;53:195–203. doi: 10.1016/j.jinsphys.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Motobu M, Tsuji N, Miyoshi T, Huang X, Islam MK, Alim MA, Fujisaki K. Molecular characterization of a blood-induced serine carboxypeptidase from the ixodid tick Haemaphysalis longicornis. FEBS J. 2007;274:3299–3312. doi: 10.1111/j.1742-4658.2007.05852.x. [DOI] [PubMed] [Google Scholar]
- Murata M, Miyashita S, Yokoo C, Tamai M, Hanada K, Hatayama K, Towatari T, Nikawa T, Katunuma N. Novel epoxysuccinyl peptides. Selective inhibitors of cathepsin B, in vitro. FEBS Lett. 1991;280:307–310. doi: 10.1016/0014-5793(91)80318-w. [DOI] [PubMed] [Google Scholar]
- Nicklin MJ, Barrett AJ. Inhibition of cysteine proteinases and dipeptidyl peptidase I by egg-white cystatin. Biochem. J. 1984;223:245–253. doi: 10.1042/bj2230245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl J, Davinic S, Blaha I, Strop P, Kostka V. Chromophoric and fluorophoric peptide substrates cleaved through the dipeptidyl carboxypeptidase activity of cathepsin B. Anal. Biochem. 1987;165:96–101. doi: 10.1016/0003-2697(87)90205-3. [DOI] [PubMed] [Google Scholar]
- Ranjit N, Zhan B, Hamilton B, Stenzel D, Lowther J, Pearson M, Gorman J, Hotez P, Loukas A. Proteolytic degradation of hemoglobin in the intestine of the human hookworm Necator americanus. J. Infect. Dis. 2009;199:904–912. doi: 10.1086/597048. [DOI] [PubMed] [Google Scholar]
- Renard G, Garcia JF, Cardoso FC, Richter MF, Sakanari JA, Ozaki LS, Termignoni C, Masuda A. Cloning and functional expression of a Boophilus microplus cathepsin L-like enzyme. Insect Biochem. Mol. Biol. 2000;30:1017–1026. doi: 10.1016/s0965-1748(00)00070-9. [DOI] [PubMed] [Google Scholar]
- Rotari VI, Dando PM, Barrett AJ. Legumain forms from plants and animals differ in their specificity. Biol. Chem. 2001;382:953–959. doi: 10.1515/BC.2001.119. [DOI] [PubMed] [Google Scholar]
- Schagger H. Tricine-SDS-PAGE. Nat. Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- Sexton KB, Witte MD, Blum G, Bogyo M. Design of cell-permeable, fluorescent activity-based probes for the lysosomal cysteine protease asparaginyl endopeptidase (AEP)/legumain. Bioorg. Med. Chem. Lett. 2007;17:649–653. doi: 10.1016/j.bmcl.2006.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sforca ML, Machado A, Figueredo RC, Oyama S, Jr, Silva FD, Miranda A, Daffre S, Miranda MT, Spisni A, Pertinhez TA. The micelle-bound structure of an antimicrobial peptide derived from the alpha-chain of bovine hemoglobin isolated from the tick Boophilus microplus. Biochemistry. 2005;44:6440–6451. doi: 10.1021/bi0475323. [DOI] [PubMed] [Google Scholar]
- Sojka D, Franta Z, Horn M, Hajdusek O, Caffrey CR, Mares M, Kopacek P. Profiling of proteolytic enzymes in the gut of the tick Ixodes ricinus reveals an evolutionarily conserved network of aspartic and cysteine peptidases. Parasit. Vectors. 2008;1:7. doi: 10.1186/1756-3305-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka D, Hajdusek O, Dvorak J, Sajid M, Franta Z, Schneider EL, Craik CS, Vancova M, Buresova V, Bogyo M, Sexton KB, McKerrow JH, Caffrey CR, Kopacek P. IrAE: an asparaginyl endopeptidase (legumain) in the gut of the hard tick Ixodes ricinus. Int. J. Parasitol. 2007;37:713–724. doi: 10.1016/j.ijpara.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshine DE. Biology of the Tick. vol. 1. Oxford University Press; New York: 1991. [Google Scholar]
- Sorgine MH, Logullo C, Zingali RB, Paiva-Silva GO, Juliano L, Oliveira PL. A heme-binding aspartic proteinase from the eggs of the hard tick Boophilus microplus. J. Biol. Chem. 2000;275:28659–28665. doi: 10.1074/jbc.M005675200. [DOI] [PubMed] [Google Scholar]
- Tsuji N, Miyoshi T, Battsetseg B, Matsuo T, Xuan X, Fujisaki K. A cysteine protease is critical for Babesia spp. transmission in Haemaphysalis ticks. PLoS. Pathog. 2008;4:e1000062. doi: 10.1371/journal.ppat.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk V, Stoka V, Turk D. Cystatins: biochemical and structural properties, and medical relevance. Front Biosci. 2008;13:5406–5420. doi: 10.2741/3089. [DOI] [PubMed] [Google Scholar]
- Williamson AL, Lecchi P, Turk BE, Choe Y, Hotez PJ, McKerrow JH, Cantley LC, Sajid M, Craik CS, Loukas A. A multi-enzyme cascade of hemoglobin proteolysis in the intestine of blood-feeding hookworms. J. Biol. Chem. 2004;279:35950–35957. doi: 10.1074/jbc.M405842200. [DOI] [PubMed] [Google Scholar]
- Yuan F, Verhelst SH, Blum G, Coussens LM, Bogyo M. A selective activity-based probe for the papain family cysteine protease dipeptidyl peptidase I/cathepsin C. J. Am. Chem. Soc. 2006;128:5616–5617. doi: 10.1021/ja060835v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.